Abstract
Recently, deep-learning-based approaches have been widely studied for deformable image registration task. However, most efforts directly map the composite image representation to spatial transformation through the convolutional neural network, ignoring its limited ability to capture spatial correspondence. On the other hand, Transformer can better characterize the spatial relationship with attention mechanism, its long-range dependency may be harmful to the registration task, where voxels with too large distances are unlikely to be corresponding pairs. In this study, we propose a novel Deformer module along with a multi-scale framework for the deformable image registration task. The Deformer module is designed to facilitate the mapping from image representation to spatial transformation by formulating the displacement vector prediction as the weighted summation of several bases. With the multi-scale framework to predict the displacement fields in a coarse-to-fine manner, superior performance can be achieved compared with traditional and learning-based approaches. Comprehensive experiments on two public datasets are conducted to demonstrate the effectiveness of the proposed Deformer module as well as the multi-scale framework.
J. Chen and D. Lu—Equal contribution and the work was done at Tencent Jarvis Lab.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Deformable image registration (DIR), which aims to estimate a proper deformable field \(\phi \) that can warp the moving image \(I_{m}\) to align with the fixed image \(I_{f}\), is an essential procedure in various medial image analysis tasks, such as surgical navigation [8], image reconstruction [18] and atlas construction [5]. Traditional registration approaches [1, 27, 28] align voxels with similar appearance through solving an optimization problem for each volume pair. Unfortunately, the computational intensive optimization limits their usage in practical clinical applications.
Recently, unsupervised deep-learning-based DIR approaches [2, 12, 29,30,31] have been widely studied for their computational efficiency. Many methods, such as VoxelMorph [2], Dual-PRNet [12] and FAIM [17], adopt the convolutional neural network (CNN) as their backbone because of its superior performance for various vision tasks [13, 25]. However, CNN shows limited capability in capturing spatial relationship [24], which becomes a bottleneck for these methods. With the recent development of vision Transformer, some efforts have also been made to explore its effectiveness in DIR task [3]. Although its attention mechanism [24] is potentially more suitable to characterize spatial relationship, directly applying it for DIR task may lead to inferior performance due to its long-range dependency. Because of the fixed anatomical structure of medical scans, preserving tissue discontinuity [4] is essential for medical image registration task. Therefore, the corresponding voxel should only be found in a limited local range, which could be undermined by the long-range dependency.
To this end, we propose a Deformer module to explicitly exploit the intrinsic property of registration task for facilitating the mapping from image representation to spatial transformation. We make a simple yet critical formulation that the displacement between a pair of voxels can be considered as the weighted summation of several basic vectors (referred as displacement bases as well) with three elements, representing the x, y and z components, respectively. Leveraging attention mechanism’s ability to capture spatial relationship from image representation, the proposed Deformer module adopts two separate branches to implement such a paradigm. The first branch learns the displacement bases, denoting the potential deformable directions, while the other one predicts the attention weight for each basis, representing the offset length along each direction. Thus, the voxel-wise displacement can be obtained via matrix product of the basis vectors and the attention weights. In addition, the multi-head strategy is applied to enable the Deformer module to extract latent information from different representation subspaces. Furthermore, a multi-scale framework, namely Deformer-based Multi-scale Registration (DMR) is customized to further boost the performance. Unlike previous methods, which either successively predict the displacement fields at different scales [1, 21, 28] or only exploit multi-scale feature maps through U-Net architecture [2, 22], we propose to introduce the Deformer module along with an auxiliary loss at each scale to learn the displacement fields in a coarse-to-fine manner and automatically fuse them through the tailored refining network, such that the information at different scales can be fully exploited without intensive successive computation.
2 Method
Problem Setting. Given a pair of moving and fixed medical 3D scans (called images or volumes as well) \(\{M,F\}\in \mathbb {R}^{D\times W \times H}\), the objective of image registration is to estimate a deformation field \(\phi \in \mathbb {R}^{3\times D\times W\times H}\) such that the warped moving scan \(\mathcal {T}(M,\phi )\in \mathbb {R}^{D\times W\times H}\) can be aligned with the fixed scan F, where D denotes the depth, W the width, H the height and 3 for the 3D spatial dimension. Specifically, the \(\phi \) can be represented as \(Id+u\), where Id denotes identity transformation and u represents the displacement field. To be consistent with previous registration studies [2, 3, 15], a spatial transformation network [14] is adopted to map the moving image to the fixed image with the estimated displacement field.
Method Overview. As shown in Fig. 1, the proposed DMR framework consists of three main components, the multi-scale encoder to extract L (\(L=4\) in this study) pairs of feature maps \(\{f_M^l,f_F^l\}_{l=1}^L\) from both the moving image M and the fixed image F, the proposed Deformer modules to exploit the interaction between M and F to deliver the displacement vector fields from coarse to fine, and the refining network to combine the latent information from different representation subspaces to enable high-resolution large-deformable registration. The detailed explanations of the Deformer module, the DMR network, as well as the loss function are stated in the following sections.
Deformer Module. To facilitate the mapping of the image representation to spatial relationship and increase the interpretability of the network, a Deformer module is designed to learn the displacement field. Based on the linear algebra theory, a displacement vector between two voxels can be formulated as the weighted summation of several basic vectors. For implementing this paradigm through network, the proposed Deformer module adopts attention mechanism with two separate branches, as displayed in Fig. 1b.
The first branch, i.e., the left branch in Fig. 1b, adopts a linear projection to convert every voxel-wise feature vector into N bases, each of which consists of three elements to represent the displacement values in three spatial directions, i.e., the x, y and z axes, respectively. In addition, we adopt the multi-head strategy [26] so that the information from different representation subspaces can be obtained, leading to \(K\times N\) bases for each voxel, where K represents the number of heads in each Deformer module. Note that only the feature maps of the moving images \(f_M^l\) are employed in this branch, which is sufficient to extract the most representative displacement bases for denoting the most likely deformation directions as demonstrated by the ablation study. On the other hand, the second branch, i.e., the right branch in Fig. 1b, adopts the concatenated feature map \([f_M^l, f_F^l]\) from both the moving and fixed images to learn the attention weight for each displacement basis. The similarity between the moving image and the fixed image is measured by a linear layer followed by a softmax function to impose the non-linearity of the module. It is worth mentioning that both linear projections are performed on each pair of voxel-wise feature vectors independently, resulting in a small number of parameters comparing with standard CNN or Transformer block. Besides, the latent information of the nearby voxels has already been incorporated into the feature vector through the CNN-based encoder. Therefore, the similarity measurement is not limited to the voxels at the same location, and the field-of-view is expanded with the decreasing of the feature map resolution. Finally, the attention weights are multiplied to the displacement bases followed by a head-wise average to obtain the displacement field \(u^l\) at scale l. The overall process can be formulated as:
where \(v^l_{i,j}\) denotes the displacement basis, \(w_{i,j}\) is the attention weight, \([\cdot ]\) represents the concatenation operation, while \(fc_1\) and \(fc_2\) denote the mapping functions of two branches, respectively.
Network Architecture. In order to fully exploit the latent information at different representation subspaces without introducing the intensive computation of successive network, we propose a DMR framework to learn displacement fields at different scales via the proposed Deformer module along with auxiliary loss to provide additional guidance.
As shown in Fig. 1, we first adopt a multi-scale CNN encoder to extract the latent representations at different scales from both the moving and fixed images. Specifically, sharing a similar architecture as [2], the encoder is composed of L 3D convolution blocks, each of which consists of a convolutional layer with a stride of 2 and a kernel size of 4, and a batch normalization layer followed by a leaky rectified linear unit (LeakyReLU) with a negative slope of 0.2. Therefore, the encoder can extract a sequence of L pairs of intermediate feature maps \(\{f_M^l,f_F^l\}_{l=1}^L\) with different scales through different convolution blocks. Note that both the moving and the fixed images are fed into the same encoder for feature extraction.
The refining network consists of repeated application of the fusion block followed by a convolution head to convert the last feature map to the final displacement field at the original resolution. Specifically, we denote the output feature map of fusion block l as \(g^l\), then the displacement field \(u^l\) at scale l is converted to a feature map \(h^l\) with a fixed number of channels C via a convolution block. Subsequently, the feature map of the last fusion block \(g^{l+1}\) is upsampled by a bilinear interpolation and added to \(h^l\) followed by the concatenation of the latent representations \(f_M^l\) and \(f_F^l\) to provide image information, formulated as:
The final component of the fusion block is another convolution block, i.e., Conv3d reducer, for reducing the channel of output high-level latent representations to C. After three cascaded fusion blocks, a convolution head including three decoding blocks, each of which has two successive structures with a convolutional layer and a ReLU activation function, is applied to deliver the final displacement field. An upsample layer is introduced after the first decoding block to restore the original resolution. More details about the refining network can be found in the supplementary materials.
Loss Function. To validate the effectiveness of the proposed DMR framework and Deformer module, we adopt the most commonly used objective function [2] for a fair comparison, which is defined as:
where \(\lambda \) is the regularization trade-off parameter, setting as 1 empirically. The first term \(\mathcal {L}_{sim}(\mathcal {T}(M,\phi ), F)\) measures the similarity between the warped moving images and the fixed scans using local normalized cross-correlation. The second term \( \mathcal {L}_{reg}(\phi )\) is a regularization imposed on the displacement fields to penalize local spatial variation.
As stated above, we propose to impose penalty on the intermediate displacement field at each scale to provide direct guidance for each level of Deformer module as well as the feature extractor. It is worth mentioning that the intermediate displacement fields should be upsampled to the original scale to warp the moving image for the computation of objective function. The overall loss function can be written as:
where \(\beta _{l}\) is the weight for the intermediate loss function at scale l.
3 Experiments and Discussion
Datasets. To validate the performance of the proposed approach, we conduct experiments on two publicly available datasets, i.e., the LONI Probabilistic Brain Atlas (LPBA40) dataset [23] and the Neurite subset of the Open Access Series of Imaging Studies (Neurite-OASIS) dataset [19]. The LPBA40 dataset [23] contains 40 3D volumes of whole-brain Magnetic Resonance Imaging (MRI) scans from normal volunteers, which is divided into 29, 3 and 8 scans for training, validation and testing, respectively. Skull stripping and spatial normalization are performed on each scan, followed by padding and cropping to ensure the same size of 160 \(\times \) 192 \(\times \) 160 voxels for each scan. Manual annotation of 56 structures are provided as the ground truth. The Neurite-OASIS datasetFootnote 1 is from the learn2reg 2021 challenge [11] and is a part of the OASIS Dataset Project [19], It contains 414 inter-patient 3D T1-weighted MRI brain scans from abnormal subjects with various stages of cognitive decline, which is split into 374, 20 and 20 scans for training, validation and testing, respectively. The scans are pre-processed by the challenge organizer, including skull stripping and spatial normalization via FreeSurfer and SAMSEG, and then cropped to 224 \(\times \) 192 \(\times \) 160 voxels. The subcortical segmentation maps of the 35 anatomical structures are provided as the ground truth for evaluation.
Evaluation Criteria. In the experiments of both datasets, we use subject-to-subject registration for optimization, where each pair of volume is selected randomly from the training sets. For evaluation, 8 LPBA40 and 20 Neurite-OASIS scans are mapped to a standard atlas [1]. Following previous works [2, 15], we adopt the commonly used average Dice score of the region-of-interest (ROI) masks between the warped images and fixed images as the main evaluation metric. To quantify the diffeomorphism and smoothness of the deformation fields, the average percentage of voxels with non-positive Jacobian determinant (\(|J_{\phi }| \le 0\)) in the deformation fields, the standard deviation of the Jacobian determinant (\(std(|J_{\phi }|)\)) the number of parameters, GPU memory and average running time to register each pair of scans on Neurite-OASIS dataset are also provided as supplementary metrics.
Qualitative results with segmentation labels of the example axial MR slices from the moving, fixed and warped images from different methods. The color curves represent the boundaries of several structures, including caudate (pink/red), putamen (brown/green), and lingual gyrus (blue/purple). The last image in the second row is the visualization of the final displacement field of our method. (Color figure online)
Implementation Details. Our method is implemented with PyTorch 1.4 and optimized using Adam optimizer [16] with mini-batch stochastic gradient descent. The model is trained on 4 NVIDIA V100 GPUs for 2,000 epochs. The batch size is set as 4 pairs. The learning rate is set as \(4\times 10^{-4}\) for the LPBA40 dataset, and \(10^{-3}\) for the Neurite-OASIS dataset to reduce the time consumption as it contains more images with larger size compared to LPBA40. Similar to previous studies [2, 21], no data augmentation is employed in our experiments. As for the Deformer modules, the head number K is 8 for all scales and the number N of displacement bases for each head is 64. In the refining network, C is set to 128. For optimization, we measure image similarity using local normalized cross-correlation with windows size of \(9 \times 9 \times 9\) in all the loss functions. The regularization parameter \(\lambda \) is empirically set as 1.0 and the weights \(\beta _l\) are all set to 1. Our implementation is publicly available at https://github.com/CJSOrange/DMR-Deformer.
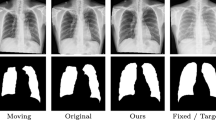
Comparison Study. For a quantitative comparison, we first compare with five state-of-the-art methods. Two of them are traditional methods, i.e., SyN [1] and NiftyReg [20], two of them utilize convolution neural networks, including VoxelMorph [2] and CycleMorph [15], and the rest two approaches incorporate vision Transformer or attention mechanism into the network, i.e., VIT-V-Net [3] and Attention-reg [24]. The results of these approaches are obtained based on the official codes released by the authors. As shown in Table 1, the proposed approach achieves 68.4% in Dice on the LPBA40 dataset and 80.4% in Dice on Neurite-OASIS, which outperforms the second best methods by 1.5% and 1.6%, respectively. Our method only generates a small percentage of folding voxels (0.624% and 1.024%) on both datasets, indicating reasonable smooth deformation fields. Moreover, we can reduce last three metrics to 7.2M/46.7G/0.61s by using a single head with little performance degradation, as shown in Table 1 of the supplementary. Figure 2 illustrates the registration results of various methods for qualitative analysis. Similar to the quantitative comparison, the proposed DMR achieves the most appealing qualitative results with better alignment between the warped image and the anatomical structure boundaries. For comparing with other multiple cascaded networks on Neurite-OASIS, we add a weakly-supervised Dice loss. DMR achieves 84.2% Dice which is comparable to Learn2Reg winner LapIRN [21] (86.2% Dice), and is superior to DLIR [6] (82.9%) and mIVIRNET [10] (83.4%). For detailed evaluation of the Dice scores for individual anatomical structures and more visualization of segmentation results as well as the displacement fields, please refer to the supplementary materials.
Ablation Study. For ablation study, we first conduct experiments to verify the effectiveness of the proposed Deformer module along with the auxiliary losses. Specifically, we compare the Deformer module with four variants: 1) Deformer-A: feeding the concatenated feature maps to the left branch for displacement basis extraction, instead of using only the feature maps of the moving images; 2) Deformer-B: removing the left branch of the Deformer module, which is equivalent to fix the displacement bases as (0, 0, 1), (0, 1, 0) and (1, 0, 0); 3) CNN: replacing the Deformer module with a CNN block, i.e., 3D ResNet [9]; 4) Transformer: replacing the Deformer module with a vision Transformer block [7]. It is worth mentioning that with only two fully connected layers performed on voxel-wise feature vectors, the proposed Deformer module has fewer parameters compared with CNN or Transformer (7.98M vs. 12.35M and 132M). The results in Table 2 demonstrate that the proposed Deformer module can better characterize the spatial transformation from image representation and using the moving images alone is sufficient to find optimal displacement bases.
We further evaluate the impact of Deformer modules at different scales by gradually removing the Deformer modules from fine to coarse scales (1/2, 1/4, 1/8 and 1/16). Naturally, the auxiliary loss at the same scale is removed along with the Deformer module. As shown in Table 3, with the removing of Deformer modules, we can observe steadily degeneration of registration performance from 68.4% to 66.2% in Dice on the LPBA40 dataset, supporting the assumption that the multi-scale Deformer module and the corresponding auxiliary loss can effectively improve the registration ability of the network. The impact of more hyper-parameters can be found in the supplementary materials.
4 Conclusion
In this paper, we proposed a novel Deformer module along with a multi-scale framework for unsupervised deformable registration. The Deformer module was designed to formulate the prediction of displacement vector as the learning of most likely deformation directions and the offset length via attention mechanism. With the two fully connected layers applied on each pair of voxel-wise feature vectors independently, the Deformer module could better characterize the local spatial correlation with fewer parameters comparing with CNN or Transformer. Further, we showed that introducing the Deformer module along with auxiliary loss in a multi-scale manner to learn the displacement fields from coarse to fine could substantially boost the registration performance. Experiments on two publicly available datasets demonstrated that our strategy outperformed the traditional and learning-based benchmark methods.
References
Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C.: Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12(1), 26–41 (2008)
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38(8), 1788–1800 (2019)
Chen, J., He, Y., Frey, E.C., Li, Y., Du, Y.: ViT-V-Net: vision transformer for unsupervised volumetric medical image registration. arXiv preprint arXiv:2104.06468 (2021)
Chen, X., Xia, Y., Ravikumar, N., Frangi, A.F.: A deep discontinuity-preserving image registration network. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12904, pp. 46–55. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87202-1_5
Dalca, A.V., Rakic, M., Guttag, J., Sabuncu, M.R.: Learning conditional deformable templates with convolutional networks. arXiv preprint arXiv:1908.02738 (2019)
De Vos, B.D., Berendsen, F.F., Viergever, M.A., Sokooti, H., Staring, M., Išgum, I.: A deep learning framework for unsupervised affine and deformable image registration. Med. Image Anal. 52, 128–143 (2019)
Dosovitskiy, A., et al.: An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
Gou, S., Chen, L., Gu, Y., Huang, L., Huang, M., Zhuang, J.: Large-deformation image registration of CT-TEE for surgical navigation of congenital heart disease. Comput. Math. Methods Med. 2018 (2018)
Hara, K., Kataoka, H., Satoh, Y.: Can spatiotemporal 3D CNNs retrace the history of 2D CNNs and ImageNet? In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 6546–6555 (2018)
Hering, A., van Ginneken, B., Heldmann, S.: mlVIRNET: multilevel variational image registration network. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 257–265. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_29
Hering, A., et al.: Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. arXiv preprint arXiv:2112.04489 (2021)
Hu, X., Kang, M., Huang, W., Scott, M.R., Wiest, R., Reyes, M.: Dual-stream pyramid registration network. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 382–390. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_43
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Jaderberg, M., Simonyan, K., Zisserman, A., et al.: Spatial transformer networks. In: Advances in Neural Information Processing Systems, vol. 28, pp. 2017–2025 (2015)
Kim, B., et al.: CycleMorph: cycle consistent unsupervised deformable image registration. Med. Image Anal. 71, 102036 (2021)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Kuang, D., Schmah, T.: FAIM – a ConvNet method for unsupervised 3D medical image registration. In: Suk, H.-I., Liu, M., Yan, P., Lian, C. (eds.) MLMI 2019. LNCS, vol. 11861, pp. 646–654. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32692-0_74
Li, R., et al.: Real-time volumetric image reconstruction and 3D tumor localization based on a single X-ray projection image for lung cancer radiotherapy. Med. Phys. 37(6Part1), 2822–2826 (2010)
Marcus, D.S., Wang, T.H., Parker, J., Csernansky, J.G., Morris, J.C., Buckner, R.L.: Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 19(9), 1498–1507 (2007)
Modat, M., et al.: Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 98(3), 278–284 (2010)
Mok, T.C.W., Chung, A.C.S.: Large deformation diffeomorphic image registration with laplacian pyramid networks. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12263, pp. 211–221. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59716-0_21
Rohé, M.-M., Datar, M., Heimann, T., Sermesant, M., Pennec, X.: SVF-Net: learning deformable image registration using shape matching. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 266–274. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_31
Shattuck, D.W., et al.: Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39(3), 1064–1080 (2008)
Song, X., et al.: Cross-modal attention for MRI and ultrasound volume registration. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12904, pp. 66–75. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87202-1_7
Tan, M., Le, Q.: EfficientNet: rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114. PMLR (2019)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, pp. 5998–6008 (2017)
Vercauteren, T., Pennec, X., Perchant, A., Ayache, N.: Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 45(1), S61–S72 (2009)
Wang, H., et al.: Validation of an accelerated ‘demons’ algorithm for deformable image registration in radiation therapy. Phys. Med. Biol. 50(12), 2887 (2005)
Xu, Z., et al.: Double-uncertainty guided spatial and temporal consistency regularization weighting for learning-based abdominal registration. arXiv preprint arXiv:2107.02433 (2021)
Xu, Z., Luo, J., Yan, J., Li, X., Jayender, J.: F3RNET: full-resolution residual registration network for deformable image registration. Int. J. Comput. Assist. Radiol. Surg. 16(6), 923–932 (2021)
Xu, Z., et al.: Adversarial uni- and multi-modal stream networks for multimodal image registration. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12263, pp. 222–232. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59716-0_22
Acknowledgements
This work was funded by the Scientific and Technical Innovation 2030-“New Generation Artificial Intelligence” (No. 2020AAA0104100), Key R &D Program of China (2018AAA0100104, 2018AAA0100100) and Natural Science Foundation of Jiangsu Province (BK20211164).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, J. et al. (2022). Deformer: Towards Displacement Field Learning for Unsupervised Medical Image Registration. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13436. Springer, Cham. https://doi.org/10.1007/978-3-031-16446-0_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-16446-0_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16445-3
Online ISBN: 978-3-031-16446-0
eBook Packages: Computer ScienceComputer Science (R0)