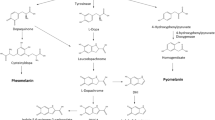
Abstract
Most organisms can synthesize a variety of natural polymers called melanins. These substances serve protective roles against physical and chemical stressors. These products result from the enzyme-catalyzed oxidation of phenolic and indolic substrates which polymerize to produce melanins such as eumelanin, pheomelanin, pyomelanin, and allomelanins. Tyrosinase and laccase protein families are primarily involved in the production of melanin. The pharmaceutical, cosmetic, optical, and electrical industries all use melanins as functional polymeric materials. The development of biotechnological processes to produce melanins is becoming an attractive alternative compared to their extraction from plant or animal matter, in which they are only present at low concentrations. Numerous types of bacteria are naturally capable of producing melanin. Using genetic engineering techniques, it is currently possible to overexpress in microorganisms the genes for melanin-producing enzymes. These advancements have allowed increasing the productivity of melanogenic organisms and have enabled the creation of novel recombinant microbial strains that can synthesize a variety of melanins. Furthermore, strains capable of completely synthesizing melanins from basic carbon sources on a gram-scale basis have been developed by metabolic engineering of microbial hosts through altering pathways relevant to the availability of melanogenic precursors. The most recent discoveries in the development of recombinant melanin-producing strains and manufacturing methods are compiled and reviewed in this chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Melanins are a family of polymeric pigments that are abundantly present in nature (d’Ischia et al. 2015). These polymers are the end products of the enzymatic oxidation of phenolic or indolic substrates. Additionally, it is thought that some of the oldest pigments found in nature are melanins, since this type of compound has been found in dinosaur and bird fossils (Zhang et al. 2010). Furthermore, intact melanin was also found in Jurassic-era squid ink sacs (Glass et al. 2012). Therefore, melanin has been proposed as a biomarker in the study of evolution (Wogelius et al. 2011). The four main types of melanin are eumelanin, pheomelanin, allomelanins, and pyomelanin. Eumelanin is produced by the oxidation of the amino acid L-tyrosine and/or L-dihydroxyphenylalanine (L-DOPA), resulting in a brown or black polymer. Alternatively, pheomelanin is formed when L-tyrosine and/or L-DOPA are oxidized in the presence of L-cysteine, resulting in a red-yellow pigment. Moreover, the oxidation of 4-hydroxyphenylacetic acid, catechols, dihydroxynaphthalene (DHN), -glutaminyl-4-hydroxybenzene, protocatechualdehyde, or tetrahydroxynaphthalene results in the formation of allomelanins. Finally, a particular kind of melanin called pyomelanin is produced when homogentisic acid is oxidized (HGA) (Fig. 1) (Lindgren et al. 2015).
Due to their chemical makeup, melanins exhibit unique physicochemical features, allowing them to function as cation exchangers, amorphous semiconductors, X-ray, γ-ray, and ultraviolet light absorbers (della-Cioppa et al. 1990; Krol and Liebler 1998; Różanowska et al. 1999; Sarna et al. 1976; Ambrico et al. 2014). Additionally, it has been demonstrated that melanins contain antioxidant and antiviral properties (Nofsinger et al. 2002; Montefiori and Zhou 1991). Therefore, the ability to obtain these polymers in large quantities and at an affordable cost is necessary for the creation of a variety of valuable products and applications. Melanins can be chemically produced or isolated from animal and plant tissues. Nevertheless, these procedures can be rather expensive and are occasionally not sustainable (Saini and Melo 2015). An innovative and viable alternative to obtaining melanins is based on the development of biotechnological processes that use melanogenic microorganisms. Two advantages of this approach are its scalability and good yields for melanin production. Furthermore, this strategy can be strengthened by using genetic engineering techniques to boost the inherent melanogenic potential of some organisms or by creating new melanin-producing strains. In this context, the expression of genes encoding the enzymes involved in the oxidation of melanin precursors is the most typical genetic modification used to improve or generate a production strain (Martínez et al. 2019).
2 Enzymes Involved in Melanin Formation: Classification and Evolution
The initial process leading to the production of melanins is the enzyme-dependent oxidation of phenolic or indolic chemicals. Tyrosinases are the most common type of enzyme associated with melanogenesis. Tyrosinases are copper enzymes that catalyze both the ortho-hydroxylation of monophenols (cresolase activity) and the oxidation of catechols (catecholase activity), generating ortho-quinone products (Garcia-Molina et al. 2007) (Fig. 1). More specifically, the enzyme tyrosinase catalyzes the hydroxylation of L-tyrosine to L-DOPA using molecular oxygen and then oxidizes this compound to dopachrome, which in turn nonenzymatically polymerizes to yield melanin (Ito 2003).
Copper proteins are classified into three categories according to their spectroscopic and structural properties. These are (1) mononuclear type-1 or blue copper proteins, which are mostly involved in electron transfer; (2) type-2 or non-blue copper proteins, typically found in enzymes that activate molecular oxygen and; (3) type-3 or binuclear copper proteins, which are part of the Di-copper superfamily or clan (CL0205) which includes phenoloxidases (i. e. tyrosinases and catechol oxidases) and hemocyanins (Jaenicke and Decker 2004; Aguilera et al. 2013; Kaintz et al. 2014). Phenoloxidases are enzymes involved in the oxidation of phenolic compounds. On the one hand, catechol oxidases have diphenolase activity, while on the other hand, tyrosinases are bifunctional enzymes with monophenolase and diphenolase activities (Aguilera et al. 2013; Kaintz et al. 2014). Hemocyanins are oxygen carriers found in arthropods and mollusks, although some may also have weak phenoloxidase activity (van Holde et al. 2001).
Type-3 copper proteins bind oxygen through two copper atoms (CuA and CuB) whose oxidation state changes from Cu+ (reduced) to Cu2+ (oxidized) upon oxygen binding. Each Cu atom is coordinated by three histidine residues provided by two pairs of α helices, forming a four α helix bundle motif (Jaenicke and Decker 2004). The CuA binding site is typically characterized by an H1(n)-H2(8)-H3 motif and the CuB binding site by an H1(3)-H2(n)-H3 motif, where n is a variable number of residues between histidines (Aguilera et al. 2013). Sequence differences in the copper binding sites and domain architecture have been used as criteria for the classification of these proteins into three subclasses: (1) α-subclass with N-terminal signal peptide, suggesting secretion or vesicle localization (includes tyrosinases in the three domains of life, plant catechol oxidases and molluscan and urochordate hemocyanins); (2) β-subclass without a signal peptide and, therefore, presumably localized in the cytosol (includes arthropod tyrosinases and hemocyanins) and; (3) γ-subclass with a signal peptide, which is a cysteine-rich region upstream of the binuclear copper center and a transmembrane domain downstream of the binuclear copper center (includes various metazoan tyrosinases such as human ones) (Aguilera et al. 2013). Among these, the β-subclass appears as the most evolutionary divergent group, presenting a motif variation in the CuA binding site as H1(2)-H2(n)-H3 (Aguilera et al. 2013) (Fig. 2).
Schematic representation of the taxonomic distribution of type-3 copper proteins. Domain architectures and sequence motifs characteristic of each subclass are represented to the right, as well as the type of enzymes present in each of them. SP signal peptide, CYS cysteine-rich region, CuA copper binding site A, CuB copper binding site B, TM transmembrane domain. The Eukaryota branch represents the branching order of α, β, and γ type-3 copper proteins. See the text for further detail on the taxonomic distribution and evolutionary processes related to type-3 copper proteins
The presence of type-3 copper proteins in the three domains of life suggests an early origin probably dating back to the times of the last universal common ancestor to all cellular life (LUCA) (Jaenicke and Decker 2004; Aguilera et al. 2013) (Fig. 2), where a common ancestral protein with a mononuclear copper center (probably CuB binding site) duplicated resulting in a protein with a binuclear copper center (CuB + CuA binding sites) from which all type-3 copper proteins evolved. This ancestral protein may have protected primitive organisms from increasing levels of oxygen, which began to accumulate 3.5 billion years ago as a byproduct of photosynthesis (van Holde et al. 2001; Jaenicke and Decker 2004).
Phylogenetic analyses have shown that type-3 copper proteins have a complex evolutionary history including multiple lineage-specific gene expansions and losses. Given the wide distribution of α-subclass proteins, it has been hypothesized that the first type-3 copper proteins belonged to this group. β and γ-subclasses appear to have originated from gene duplication within the Eukaryota domain. Among these, the most ancient one is the β-subclass, which seems to have originated before the divergence of amoebozoan and opisthokont lineages (present in amoebae, fungi, and some animals) where they lost the signal peptide and their sequences began to diverge at the CuA binding site. More recently, γ-subclass appears to have originated within the metazoan lineage (only found in animals), where they acquired the cysteine-rich region and the transmembrane domain (Aguilera et al. 2013) (Fig. 2).
Metazoan tyrosinases are mainly members of the γ-subclass (where α and β-subclass tyrosinases were lost); although α-subclass tyrosinases are maintained in Cnidaria, Nematoda and Mollusca; whereas β-subclass tyrosinases are found in Porifera, Urochordata, and Arthropoda. Regarding hemocyanins, α-subclass hemocyanins were probably duplicated before the divergence of bilaterians but were lost in most lineages except for urochordates and molluscans, whereas β-subclass hemocyanins were duplicated more recently within the Arthropoda lineage (van Holde et al. 2001; Aguilera et al. 2013). Although metazoans are the only kingdom that possesses proteins from the α, β, and γ-subclasses, gene loss has occurred to such an extent that Ciona intestinalis is the only organism possessing the three types in its genome (α-subclass hemocyanin and, β and γ-subclass tyrosinases) (Aguilera et al. 2013).
In vertebrates, in addition to tyrosinase, two tyrosinase-related proteins are also involved in different steps of melanin biosynthesis: tyrosinase-related protein-1 (tyrp1) and tyrosinase-related protein-2 (tyrp2) also known as DOPAchrome tautomerase (dct). These two enzymes influence the quantity and quality of melanins produced and are involved in the stabilization of the tyrosinase structure (Esposito et al. 2012). Phylogenetic analyses suggest a duplication event before the divergence of urochordates and vertebrates resulting in tyrosinases and a tyrosinase-related protein, and a second duplication event before the divergence of teleost fishes gave rise to the two tyrosinase-related proteins tyrp1 and tyrp2. Interestingly, it may be possible that these enzymes do not coordinate Cu atoms at the metal binding sites, as tyrp2 is known to coordinate two zinc atoms instead (Esposito et al. 2012).
To the best of our knowledge, the evolution of bacterial and archaeal tyrosinases has not been extensively explored. Thus, for the time being, a tentative scheme proposes at least three unambiguous types of bacterial tyrosinases based on their domain architecture. For example, the first type includes the well-studied tyrosinases from Streptomyces sp. which are produced as a heterodimer where tyrosinase (melC2 gene product) is bound to a caddy protein (melC1 gene product) involved in heterodimer secretion and delivery of copper atoms to the active site of tyrosinase. A second type is not associated with a caddy protein for copper incorporation and includes tyrosinases such as that of Bacillus megaterium. Finally, a third type includes tyrosinases produced as zymogens that, similarly to plant and fungal tyrosinases/catechol oxidases, require proteolytic removal of a C-terminal domain for activation (Fairhead and Thöny-Meyer 2012; Pretzler and Rompel 2018). Interestingly, a preliminary analysis performed in our group suggests that tyrosinases from plant-associated bacteria (e. g. those from Rhizobium etli or Ralstonia solanacearum) are more closely related to plant catechol oxidases, which implies horizontal gene transfers that would further increase the complexity of the already intricated evolutionary history of this protein family. Additionally, a cold-adapted tyrosinase from the archaeon Candidatus Nitrosopumilus koreensis is distinct from other known bacterial tyrosinases (Kim et al. 2016).
Despite efforts to classify type-3 copper proteins based on their domain architecture, conserved sequence motifs, and tertiary structure elements, it is still unclear what changes have driven the functional differentiation of these proteins throughout evolution. However, besides the six conserved histidines, mutagenesis studies have shed some light on some amino acids of great catalytic importance that could explain the functional differentiation between catechol oxidases and tyrosinases. These include a variable gatekeeper residue which may support or inhibit substrate entry; a waterkeeper residue (Glu for tyrosinase activity) required to maintain a stable water network around the active center; variable residues (Asn and Asp for tyrosinase activity) one position ahead of the first and second histidines in CuB copper binding site, which are involved in increased basicity of adjacent histidines required for substrate deprotonation; a thioether bond (absent in tyrosinases) between a cysteine residue and the second histidine in the CuA copper binding site, which upon breaking allows the histidine to move more freely within the active site where it can also intervene in substrate deprotonation and; a seventh histidine one position before the third histidine in the CuB copper binding site and disulfide bonds that may stabilize the enzyme’s catalytic activity (Kampatsikas and Rompel 2021).
Lacasses are another group of enzymes involved in melanogenesis. These enzymes which have been found in bacteria, fungi, and plants, are not related to tyrosinases but are also copper-dependent oxidoreductases (Valderrama et al. 2003). For example, the enzyme 4-hydroxyphenylacetic acid (4-HPA) hydroxylase is involved in the catabolism of 4-HPA in bacteria. This enzyme is a two-component flavin adenine dinucleotide (FAD)-dependent monooxygenase (Gibello et al. 1995). This group of enzymes displays a broad substrate range, they can hydroxylate various monohydric and dihydric phenols (Prieto et al. 1993).
3 Production of Melanins with Genetically Engineered Microorganisms
3.1 Eumelanin Production
The ability to obtain these pigments from plentiful and affordable sources is a prerequisite for the current and future uses of melanin. By using relatively simple procedures, these products can be recovered from natural sources such as the tissues of animals or plants. However, the mixture of various melanin types and related compounds found in these sources frequently makes purifying processes difficult and could result in a product with varied compositions. Furthermore, these polymers can be produced chemically or enzymatically by oxidizing phenolic or indolic substrates (Saini and Melo 2015). These processes can produce melanins that are highly pure, but they come at a high price (d’Ischia et al. 2015). Alternatively, culturing melanin-producing microorganisms or microbes that have been genetically modified to produce melanin is another method for obtaining these polymers. This last method allows for the production of melanins at a high yield and relatively low cost.
The experimental procedures collectively referred to as “genetic engineering “ allow altering the genetic make-up of bacteria to increase or give them the capacity to generate chemicals. An increasing number of microbes may be genetically engineered for producing melanin, and the identification of individual genes and melanin-producing pathways has been possible through the application of DNA sequencing technologies in conjunction with biochemical investigations. This knowledge and technologies are the basis for creating recombinant microbes with improved melanin production and the capacity to transfer this capability to non-melanogenic bacteria.
The latest developments in the design of recombinant microbial strains and the production techniques used for the synthesis of melanins are reviewed and analyzed in the following sections. The bacterium Escherichia coli was the first documented recombinant melanogenic microbe. Genes from the actinomycete Streptomyces antibioticus were altered to be expressed in E. coli. The two genes in question, mel and ORF438 at the mel locus in S. antibioticus are necessary for the synthesis of melanin. In agar plates and liquid cultures, the recombinant E. coli strain was capable of producing eumelanin from L-tyrosine. Furthermore, it has been demonstrated that the S. antibioticus tyrosinase enzyme could use synthetic, non-natural amino acids such N-acetyl-L-tyrosine and L-tyrosine ethyl ester, as substrates, to produce synthetic melanins (Della-Cioppa et al. 1990). In a different report, a recombinant E. coli strain derived from JM109 was created using the mel locus from S. antibioticus. In this case, the phage T5 promoter and two lac operators were used to control the expression of the mel gene. The resulting recombinant strain was cultivated in LB medium, producing 0.4 g/L of eumelanin (Table 1). Eumelanin was removed from the culture medium by precipitation at a pH of 3.0, followed by dissolution at a pH of 8.0 in distilled water. Afterward, liquid chromatography on Sephadex LH-20 was performed. To investigate how the presence of this polymer affected the antibacterial activity of several antibiotics, pure eumelanin was used. It was found that eumelanin dose-dependently decreased the antibacterial effect of ampicillin, kanamycin, polymyxin B, and tetracycline against E. coli (Lin et al. 2005). In addition to the potential therapeutic significance of such data, the observed results could be used to select for greater melanin-producing recombinant strains based on antibiotic resistance.
Another early study demonstrated that the Bacillus thuringiensis strain 4D11 was able to produce melanin when cultivated with L-tyrosine at 42 °C (Ruan et al. 2004). These findings suggested that in the genome of this organism there should be a gene encoding for tyrosinase. Considering that the genome sequence of B. thuringiensis 4D11 was unknown, a cloning technique was then developed based on the predicted sequence similarities with a tyrosinase gene from Bacillus cereus 10987. Based on the tyrosinase gene sequence from B. cereus 10987, a set of PCR primers was created and used to amplify an 1179 bp DNA fragment from B. thuringiensis 4D11 DNA. This DNA fragment shared 99% of its encoded amino acid sequence with the tyrosinase from B. cereus 10987. The lac promoter was used to express the PCR product in plasmid pGEM-7zf. This plasmid was then used to transform E. coli DH5. The resulting recombinant strain produced eumelanin at a titer of 5.6 g/L when cultured in a casein liquid medium (Table 1). Intriguingly, it was found that in trials involving exposure to UV radiation, this recombinant strain outperformed DH5 in terms of survival rates (Ruan et al. 2005). These findings demonstrate how, in addition to enabling the biotechnological production of melanin, the heterologous expression of a gene encoding a tyrosinase might improve the host’s UV radiation resistance, a characteristic that might be advantageous in the case of microorganisms used in the field, such as B. thuringiensis. Moreover, microorganisms that can endure intense UV exposure could be of benefit in future long-term space missions and planet-colonization initiatives, as microbes are thought to be crucial to help maintain human existence by producing food, valuable compounds, and recycling trash (Horneck et al. 2010; https://blogs.scientificamerican.com/observations/microbes-might-be-key-to-a-mars-mission/). Furthermore, melanin can absorb X and γ rays, which may help engineered bacteria survive in conditions outside of our planet.
Rhizobium etli, a soil bacterium, is particularly significant for agriculture, as it can fix nitrogen by forming nodules in the roots of Phaseolus vulgaris plants. This bacterium can also synthesize melanin in the symbiotic nodules through a symbiotic plasmid that contains a gene encoding tyrosinase (melA) (González et al. 2003; Piñero et al. 2007). The expression vector pTrc99A was used to clone the melA gene under the strong trc promoter, resulting in pTrcmelA, which was used to transform E. coli. When L-tyrosine was used as a substrate, the recombinant E. coli strain was able to produce eumelanin at 30 °C, yet, increasing the temperature to 37 °C significantly lowered melanin yields (Cabrera-Valladares et al. 2006). Moreover, it was also discovered that melanin production occurred only in the stationary culture phase and, when compared to other colonies, a recombinant E. coli colony on media containing L-tyrosine showed a deeper color. Following DNA sequencing of this clone’s melA gene, it was discovered that it had undergone a spontaneous mutation, changing the Asp535 residue in the MelA tyrosinase enzyme to a Gly residue. Therefore, MutMelA was the name given to this mutant version of MelA. When compared to a strain that expresses the wild-type enzyme, it was observed that eumelanin synthesis in liquid cultures begins sooner in cultures of E. coli expressing MutmelA. A study was performed to find the best conditions for pigment synthesis in liquid cultures utilizing a recombinant E. coli strain expressing MutmelA. The influence of culture temperature, pH, isopropyl-d-thio-galactopyranoside (IPTG) as a gene inducer, antibiotic concentration for plasmid selection pressure, and eumelanin concentration was assessed. It was found that the ideal bioreactor conditions were: 0.1 mmol/L of IPTG, a culture temperature of 30 °C, and a change in medium pH from 7.0 to 7.5 at the beginning of the eumelanin production phase. Additionally, L-tyrosine was added to the culture medium at 6 g/L as a eumelanin precursor. With a final titer of 6 g/L under these conditions, a 100% conversion yield of L-tyrosine to eumelanin was observed (Table 1) (Lagunas-Muñoz et al. 2006). These findings emphasize the significance of optimizing culture conditions as a factor in achieving the highest yield and productivity while using a recombinant melanogenic strain.
As part of a bioprospecting investigation, microorganisms capable of producing melanin were identified from soil samples in China. Among these isolates, Streptomyces kathirae SC-1 had the best potential for melanin synthesis among all isolates. Employing this organism and a surface response approach to enhance the medium and growth conditions, 13.7 g/L of melanin was produced (Guo et al. 2014). It is significant to note that yeast extract, which provides a variety of melanin precursors, was a component of the culture medium used in this investigation. Therefore, to identify the type of melanin generated, the resultant polymer needs to be analyzed to ascertain its chemical makeup. To provide further light on this organism’s melanogenesis process, a tyrosinase was homogeneously purified, which is a 30-kDa enzyme (called tyrosinase MelC) that has a Km of 0.42 mM for L-DOPA and 0.25 mM for L-tyrosine. The primers used to amplify the melC gene and its promoter region were designed using the partial amino acid sequence of this tyrosinase. Two potential promoters, Pskmel and P135, were discovered by sequence analysis. In the replicative plasmid pIJ86, the gene melC was cloned under the transcriptional control of either the putative promoter or the constitutive promoter PermE*. The resultant constructs were then transformed into S. lividans and S. kathirae. After characterizing the recombinant strains of S. lividans, it was demonstrated that Pskmel is the functional promoter for melC. S. kathirae recombinant strains that were grown in melanin-producing conditions. It was also found that the amounts of melanin generated by strains expressing melC from PermE* or Pskmel were 24.9 and 28.8 g/L, respectively (Table 1) (Guo et al. 2015). It should be emphasized that these are the highest reported melanin titers, showing the possibility of using genetic engineering methods to significantly increase a melanogenic organism’s production capability (Table 1). This manufacturing method can be further optimized, particularly in terms of the culture medium composition, as yeast extract, at the concentration used in the referenced work (37 g/L), becomes a costly ingredient. Moreover, some of the components of yeast extract can interact with melanin precursors to produce a polymer that isn’t completely derived from L-tyrosine. To enhance the existing manufacturing method, future research should aim at finding a growth medium that only contains salts and a simple carbon source.
3.2 Pyomelanin Production
When cultured on a medium containing L-tyrosine, the soil bacterium Pseudomonas putida strain F6 exhibits the ability to produce a dark pigment. Thus, transposon mutagenesis was done to learn more about the function of the genes involved in the production of this pigment. Two mutants with improved synthesis were obtained by this approach. When compared to P. putida F6, one of these mutants (F6-HDO) generated 0.35 g/L of the pigment, which was identified as a type of melanin (Table 1). It is interesting to note that, when compared to the wild-type strain, this mutant showed greater resistance to UV radiation and H202. Genetic testing revealed that a gene encoding HGA 1,2-dioxygenase (HGO) was disrupted by transposon mutagenesis. This enzyme participates in the degradation process that turns HGA into 4-maleylacetoacetate. As a result, it is expected that this mutation will cause HGO to use less HGA. This finding suggests that in this mutant strain, HGA is the pyomelanin precursor (Fig. 3) (Nikodinovic-Runic et al. 2009). Precursors for HGA synthesis originate from the L-tyrosine biosynthetic pathway. Furthermore, the enzyme hydroxyphenylpyruvate dehydrogenase (HPPD) converts the intermediate 4-hydroxyphenylpyruvate (HPP) into HGA (Fig. 3).
Metabolic pathways and expressed genes related to the synthesis of melanins with engineered microorganisms. Dashed arrows indicate two or more enzyme reactions. Abbreviations: PTS phosphotransferase system glucose transport protein, Gly glycerol, Gly3P glycerol-3-phosphate, G6P glucose-6-phosphate, E4P D-erythrose 4-phosphate, PEP phosphoenolpyruvate, DAHP 3-deoxy-D-arabino-heptulosonate 7-phosphate, HPP 4-hydroxyphenylpyruvate, CHA chorismate, ANT anthranilate, PPA phenylpyruvate, HPPD hydroxyphenylpyruvate dehydrogenase, L-Tyr L-tyrosine, L-Trp L-tryptophan, aroGfbr feedback inhibition resistant DAHP synthase, CadA L-lysine decarboxylase, trpEG anthranilate synthase component I, trpD9923 mutant version of TrpD causing the loss of anthranilate phosphoribosyl transferase activity and retaining anthranilate synthase activity, tyrC cyclohexadienyl dehydrogenase, C3H p-coumarate 3-hydroxylase, TAL tyrosine ammonia-lyase, FCS feruloyl-CoA synthetase, ECH enoyl-CoA hydratase/aldolase, AntABC terminal oxygenase and the reductase components of anthranilate 1,2-dioxygenase, pheACM chorismate mutase domain from chorismate mutase-prephenate dehydratase, MutmelA mutant version of tyrosinase (Martínez et al. 2019)
A recombinant strain of E. coli for pyomelanin production was developed by cloning the chromosomal hpd gene that encodes the 4 hydroxyphenylpyruvate dioxygenase from Pseudomonas auruginosa PAO1, and then placing it in the pVO vector under transcriptional control of the arabinose promoter pBAD. A biotransformation protocol for pyomelanin synthesis with the recombinant E. coli strain was developed by defining the parameters influencing growth and production. The optimal growth conditions were determined to be as follows: use of mineral medium with glucose as carbon and energy source, arabinose 0.1% to induce gene hpd, casamino acids at 0.2%, and tyrosine 1 mM. Under these conditions, after 6 days of culture, 213 mg/L of pyomelanin was produced (Bolognese et al. 2019).
In another report, a strain of Ralstonia pickettii that produces pyomelanin was isolated from soil samples. From this strain, gene hppD encoding 4-hydroxyphenyl pyruvate dioxygenase was amplified and cloned in plasmid pET-28a(+). This plasmid was then transformed into E. coli that in turn was cultured to produce pyomelanin. In these experiments, a dependency on metal ions was observed. In this case, the highest pyomelanin production was achieved while supplementing Cu2+, resulting in a titer of 315 mg/L (Seo and Choi 2020).
A new strain of Flavobacterium kingsejongi could produce a dark pigment when L-tyrosine was present in the culture medium. The chemical characterization of such pigment showed properties consistent with melanin. However, it was not clear if this pigment was eumelanin, pyomelanin or a mixture of both polymers. Regardless, the hpd gene encoding 4-hydroxyphenyl pyruvate dioxygenase from F. kingsejongi was amplified and cloned in expression vector pET-21a (+). The resulting plasmid was then transformed into E. coli. Afterward, this recombinant strain was cultured in a 5-L bioreactor containing TB medium supplemented with 10 g/L L-tyrosine and 1 mM IPTG. Under these conditions, 3.76 g/L of melanin was produced (Lee et al. 2022).
Random mutagenesis is a reasonably simple approach for strain enhancement; however, it can only be used with organisms that naturally produce melanin. A drawback of this approach is that the site and type of mutation are not easily determined. This restricts the application of genetic engineering methodologies for strain enhancement. Additionally, the strain may return to a low producer phenotype since the genetic alterations caused by random mutagenesis might be unstable. The enhanced strain’s genome may be sequenced to learn more about the type of mutation that caused it as well as the genes and pathways that contributed to the observed phenotype. Knowing this, the melanogenic organism can be “reverse-engineered” by reintroducing the detected mutations through genetic engineering techniques. Furthermore, by using this method, it is possible to distinguish between genetic alterations that are connected to an enhanced phenotype from those that might be harmful to production purposes. In the preceding examples, recombinant strains and procedures for converting various aromatic chemicals into melanins were detailed. Tyrosinases may use different precursors as substrates to produce melanins with different colors (Table 1). Despite these benefits, several disadvantages might be thought. One of them is the use of expensive of using pure melanin precursors. Therefore, to decrease expenses, less pure melanin precursors such as protein hydrolysates or yeast extract are used. Yet, using these more affordable precursors causes new issues. For example, these culture media may contain a variety of substances that can act as substrates of tyrosinases or that can react with melanin precursor molecules, therefore, their usage can lead to diversity in the composition of the generated melanins. Additionally, techniques for purifying melanin become more complicated and costly when non-defined media are used.
4 Production of Allomelanin and Novel Types of Melanin
In the chemical and food industries, chemicals known as phenolic aldehydes are commonly used. Thus, a project was launched that aimed to synthesize this class of compounds through engineered E. coli strains that have been altered to produce caffeic acid from L-tyrosine. To do so, the expression of the enzyme tyrosine ammonia-lyase (TAL) converted L-tyrosine to p-coumaric acid and p-coumarate 3-hydroxylase (C3H) converted p-coumaric acid to caffeic acid (Fig. 3). The resulting product was a black pigment with melanin-like properties, which was probably created by some of the oxidases encoded in the E. coli genome that can oxidize the catechol moiety of various molecules. Additionally, it was shown that by culturing E. coli with protocatechualdehyde in the media, a brown melanin pigment was produced, whereas when caffeic acid was added, a black pigment was generated. Additionally, the genes from Burkholderia glumae BGR1’s encoding for feruloyl-CoA synthetase (FCS) and enoyl-CoA hydratase/aldolase (ECH) were expressed in E. coli (Fig. 3), resulting in a recombinant strain with the capacity to transform caffeic acid into protocatechualdehyde. Furthermore, this study found that wild-type E. coli BL21(DE3) generated 0.15 g/L of melanin when exposed to 5 mM caffeic acid. (Table 1). The production of melanin increased when the same quantity of caffeic acid was given to a culture of a recombinant strain expressing fcs and ech, reaching a titer of 0.2 g/L (Jang et al. 2018). Although the produced melanin chemical makeup has not been determined, it is most likely a polymer made up of a combination of protocatechualdehyde and caffeic acid moieties. These findings show that recombinant E. coli can produce caffeic acid and protocatechualdehyde melanins. It can also be expected that this strain’s FCS and ECH activities affect how much melanin and/or melanin precursors are produced, but the exact processes behind these effects are not yet entirely known. Therefore, the synthesized melanin’s detailed characterization should shed further light on the chemical precursors involved in its synthesis. Additionally, finding the native E. coli enzyme responsible for the oxidation of protocatechualdehyde and caffeic acid, which results in their polymerization into melanin, should also be of interest as the development of melanin-producing strains should be possible by the cloning and overexpression of the gene encoding this yet undiscovered oxidase. In a subsequent publication, it was shown that soft contact lenses may be dyed using protocatechualdehyde-based melanin (Ahn et al. 2019), providing an advantage when compared to chemically produced dyes, as the antibacterial and antioxidant properties of melanins could benefit these products.
The synthesis of a novel melanin polymer was explored in E. coli by the co-expression of tyrosinase (MelC) and cytochrome P450 monooxygenase (CYP102G4). The tyrosinase MelC synthesizes eumelanin in the presence of L-tyrosine as substrate, whereas CYP102G4 is involved in the synthesis of indigo dye from L-tryptophan-derived indole. The gene melC was obtained from Bacillus megaterium, and cloned in the pET-24a(+) vector. Gene cyp102G4 was amplified from Streptomyces cattleya and cloned in pETduet-1. Single plasmids or both were transformed into the E. coli BL21 (DE3) strain. Cultures with a strain expressing melC in the presence of L-tyrosine and L-tryptophan resulted in the production of 1 g/L of melanin. In contrast, under the same conditions, a strain expressing both melC and cyp102G4 produced 3.5 g/L of melanin. The characterization of this novel melanin demonstrated it had noticeable dyeing capabilities on cellulose paper. In addition, it displayed electrical conductivity, rendering it a potential material for organic electrodes (Park et al. 2020).
In another study, the production of novel synthetic melanin was explored by employing C5-diamine to increase the crosslinking of melanin units. E. coli strains expressing combinations of the following enzymes were evaluated: Tyrosine ammonia lyase, p-coumarate 3-hydroxylase, feruloyl-CoA synthetase, enoyl-CoA hydratase, lysine decarboxylase, and tyrosinase. Some of these strains produced melanin derived from caffeic acid, protocatechualdehyde, or L-DOPA to a maximum level of 400 mg/L.
A strain expressing all the previous enzymes showed the capacity to produce a melanin-diamine complex. Using this strain in biotransformation experiments, 20 mg/L of a melanin-diamine complex was formed while supplying 5 mM caffeic acid, L-tyrosine, and L-lysine (Ahn et al. 2021). This relatively low production level is attributed to toxicity from the C5-diamine, which is formed by lysine decarboxylase. Therefore, to improve the production of melanin-diamine, further studies must be done to contend with C5-diamine toxicity.
5 Metabolic Engineering Applied for De Novo Synthesis of Melanins
Developing microbial strains for the complete synthesis of melanins from simple carbon sources is one possible solution for the challenges stated previously. Based on metabolic engineering techniques, it is possible to increase flux into the shikimate pathway, which is responsible for producing the precursors of aromatic amino acids. For instance, L-tyrosine, a precursor of eumelanin, was produced from glucose by an E. coli strain modified through metabolic engineering techniques (Chávez-Béjar et al. 2008). To do so, carbon flow to the L-tyrosine biosynthetic pathway was increased by overexpressing the genes for cyclohexadienyl dehydrogenase (TyrC) from Zymomonas mobilis, the chorismate mutase domain from the original enzyme chorismate mutase-prephenate dehydratase, and a feedback-insensitive version of the enzyme 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase (aroGfbr). Additionally, this strain expressed the tyrosinase MutMelA (Fig. 3). The resulting strain was able to produce eumelanin from glucose. But it was found that the L-tyrosine pool was decreased by MutMelA activity, which led to a defect in cell growth. It was then found that Cu is required as a cofactor for the correct functioning of the enzyme tyrosinase. Thus, to prevent L-tyrosine depletion by MutMelA, Cu was left out of the medium during the first half of the culture. Then, tyrosinase was activated by the addition of CuSO4 to the medium, which began the eumelanin synthesis phase. This method was used in bioreactor cultures where the only carbon source was glucose. In this culture, 3.2 g/L of eumelanin was produced in 120 h (Table 1) (Chávez-Béjar et al. 2013). These results were the first instance of the application of metabolic engineering to create a strain for the complete synthesis of eumelanin from glucose. This work shed light on the possible detrimental effects on cell physiology brought on by tyrosinase expression at high levels. Yet, by using a delayed activation of the heterologous enzyme, this issue was solved. A different approach might be based on the precise regulation of gene induction during a particular stage of the production culture.
During the characterization of the enzyme MutMelA, it was found that in addition to L-tyrosine, catechol may be used as a substrate. Therefore, this enzyme might be used to produce catechol melanin. To put this theory to the test, a bioconversion process was created using an E. coli strain that expresses MutMelA and grows on a medium that contains catechol (0.85 g/L) as a tyrosinase substrate, and glycerol 40 g/L as a carbon source. In this experiment, 0.29 g/L of catechol melanin was generated after 54 h. To further enhance this process, metabolic engineering was evaluated to create a strain capable of producing catechol melanin from a simple carbon source. The method used was based on employing an engineered strain of E. coli capable of generating catechol from a simple carbon source (Balderas-Hernández et al. 2014). The chosen mutant strain was E. coli W3110 trpD9923, which overproduces the intermediate anthranilate in the L-tryptophan biosynthesis pathway (Yanofsky et al. 1971). This strain was altered to enhance carbon flow to anthranilate by overexpressing the aroGfbr and tktA genes that encode for the feedback-insensitive form of the enzyme DAHP synthase and transketolase, respectively (Fig. 3). These modifications resulted in a twofold increase of the anthranilate titer in flask cultures (Balderas-Hernández et al. 2009). This strain was further modified through the expression of the genes antABC which encodes anthranilate 1,2-dioxygenase from P. aeruginosa PAO1, granting it the capacity to convert anthranilate to catechol (Fig. 3). Lastly, the strain was further modified by integrating the gene MutmelA into the chromosome at the location of the lacZ gene. The resultant strain was assessed in 1-liter bioreactor cultures. The carbon source in the culture medium was glycerol, 40 g/L. This carbon source was chosen instead of glucose because it does not utilize the aromatics precursor PEP during its internalization and phosphorylation. Furthermore, glycerol is a very affordable, plentiful, and renewable carbon source, as it is a byproduct of biodiesel and soap manufacturing (Tan et al. 2013). Additionally, 2 g/L of yeast extract was added to the culture medium because the employed strain is an L-tryptophan auxotroph. Under these conditions, the modified strain showed growth for 17 h before entering the stationary phase, which lasted 72 h. At 18 h, just at the beginning of the stationary phase, catechol melanin started to accumulate. A total of 1.21 g/L of catechol melanin were recovered from the culture medium, while 0.73 g/L of catechol was accumulated at the end of the culture (Table 1) (Mejía-Caballero et al. 2016). This finding suggests that the rate of synthesis of this precursor is greater than the capacity of MutMelA to metabolize it. Therefore, tyrosinase activity should be increased as a goal to improve strain performance.
The yeast Yarrowia lipolytica displays a natural capacity to produce pyomelanin. To improve the de novo pyomelanin synthesis capacity in this yeast, a strategy was followed based on the utilization of Y. lipolytica strain JMY8032 which was engineered for aromatic amino acid production. In this strain, flow into the L-tyrosine and L-phenylalanine pathways was increased by expressing unregulated mutant versions of enzymes DAHP synthase, chorismate mutase, and aromatic aminotransferase I. Meanwhile, gene 4HPPD encoding 4-hydroxyphenyl pyruvate dioxygenase was disrupted. The resulting mutant strain did not produce pyomelanin, confirming that this gene is involved in HGA synthesis. Thus, to generate a strain that overexpresses 4HPPD, this gene was cloned under the control of the strong constitutive pTEF promoter. The Golden Gate cloning procedure was employed to generate strain JMY8208 with 4HPPD integrated into the chromosome (Larroude et al. 2021). Genetic characterization of this strain showed that it contained three copies of the 4HPPD overexpression cassette. Then, this strain was grown in YNB medium supplemented with glucose at 28 °C in shake flasks, and, after 5 days of culture, 4.5 g/L of pyomelanin was synthesized (Larroude et al. 2021).
6 Conclusions and Perspectives
Melanins can be considered functional polymers with multiple potential industrial applications. Some significant technological problems related to melanins are achieving their large-scale synthesis while obtaining specific chemical composition and maintaining affordable production costs. These challenges can be overcome by isolating and employing naturally occurring melanogenic bacteria. This proposal presents various benefits, such as the potential to rapidly establish a production process. However, using naturally occurring melanogenic organisms might have certain downsides, such as the need to use complicated conditions to activate the synthesis of melanin. Utilizing complex media makes purifying processes more difficult and increases the risk of producing melanin with undesirable chemical components. In turn, these issues can be solved by using genetic engineering techniques to create new melanogenic strains or to enhance the expression of naturally occurring genes involved in melanogenesis. Furthermore, it is increasingly easier to do so thanks to the growing body of information available regarding the biochemistry and genetics of melanin synthesis in many species. Through these innovative techniques, new melanogenic strains have been developed that can produce melanin from simple carbon sources. These efforts have resulted in strains and processes capable of producing melanin polymers at the gram scale (Table 1).
Tyrosinase gene overexpression is the primary genetic modification used to create or enhance melanogenic organisms. This process generally involves cloning the tyrosinase gene in a replicative plasmid vector controlled by an inducible promoter. By adding inducers, this method allows exact control of the amount and timing of gene expression, enabling the optimization of the production process. Furthermore, antibiotics must be added as a selective pressure when employing expression plasmids, for example, in cases when it is necessary to prevent the development of plasmid-less cells. The requirement for a chemical inducer to be present in the culture medium is another issue. Antibiotics and inducers drive up manufacturing costs and make purifying processes more challenging. Hence, alternative antibiotic-free plasmid selection methods, as well as non-chemically based gene induction techniques, need to be developed, including process optimization, to produce melanins at grams per liter level to simplify purification processes and to reduce production costs. (Vidal et al. 2008).
When comparing the strains that transform melanin precursors present in the culture medium, it can be observed that in several of the reports reviewed so far, the melanin titers and volumetric productivities are lower in processes where the production strain used was modified by metabolic engineering where the objective was challenging to convert the carbon sources into melanins (Table 1). In these cases, the reported titers and productivities for eumelanin are lower than those that have been seen when L-tyrosine was used as a precursor (Santos et al. 2012). This fact demonstrates that there may still be room for improvement in the production process and strain engineering.
To improve the currently available melanin production strains, the use of synthetic biology, mutagenesis techniques, and adaptive laboratory evolution (ALE), should be assessed (Bassalo et al. 2016). Specifically, the engineering of complicated phenotypes may be possible with the application of ALE. In one study, ALE was paired with a synthetic biosensor module that reacts to the intracellular concentration of aromatic amino acids, which resulted in an improved strain of S. cerevisiae able to produce muconic acid (Leavitt et al. 2017). This last strain displayed an increased flux in the common aromatic amino acid pathway; thus, it could be further modified to enhance L-tyrosine synthesis by following established methods. The S. cerevisiae strain created in the such study could also serve as a platform for eumelanin production if such modifications are made. In a different publication, an E. coli strain expressing the MelA tyrosinase from R. etli was used to create a high-throughput screen for the manufacture of L-tyrosine by linking the synthesis of this amino acid to the production of melanin (Santos and Stephanopoulos 2008). This technique was used to find E. coli strains that have increased L-tyrosine synthesis capacity. In said work, E. coli was genetically modified using metabolic engineering techniques which resulted in high-level L-tyrosine synthesis. Global transcription machinery engineering (gTME) was applied to this strain to increase its capacity to synthesize L-tyrosine (Alper et al. 2006). This technique was performed by producing two different gTME libraries of the RNA polymerase rpoA and rpoD subunits in the modified strain of E. coli. Based on the melanin color intensity of colonies from these libraries, improved L-tyrosine producers from these two libraries were identified on agar plates. When compared to the modified parent strain, three mutant isolates showed a twofold increase in L-tyrosine titer (Santos et al. 2012). Additionally, it should be highlighted that these strains may be used for eumelanin production from glucose.
Tyrosinase activity is one of the factors limiting production in strains engineered to synthesize melanin from a simple carbon source (Chávez-Béjar et al. 2013; Mejía-Caballero et al. 2016), and it could also be limiting the synthesis of melanin in other modified strains. Thus, it is crucial to evaluate tyrosinase enzymes from various biological sources to identify those with the required characteristics for biotechnological application. The extensive genome and metagenome data that is currently available should provide many genes encoding tyrosinases that can be evaluated experimentally for melanin production. Meanwhile, protein engineering could be a practical way to enhance this family of enzymes. Furthermore, an advantage of working with tyrosinases is the simple activity assay based on visual detection of melanin production, which allows for high-throughput selection methods (Santos and Stephanopoulos 2008).
Despite the technological advancements in the design of strains and methods for the synthesis of melanin, there are still many fundamental concerns that need to be resolved. For instance, the kinetics of melanin polymerization is a significant issue, as it is believed that after being produced in the cytosol, melanin precursors leave the cell and begin to polymerize in the culture media. Consequently, as the polymer increases in size, a wide variety of melanin molecules are produced. These macromolecules should have unique physical characteristics. For example, distinct eumelanin isolates from different manufacturing cultures exhibit a range of hues, from yellow to black (Chávez-Béjar et al. 2013). Therefore, it is crucial to research the kinetics of melanin polymerization in production cultures and the features of polymers of specific sizes as this information could help isolate products with specific attributes.
As observed in the publications examined in this chapter, most of the published research on microbial melanin production is focused on eumelanin. This makes sense, as it is a polymer that has undergone considerable characterization since it is the type of melanin found in humans. Therefore, the produced eumelanin may be applied in the medical and cosmetic fields. Additionally, it should be emphasized that in terms of their chemical composition, melanins are a diverse set of polymers. In consequence, only a small portion of this chemical diversity has been studied thus far. Yet, processes used for the production of catechol, caffeic acid, and protocatechualdehyde melanins have been reported, in addition to eumelanin. Therefore, it stands to reason that different forms of melanin would operate differently depending on the application. For example, a recent study found that protocatechualdehyde-based melanin performed better than eumelanin or caffeic acid melanin when used as a pigment in soft contact lenses (Ahn et al. 2019). Additionally, synthetic non-natural amino acids and other substances that tyrosinases can use as substrates can be used to create non-natural melanins (Della-Cioppa et al. 1990). As a result, a wide variety of this class of polymers is expected to be produced soon. Finally, it is expected that the range of applications for these aromatic polymers should significantly increase when new strains and production techniques for novel natural and synthetic melanins are developed.
References
Aguilera F, McDougall C, Degnan BM (2013) Origin, evolution and classification of type-3 copper proteins: lineage-specific gene expansions and losses across the Metazoa. BMC Evol Biol 13:96
Ahn SY, Choi M, Jeong DW, Park S, Park H, Jang KS, Choi KY (2019) Synthesis and chemical composition analysis of protocatechualdehyde-based novel melanin dye by 15T FT-ICR: high dyeing performance on soft contact lens. Dyes Pigments 160:546–554
Ahn SY, Jang S, Sudheer PD, Choi KY (2021) Microbial production of melanin pigments from caffeic acid and L-tyrosine using Streptomyces glaucescens and FCS-ECH-expressing Escherichia coli. Int J Mol Sci 22(5):2413
Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314(5805):1565–1568
Ambrico M, Vecchia NFD, Ambrico PF, Cardone A, Cicco SR, Ligonzo T et al (2014) A Photoresponsive red-hair-inspired polydopamine-based copolymer for hybrid photocapacitive sensors. Adv Funct Mater 24(45):7161–7172
Balderas-Hernández VE, Sabido-Ramos A, Silva P, Cabrera-Valladares N, Hernández-Chávez G, Báez-Viveros JL et al (2009) Metabolic engineering for improving anthranilate synthesis from glucose in Escherichia coli. Microb Cell Factories 8(1):19
Balderas-Hernández VE, Treviño-Quintanilla LG, Hernández-Chávez G, Martinez A, Bolívar F, Gosset G (2014) Catechol biosynthesis from glucose in Escherichia coli anthranilate-overproducer strains by heterologous expression of anthranilate 1, 2-dioxygenase from Pseudomonas aeruginosa PAO1. Microb Cell Factories 13(1):136
Bassalo MC, Liu R, Gill RT (2016) Directed evolution and synthetic biology applications to microbial systems. Curr Opin Biotechnol 39:126–133
Bolognese F, Scanferla C, Caruso E, Orlandi VT (2019) Bacterial melanin production by heterologous expression of 4-hydroxyphenylpyruvate dioxygenase from Pseudomonas aeruginosa. Int J Biol Macromol 133:1072–1080
Cabrera-Valladares N, Martínez A, Pinero S, Lagunas-Munoz VH, Tinoco R, De Anda R et al (2006) Expression of the melA gene from Rhizobium etli CFN42 in Escherichia coli and characterization of the encoded tyrosinase. Enzym Microb Technol 38(6):772–779
Chávez-Béjar MI, Lara AR, López H, Hernández-Chávez G, Martinez A, Ramírez OT et al (2008) Metabolic engineering of Escherichia coli for L-tyrosine production by expression of genes coding for the chorismate mutase domain of the native chorismate mutase-prephenate dehydratase and a cyclohexadienyl dehydrogenase from Zymomonas mobilis. Appl Environ Microbiol 74(10):3284–3290
Chávez-Béjar MI, Balderas-Hernández VE, Gutiérrez-Alejandre A, Martinez A, Bolívar F, Gosset G (2013) Metabolic engineering of Escherichia coli to optimize melanin synthesis from glucose. Microb Cell Factories 12(1):108
d’Ischia M, Wakamatsu K, Cicoira F, Di Mauro E, Garcia-Borron JC, Commo S et al (2015) Melanins and melanogenesis: from pigment cells to human health and technological applications. Pigment Cell Melanoma Res 28(5):520–544
Della-Cioppa G, Garger SJ, Sverlow GG, Turpen TH, Grill LK (1990) Melanin production in Escherichia coli from a cloned tyrosinase gene. Bio/Technology 8(7):634
Esposito R, D’Aniello S, Squarzoni P, Pezzotti MR, Ristoratore F, Spagnuolo A (2012) New insights into the evolution of metazoan tyrosinase gene family. PLoS One 7(4):e35731
Fairhead M, Thöny-Meyer L (2012) Bacterial tyrosinases: old enzymes with new relevance to biotechnology. New Biotechnol 29(2):183–191
Garcia-Molina F, Munoz JL, Varon R, Rodriguez-Lopez JN, Garcia-Canovas F, Tudela J (2007) A review on spectrophotometric methods for measuring the monophenolase and diphenolase activities of tyrosinase. J Agric Food Chem 55(24):9739–9749
Gibello A, Ferrer E, Sanz J, Martin M (1995) Polymer production by Klebsiella pneumoniae 4-hydroxyphenylacetic acid hydroxylase genes cloned in Escherichia coli. Appl Environ Microbiol 61(12):4167–4171
Glass K, Ito S, Wilby PR, Sota T, Nakamura A, Bowers CR et al (2012) Direct chemical evidence for eumelanin pigment from the Jurassic period. Proc Natl Acad Sci 109(26):10218–10223
González V, Bustos P, Ramírez-Romero MA, Medrano-Soto A, Salgado H, Hernández-González I et al (2003) The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol 4(6):R36
Guo J, Rao Z, Yang T, Man Z, Xu M, Zhang X (2014) High-level production of melanin by a novel isolate of Streptomyces kathirae. FEMS Microbiol Lett 357(1):85–91
Guo J, Rao Z, Yang T, Man Z, Xu M, Zhang X, Yang ST (2015) Cloning and identification of a novel tyrosinase and its overexpression in Streptomyces kathirae SC-1 for enhancing melanin production. FEMS Microbiol Lett 362(8):fnv041
Horneck G, Klaus DM, Mancinelli RL (2010) Space microbiology. Microbiol Mol Biol Rev 74(1):121–156
Ito S (2003) A chemist’s view of melanogenesis. Pigment Cell Res 16(3):230–236
Jaenicke E, Decker H (2004) Functional changes in the family of type 3 copper proteins during evolution. Chembiochem 5:163–169
Jang S, Gang H, Kim BG, Choi KY (2018) FCS and ECH dependent production of phenolic aldehyde and melanin pigment from l-tyrosine in Escherichia coli. Enzym Microb Technol 112:59–64
Kaintz C, Mauracher SG, Rompel A (2014) Type-3 copper proteins: recent advances on polyphenol oxidases. In: Christov CZ (ed) Advances in protein chemistry and structural biology: metal-containing enzymes. Academic, pp 1–35
Kampatsikas I, Rompel A (2021) Similar but still different: which amino acid residues are responsible for varying activities in type-iii copper enzymes? ChemBioChem 22:1161–1175
Kim H, Yeon YJ, Cho YR, Song W, Pack SP, Choi YS (2016) A cold-adapted tyrosinase with an abnormally high monophenolase/diphenolase activity ration originating from the marine archaeon Candidatus Nitrosopumilus koreensis. Biotechnol Lett 38:1535–1542
Krol ES, Liebler DC (1998) Photoprotective actions of natural and synthetic melanins. Chem Res Toxicol 11(12):1434–1440
Lagunas-Muñoz VH, Cabrera-Valladares N, Bolívar F, Gosset G, Martinez A (2006) Optimum melanin production using recombinant Escherichia coli. J Appl Microbiol 101(5):1002–1008
Larroude M, Onésime D, Rué O, Nicaud JM, Rossignol T (2021) A Yarrowia lipolytica strain engineered for pyomelanin production. Microorganisms 9(4):838
Leavitt JM, Wagner JM, Tu CC, Tong A, Liu Y, Alper HS (2017) Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae. Biotechnol J 12(10):1600687
Lee HS, Choi JY, Kwon SJ, Park ES, Oh BM, Kim JH, Lee PC (2022) Melanin biopolymer synthesis using a new melanogenic strain of Flavobacterium kingsejongi and a recombinant strain of Escherichia coli expressing 4-hydroxyphenylpyruvate dioxygenase from F. kingsejongi. Microb Cell Factories 21(1):1–15
Lin WP, Lai HL, Liu YL, Chiung YM, Shiau CY, Han JM et al (2005) Effect of melanin produced by a recombinant Escherichia coli on antibacterial activity of antibiotics. J Microbiol Immunol Infect 38(5):320–326
Lindgren J, Moyer A, Schweitzer MH, Sjövall P, Uvdal P, Nilsson DE et al (2015) Interpreting melanin-based coloration through deep time: a critical review. Proc R Soc B Biol Sci 282(1813):20150614
Martínez LM, Martinez A, Gosset G (2019) Production of melanins with recombinant microorganisms. Front Bioeng Biotechnol 7:285
Mejía-Caballero A, de Anda R, Hernández-Chávez G, Rogg S, Martinez A, Bolívar F et al (2016) Biosynthesis of catechol melanin from glycerol employing metabolically engineered Escherichia coli. Microb Cell Factories 15(1):161
Montefiori DC, Zhou J (1991) Selective antiviral activity of synthetic soluble L-tyrosine and L-dopa melanins against human immunodeficiency virus in vitro. Antivir Res 15(1):11–25
Nikodinovic-Runic J, Martin LB, Babu R, Blau W, O’Connor KE (2009) Characterization of melanin-overproducing transposon mutants of Pseudomonas putida F6. FEMS Microbiol Lett 298(2):174–183
Nofsinger JB, Liu Y, Simon JD (2002) Aggregation of eumelanin mitigates photogeneration of reactive oxygen species. Free Radic Biol Med 32(8):720–730
Park H, Yang I, Choi M, Jang KS, Jung JC, Choi KY (2020) Engineering of melanin biopolymer by co-expression of MelC tyrosinase with CYP102G4 monooxygenase: structural composition understanding by 15 tesla FT-ICR MS analysis. Biochem Eng J 157:107530
Piñero S, Rivera J, Romero D, Cevallos MA, Martínez A, Bolívar F, Gosset G (2007) Tyrosinase from Rhizobium etli is involved in nodulation efficiency and symbiosis-associated stress resistance. J Mol Microbiol Biotechnol 13(1–3):35–44
Pretzler M, Rompel A (2018) What causes the different functionality in type-III-copper enzymes? A state of the art perspective. Inorg Chim Acta 481:25–31
Prieto MA, Perez-Aranda A, Garcia JL (1993) Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J Bacteriol 175(7):2162–2167
Różanowska M, Sarna T, Land EJ, Truscott TG (1999) Free radical scavenging properties of melanin: interaction of eu-and pheo-melanin models with reducing and oxidising radicals. Free Radic Biol Med 26(5–6):518–525
Ruan L, Yu Z, Fang B, He W, Wang Y, Shen P (2004) Melanin pigment formation and increased UV resistance in Bacillus thuringiensis following high temperature induction. Syst Appl Microbiol 27(3):286–289
Ruan L, He W, He J, Sun M, Yu Z (2005) Cloning and expression of mel gene from Bacillus thuringiensis in Escherichia coli. Antonie Van Leeuwenhoek 87(4):283–288
Saini AS, Melo JS (2015) One-pot green synthesis of eumelanin: process optimization and its characterization. RSC Adv 5(59):47671–47680
Santos CNS, Stephanopoulos G (2008) Melanin-based high-throughput screen for L-tyrosine production in Escherichia coli. Appl Environ Microbiol 74(4):1190–1197
Santos CNS, Xiao W, Stephanopoulos G (2012) Rational, combinatorial, and genomic approaches for engineering L-tyrosine production in Escherichia coli. Proc Natl Acad Sci 109(34):13538–13543
Sarna T, Hyde JS, Swartz HM (1976) Ion-exchange in melanin: an electron spin resonance study with lanthanide probes. Science 192(4244):1132–1134
Seo D, Choi KY (2020) Heterologous production of pyomelanin biopolymer using 4-hydroxyphenylpyruvate dioxygenase isolated from Ralstonia pickettii in Escherichia coli. Biochem Eng J 157:107548
Tan HW, Aziz AA, Aroua MK (2013) Glycerol production and its applications as a raw material: a review. Renew Sust Energ Rev 27:118–127
Valderrama B, Oliver P, Medrano-Soto A, Vazquez-Duhalt R (2003) Evolutionary and structural diversity of fungal laccases. Antonie Van Leeuwenhoek 84(4):289–299
Van Holde KE, Miller KI, Decker H (2001) Hemocyanins and invertebrate evolution. J Biol Chem 279(19):15563–15566
Vidal L, Pinsach J, Striedner G, Caminal G, Ferrer P (2008) Development of an antibiotic-free plasmid selection system based on glycine auxotrophy for recombinant protein overproduction in Escherichia coli. J Biotechnol 134(1–2):127–136
Wogelius RA, Manning PL, Barden HE, Edwards NP, Webb SM, Sellers WI et al (2011) Trace metals as biomarkers for eumelanin pigment in the fossil record. Science 333(6049):1622–1626
Yanofsky C, Horn V, Bonner M, Stasiowski S (1971) Polarity and enzyme functions in mutants of the first three genes of the tryptophan operon of Escherichia coli. Genetics 69(4):409
Zhang F, Kearns SL, Orr PJ, Benton MJ, Zhou Z, Johnson D et al (2010) Fossilized melanosomes and the colour of cretaceous dinosaurs and birds. Nature 463(7284):1075
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Martínez, L.M., Cisneros-Martínez, A.M., Hernández-Chávez, G., Martinez, A., Gosset, G. (2023). Biotechnological Production of Melanins with Recombinant Microorganisms. In: Gosset, G. (eds) Melanins: Functions, Biotechnological Production, and Applications. Springer, Cham. https://doi.org/10.1007/978-3-031-27799-3_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-27799-3_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-27798-6
Online ISBN: 978-3-031-27799-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)