Abstract
Graph neural network (GNN) has achieved tremendous success in histological image classification, as it can explicitly model the notion and interaction of different biological entities (e.g., cell, tissue and etc.). However, the potential of GNN has not been fully unleashed for histological image analysis due to (1) the fixed design mode of graph structure and (2) the insufficient interactions between multi-level entities. In this paper, we proposed a novel spatial-hierarchical GNN framework (SHGNN) equipped with a dynamic structure learning (DSL) module for effective histological image classification. Compared with traditional GNNs, the proposed framework has two compelling characteristics. First, the DSL module integrates the positional attribute and semantic representation of entities to learn the adjacency relationship of them during the training process. Second, the proposed SHGNN can extract rich and discriminative features by mining the spatial features of different entities via graph convolutions and aggregating the semantic of multi-level entities via a vision transformer (ViT) based interaction mechanism. We evaluate the proposed framework on our collected colorectal cancer staging (CRCS) dataset and the public breast carcinoma subtyping (BRACS) dataset. Experimental results demonstrate that our proposed method yield superior classification results compared to state-of-the-arts.
W. Hou and H. Huang—Contributed equally to this work.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Histological image classification
- Graph neural network
- Dynamic structure
- Hierarchical representation
- Vision transformer
1 Introduction
Histopathological examination is considered as the “golden standard” for diagnosis and treatment planning of many diseases [1, 2]. For example, in clinical practice, gastroenterologists need to manually assess the histological images obtained by whole-slide scanning systems [3, 4]. However, due to the complex morphology and structure of human tissues and the continuum of histologic features phenotyped across the diagnostic spectrum, it is a tedious and time-consuming task to manually classify the histological images [5, 6]. Therefore, automatic histological image classification is highly demanded in clinical practice.
Over the years, convolutional neural networks (CNNs) [7, 8] have greatly promoted the development of computational pathology. For example, Bai et al. [9] employed a pretrained google inception net (GoogLeNet) for learning high-level representations and further constructed a softmax classifier for patch-level classification of NHL histological images. Li et al. [10] proposed an atrous DenseNet (ADN) to integrate atrous convolutions with the dense block to extract multiscale features for histological image classification. However, one of the main shortcomings of CNN-based approaches is that convolution kernels only deal with regular pixel-wise regions and thereby ignore the notion of biological entities (e.g., cell, tissue and etc.), which are essential for the histological image classification task and interpretability analysis [11].
Recently, graph neural networks (GNNs) [12,13,14] has shown great potential in modeling the notion and interaction of biological entities [11, 15,16,17,18,19] for histological image classification. For example, Zhou et al. [19] first extracted the nuclei in histological images and constructed a cell-level graph according to the spatial relationship, and then designed an GNN to process the cell-level graph and performed image classification. In addition to cell-level graph, Pati et al. [11] proposed a HACT for pathological image analysis, where they introduced the tissue-level graph by the superpixel technique. Although the above GNN-based methods are able to improve the performance of histological image analysis, they still have two shortcomings that prevent them from achieving more satisfactory results. On the one hand, existing methods [11, 19] connect the entities to generate a static graph representation according to the prior hypothesis, which lacks sufficient medical explanation and thus may degrade the representation capability of the graph. On the other hand, HACT [11] aligns multi-level entities and aggregates them by the add operation, resulting in the information loss and the insufficient interaction.
In this paper, we propose a novel spatial-hierarchical GNN framework (called SHGNN) equipped with dynamic structure learning (DSL) to explore the spatial topology and hierarchical dependency of the multi-level biological entities for improving histological image classification. We first design a DSL module to integrate the positional attribute and semantic feature representation of entities to automatically learn the adjacency relationship among different entities during the training process. By using such a dynamic learning scheme, the proposed framework is capable of capturing the task-related information for dynamic graph structure building, leading to more reliable message passing. More importantly, we adopt graph convolutional operations to mine the spatial features of different nodes (entities) and further design a novel vision transformer (ViT) paradigm to attentively aggregate the semantic of multi-level entities, obtaining more rich and discriminative features for high accurate classification. We conduct extensive experiments on our collected colorectal cancer staging (CRCS) dataset and the public breast carcinoma subtyping (BRACS) dataset to evaluate our proposed framework. The experimental results demonstrate that our method consistently outperforms state-of-the-art approaches on both datasets. Our code is made available at https://github.com/HeLongHuang/SHGNN.
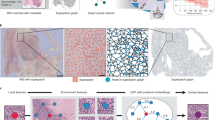
Overview of the proposed framework. An analytical scheme with \(L=3\) levels is used for illustration. Multi-level biological entities are extracted and processed by our proposed SHGNN with DSL, to construct task-related graph structure and excavate spatial-hierarchical features for classification. Note that not all nodes and hierarchical relations are shown for visual clarity.
2 Methodology
Figure 1 illustrates the pipeline of the proposed approach. We first extract the multi-level entities from the histological images, including the nuclei and the tissues with different scales (see Fig. 1(a)). Each entity is regarded as a node of the graph, for which the representation is extracted by the ImageNet [20] pretrained CNN encoder. For the DSL module (see Fig. 1(b)), we construct independent learning branches on the position attribute and feature representation of entities, and further combine their embedded representation as the judgment basis of adjacency relationship, so as to achieve the construction of dynamic multi-level graphs. The multi-level graphs are fed in parallel into the proposed spatial-hierarchical graph neural network (see Fig. 1(c)) for spatial relationship learning and hierarchical interaction to generate the final graph representation. The final graph representation of the histological image is fed into an attention pooling layer with a multi-layer perceptron (MLP) head to produce the classification result in a supervised manner. In the following subsections, we will detail the multi-level entities extraction, the design of DSL module, and the learning strategy of SHGNN, respectively.
2.1 Multi-level Entities Extraction
For a given sample, let the histological image X and classification label Y be a single observation in a dataset \(\{X_i,Y_i\}^{N}_{i=1}\). To construct a biologically meaningful representation for X, we conduct multi-level histological entity analysis in X, including (1) cell analysis and (2) multi-scale tissue analysis. Cell analysis aims to characterize the low-level cell information. Specifically, a pretrained HoVer-Net [21] is used to obtain the segmentation masks of nuclei. The feature representation of each cell entities is a 512-dimensional vector, which is extracted by processing the patches centered around nuclei centroids via a pretrained ResNet34 encoder [7]. We denote the feature set of cell-level entities as \(\mathcal {V}_{0} \in \mathbb {R}^{|\mathcal {V}_{0}| \times 512}\), where \(|\mathcal {V}_{0}|\) denote the number of cell-level entities. Multi-scale tissue analysis aims to effectively depict the high-level tissue microenvironments with different scales of shape (e.g., stroma, vessel and etc.). Specifically, the SLIC superpixel [22] algorithms with different scales (the number of superpixels per image) are employed to obtain the segmentation masks of multi-scale tissues. The feature representation of each tissue is figured out by averaging the 512-dimensional deep features [7] of the patches cropped from the tissue. We denote the feature set of tissue-level entities at scale \(s \in S\) as \(\mathcal {V}_{s} \in \mathbb {R}^{|\mathcal {V}_{s}| \times 512}\), where \(|\mathcal {V}_{s}|\) denote the number of tissue-level entities at scale s. After cell analysis and multi-scale tissue analysis, \(L= S+1\) levels of entities are extracted. According to the image size and hardware conditions, L can be appropriately increased to achieve more fine-grained hierarchical entity analysis.
2.2 Dynamic Structure Learning
Previous methods [11, 19] often link the entities to generate a static graph representation based on the prior hypothesis, such as spatial distance adjacent matrix and k-nearest neighbor adjacent matrix. However, these methods lack medical explanation, which may degrade the representation capability of the graph. As shown in Fig. 1, we propose a DSL module to dynamically learn the adjacent relations between entities. Specifically, we comprehensively consider the feature representation \(\mathcal {V}_l\) and position attribute \(\mathcal {P}_l\) of \(l_{th}\) level entities as the judgment basis, where the position attribute \(\mathcal {P}_l\in \mathbb {R}^{|\mathcal {V}_l| \times 2}\) is the 2D-spatial coordination of the centroid of the entity. We first align \(\mathcal {V}_l\) and \(\mathcal {P}_l\) to the embedding space by two projection layers and concatenate them to obtain the joint representation \(\mathcal {J}_l\). This process can be written as:
where \(\textbf{W}_1\) and \(\textbf{W}_2\) are learnable weight matrices of fully-connected (FC) layer. \(Concat[\cdot ]\) denotes the concatenation operation and \(\sigma \) denotes the activation function, such as LeakyReLU. Next, in the space \(\mathcal {J}_l\), we use an online k-Nearest Neighborhood (k-NN) criteria to build the topology for each entity. Specifically, within a threshold distance \(d_{min}\), an edge \(e_{uv} \in \mathcal {E}_l\) is built for each entity \(v \in \mathcal {V}_l\) if:
where \({||v, w||_{2}}\) is the L2 distance of v and w in the space \(\mathcal {J}_l\). \(\text {min}(\cdot )\) is the minimum operation. \(d_{k}\) is the \(k_{th}\) smallest distance in \({||v, w||_{2}}\).
The entities of each level are fed into the DSL module respectively to generate the adjacent matrix. Formally, the graph representation of multi-level entities can be represented as \(\mathcal {G}_l = \{\mathcal {V}_l, \mathcal {E}_l\}\), \( l \in \{0,1,...,L-1\}\). Specially, \(\mathcal {G}_0\) denotes the cell graph while the others denote the tissue graphs. It is worth noting that the proposed DSL module is embedded into our model end-to-end, so that the multi-level graph structure can dynamically change and is able to autonomously capture task-related information for more reliable message passing.
2.3 Spatial-Hierarchical Graph Neural Network
Spatial Graph Convolution. As shown in Fig. 1, we adopt a graph convolution to extract the spatial features in the spatial dimension of multi-level graphs. Formally, the forward propagation rule of multi-level graphs can be written as
where \( \widetilde{\mathcal {G}}_{l}\) denotes the generated graph. GraphSAGE represents the inductive graph convolution [23] used in our model, which allows the message passing in the spatial dimension of multi-level graph. It should be noted that other spatial graph convolutions with different massage passing mechanism also can be used to explore the different relationship between extracted entities. \(\sigma (\cdot )\) denotes the activation function, such as ReLU.
Attentive Hierarchical Interaction. As shown in Fig. 1, based on the biological affiliation of cell and tissues, each cell entity has subordinate relation with a determinate tissue entity at every scale, forming \(|\mathcal {V}_0|\) sequences in hierarchical dimension. Inspired by the long-range dependency modeling ability and attention mechanism of Transformer [24], we incorporate a vision transformer(ViT) paradigm [25] into our network to investigate the hierarchical interaction between the multi-level entities and selectively aggregate the interaction information to produce the final graph representation for classification. Specifically, each hierarchical sequence is tokenized and attached with positional embedding as the input of a Transformer encoder consisting of Multi-Headed Self-Attention [24], layer normalization (LN) [26] and MLP blocks. In addition, an extra learnable classification token is prepended to the hierarchical sequence, and its representation at the output layer of the Transformer encoder serves as the final representation. By inputting all hierarchical sequences to this ViT module, the multi-level graphs are transformed into a novel graph representation \(\widetilde{\mathcal {G}}_{cls} \in \mathbb {R}^{|\mathcal {V}_0| \times D} \), where D is the output dimension of ViT. This process can be written as
Classification Layer. Based on the graph \(\widetilde{\mathcal {G}}_{cls}\) with spatial and hierarchical information of histological image, a more reliable output prediction can be obtained by:
where Readout is a global attention pooling layer [27] for generating representation for the final graph. For the network training, the cross-entropy loss is adopted for classification tasks and the objective loss is defined as
where N is the number of samples, T is the number of classes.
3 Experiments
3.1 Clinical Datasets and Evaluation Protocols
CRCS Dataset. CRCS dataset contains 5610 colorectal histological images with the fixed size of 512 px \(\times \) 512 px. Based on strict proofs, all images were scanned at \(\times \)20 and manually marked by licensed clinicians, and have three types of labels: Normal, low grade intraepithelial neoplasia (LGIN) and high grade intraepithelial neoplasia (HGIN).
BRACS Dataset. BRACS [28] contains 4391 breast histological images scanned with an Aperio AT2 scanner at 0.25 \(\upmu \)m/pixel resolution. The average size of images is 1778 px \(\times \) 1723 px. The images were annotated as being Normal, Benign, Usual ductal hyperplasia (UDH), Atypical Ductal Hyperplasia (ADH), Flat Epithelial Atypia (FEA), Ductal Carcinoma In Situ (DCIS), and Invasive.
Experimental Setup. The area under the curve (AUC) is used as the evaluation metric. For each trial, five repeated 3-fold cross-validations (3-fold CVs) are adopted. All trials are conducted on a workstation with an Intel i9-9820X @ 4.1 GHz CPU and four NVIDIA GeForce RTX 2080Ti (11 GB) GPUs. The extraction of multi-level entities is implemented using Histocartography library [29]. Our GCN model is implemented by Pytorch Geometric [30]. Considering the resolution of histological images, the graph construction parameters of different data sets are set as follows. For CRCS, two-scale (200, 300 superpixels per image) tissue analysis is adopted (\(L = 3\)). For BRACS, one-scale (700 superpixels per image) tissue analysis is adopted (\(L = 2\)). The k was tuned from {3, 5, 7, 9}. The \(d_{min}\) was tuned from {1, 5, 10, 15}, respectively. The Adam optimizer was adopted, and the network was trained for 60 epochs. The learning rate is initially set to 1e−4 and decays to 1e−5 after 40 epochs.
3.2 Comparison with State-of-the-Arts
We first compare the proposed method with two CNN based methods: (1) GoogleNet [9], (2) ADN [10], as well as two GNN based methods: (3) CGC-Net [19], (4) HACT [11]. The comparative results on CRCS and BRACS datasets are shown in Table 1 and Table 2, respectively. Generally, due to the advantage of the biological entity oriented modeling, the overall performance of GNN based methods is better than that of CNN based methods. As our method not only considers the task-related information for graph structure design but also excavates the interaction information of the multi-level entities, our method outperforms the existing SOTAs on both two datasets.
3.3 Ablation Study
We first compare the DSL module with traditional fixed methods for graph structure design, including random adjacency matrix, space distance adjacency matrix and k-NN adjacency matrix [11, 19], shown in Fig. 2(a). It can be observed that our DSL module is superior to the traditional fixed methods, as DSL module introduced the task-related information for enhancing the presentation capability of the graph. We also compare the attentive hierarchical interaction of SHGNN with add, multiplication, and concatenation forms [11], shown in Fig. 2(b). Overall, the proposed method consistently outperforms the fixed non-interactive methods, since the attention mechanism can adaptively select the useful multi-level entities for the task and hierarchical interaction can produce more abundant information for the decision-making.
3.4 Visualization of Proposed Framework
Figure 3 visualizes the learning process and attention map of the proposed framework. On the one hand, the middle figures show the evolution process of the cell graph structure, which indicates the proposed DSL module can dynamically refine the graph structure. On the other hand, the attention map (see right figures) can be obtained by the global attention pooling layer conducted on \(\widetilde{\mathcal {G}}_{cls}\), which may aid clinical diagnosis and potentially lead to biomarker discoveries.
4 Conclusion
In this paper, we propose a novel deep graph neural network for automatic histological image classification. The first advantage of the proposed model is to dynamically learn the connection structure of multi-level biological entities that better serves as the input of SHGNN for the classification task. Further, our proposed SHGNN combines spatial graph convolution with an attentive hierarchical interaction mechanism to simultaneously capture the spatial-hierarchical feature of the histological images, so that the potential of multi-level entities can be fully unleashed. Experimental results on two clinical datasets demonstrate our model achieves state-of-the-art performance over the existing models. The main limitation of our method lies in the relatively larger complexity comes from the extraction of multi-level entities. In the future, we will develop more computation-efficient strategies to accelerate the computation of the framework and evaluate our framework on other tasks.
References
Yao, X.H., et al.: Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 30(6), 541–543 (2020)
Cai, Z., et al.: A greater lymph node yield is required during pathological examination in microsatellite instability-high gastric cancer. BMC Cancer 21(1), 1–9 (2021)
Kiesslich, R., et al.: Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo. Gastroenterology 127(3), 706–713 (2004)
Sterlacci, W., Vieth, M.: Early Colorectal Cancer. In: Baatrup, G. (ed.) Multidisciplinary Treatment of Colorectal Cancer, pp. 263–277. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-58846-5_28
Shi, X., et al.: Genes involved in the transition from normal epithelium to intraepithelial neoplasia are associated with colorectal cancer patient survival. Biochem. Biophys. Res. Commun. 435(2), 282–288 (2013)
Nowacki, T.M., et al.: The risk of colorectal cancer in patients with ulcerative colitis. Dig. Dis. Sci. 60(2), 492–501 (2015). https://doi.org/10.1007/s10620-014-3373-2
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Hou, W., Wang, L., Cai, S., Lin, Z., Yu, R., Qin, J.: Early neoplasia identification in Barrett’s esophagus via attentive hierarchical aggregation and self-distillation. Med. Image Anal. 72, 102092 (2021)
Bai, J., Jiang, H., Li, S., Ma, X.: NHL pathological image classification based on hierarchical local information and GoogLeNet-based representations. Biomed. Res. Int. 2019, 1065652 (2019)
Li, Y., Xie, X., Shen, L., Liu, S.: Reverse active learning based atrous DenseNet for pathological image classification. BMC Bioinform. 20(1), 1–15 (2019)
Pati, P., et al.: Hierarchical graph representations in digital pathology. Med. Image Anal. 75, 102264 (2022)
Zhou, J., et al.: Graph neural networks: a review of methods and applications. arXiv preprint arXiv:1812.08434 (2018)
Wu, Z., Pan, S., Chen, F., Long, G., Zhang, C., Philip, S.Y.: A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 32, 4–24 (2020)
Jia, Z., et al.: GraphSleepNet: adaptive spatial-temporal graph convolutional networks for sleep stage classification. In: IJCAI, pp. 1324–1330 (2020)
Lu, W., Graham, S., Bilal, M., Rajpoot, N., Minhas, F.: Capturing cellular topology in multi-gigapixel pathology images. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 260–261 (2020)
Raju, A., Yao, J., Haq, M.M.H., Jonnagaddala, J., Huang, J.: Graph attention multi-instance learning for accurate colorectal cancer staging. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12265, pp. 529–539. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59722-1_51
Anklin, V., et al.: Learning whole-slide segmentation from inexact and incomplete labels using tissue graphs. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12902, pp. 636–646. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87196-3_59
Chen, R.J., et al.: Pathomic fusion: an integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans. Med. Imaging 41, 757–770 (2022)
Zhou, Y., Graham, S., Alemi Koohbanani, N., Shaban, M., Heng, P.A., Rajpoot, N.: CGC-Net: cell graph convolutional network for grading of colorectal cancer histology images. In: Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops, pp. 1–11 (2019)
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. IEEE (2009)
Graham, S., et al.: HoVer-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 58, 101563 (2019)
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., Süsstrunk, S.: SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 34(11), 2274–2282 (2012)
Hamilton, W., Ying, Z., Leskovec, J.: Inductive representation learning on large graphs. In: Advances in Neural Information Processing Systems, pp. 1024–1034 (2017)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Dosovitskiy, A., et al.: An image is worth 16x16 words: transformers for image recognition at scale. In: International Conference on Learning Representations (2021)
Ba, J.L., Kiros, J.R., Hinton, G.E.: Layer normalization. arXiv preprint arXiv:1607.06450 (2016)
Li, Y., Tarlow, D., Brockschmidt, M., Zemel, R.: Gated graph sequence neural networks. arXiv preprint arXiv:1511.05493 (2015)
Brancati, N., et al.: BRACS: a dataset for breast carcinoma subtyping in H &E histology images. arXiv preprint arXiv:2111.04740 (2021)
Jaume, G., Pati, P., Anklin, V., Foncubierta, A., Gabrani, M.: HistoCartography: a toolkit for graph analytics in digital pathology. In: Proceedings of the MICCAI Workshop on Computational Pathology. In: Proceedings of Machine Learning Research, 27 September 2021, vol. 156, pp. 117–128. PMLR (2011)
Fey, M., Lenssen, J.E.: Fast graph representation learning with PyTorch Geometric. In: ICLR Workshop on Representation Learning on Graphs and Manifolds (2019)
Acknowledgement
This work was supported by Ministry of Science and Technology of the People’s Republic of China (2021ZD0201900) (2021ZD0201903).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Hou, W., Huang, H., Peng, Q., Yu, R., Yu, L., Wang, L. (2022). Spatial-Hierarchical Graph Neural Network with Dynamic Structure Learning for Histological Image Classification. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13432. Springer, Cham. https://doi.org/10.1007/978-3-031-16434-7_18
Download citation
DOI: https://doi.org/10.1007/978-3-031-16434-7_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16433-0
Online ISBN: 978-3-031-16434-7
eBook Packages: Computer ScienceComputer Science (R0)