Abstract
BOLD fMRI has been an established tool for studying the human brain’s functional organization. Considering the high dimensionality of fMRI data, various computational techniques have been developed to perform the dimension reduction such as independent component analysis (ICA) or sparse dictionary learning (SDL). These methods decompose the fMRI as compact functional brain networks, and then build the correspondence of those brain networks across individuals by viewing the brain networks as one-hot vectors and performing their matching. However, these one-hot vectors do not encode the regularity and variability of different brains, and thus cannot effectively represent the functional brain activities in different brains and at different time points. To bridge the gaps, in this paper, we propose a novel unsupervised embedding framework based on Transformer to encode the brain function in a compact, stereotyped and comparable latent space where the brain activities are represented as dense embedding vectors. The framework is evaluated on the publicly available Human Connectome Project (HCP) task based fMRI dataset. The experiment on brain state prediction downstream task indicates the effectiveness and generalizability of the learned embeddings. We also explore the interpretability of the embedding vectors and achieve promising result. In general, our approach provides novel insights on representing regularity and variability of human brain function in a general, comparable, and stereotyped latent space.
L. Zhao and Z. Wu–Co-first authors.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
FMRI has been an established neuroimaging technique for studying the human brain’s functional organization [10]. However, a major challenge in fMRI-based neuroscience studies is that the number of voxels in 4D spatiotemporal fMRI data is greatly larger than the number of subject brains [12], which is also known as “curse-of-dimensionality” problem [3]. To mitigate negative effects brought by this imbalance, various computational tools have been developed to select the task-relevant features and discard the redundant ones as well as the noises [1, 4, 11]. For example, principal component analysis (PCA) transforms the correlated voxels into several uncorrelated principal components, which is used as representation of the spatiotemporal fMRI data [1]. Independent component analysis (ICA) based methods assume that the fMRI signals are a “mixture” of spatially or temporally independent patterns (e.g., paradigm-related responses) that could be decomposed from brain fMRI signals [4]. In this way, the analysis can be performed on those much more compactly represented independent patterns rather than the raw voxels in 4D space. In addition to PCA and ICA based matrix decomposition methods, sparse dictionary learning (SDL) was also employed to decompose the fMRI into a over-complete dictionary (temporal activities) and a sparse representation matrix (spatial patterns) [11].
Despite the wide adoption and application of the above-mentioned matrix decomposition techniques, the resulted temporal and/or spatial patterns obtained by those methods are not intrinsically comparable across different individual brains. That is, even with the same hyper-parameters in those matrix decomposition methods like ICA/SDL for different brains, there is no correspondence among the temporal and/or spatial brain network patterns from different subjects. Moreover, even with image registration or pattern matching methods, the huge variability of brain function across individual brains cannot ensure that corresponding brain networks can be identified and matched in different brains. From our perspective, a fundamental difficulty in traditional matrix decomposition methods for fMRI data modeling is that these methods attempt to decompose and represent the brain’s functional organization as brain networks and then try to match the correspondences of those networks across individuals and populations. In this process, different brain networks are viewed as one-hot vectors and the mapping or matching are performed on those one-hot vectors. Actually, the one-hot vector representation and matching of brain networks in those matrix decomposition methods do not encode the regularity and variability of different brains, and as a consequence, these one-hot vectors of brain networks do not offer a general, comparable, and stereotyped space for brain function. To address this critical problem, an intuitive way is to encode brain function in a compact, stereotyped and comparable latent space where the brain activities measured by fMRI data in different brains and at different time points can be meaningfully embedded and compactly represented.
As an effective methodology for high-dimensional data embedding, deep learning has been widely employed in fMRI data modeling and achieved superior results over those traditional matrix decomposition methods [6, 8, 13, 17]. However, as far as we know, prior deep learning models of fMRI data were not specifically designed towards the effective embedding of human brain function for the purpose of compact representation of regularity and variability in different brains. Instead, prior methods were designed for specific tasks, such as fMRI time series classification [9], hierarchical brain network decomposition [5], brain state differentiation [15], among others. Therefore, existing deep learning models of fMRI data still do not offer a general, comparable, and stereotyped space for representing human brain function. Importantly, the compact and comparable embeddings can also be easily integrated into other deep learning frameworks, paving the road for multi-modal representation learning such as connecting the text stimuli in semantic space of Natural Language Processing (NLP) and the brain’s responses to those stimuli in brain function embedding space.
To bridge the above gaps, in this paper, we formulate the effective and general representation of human brain function as an embedding problem. The key idea is that each 3D volume of fMRI data can be embedded as a dense vector which profiles the functional brain activities at the corresponding time point. The regularity and variability of brain function at different time points and across individual brains can be effectively measured by the distance in the embedding space. That is, our embedding space offers a general, comparable, and stereotyped space for brain function. Specifically, to achieve such effective embedding space of human brain function, we designed a novel Temporal-Correlated Autoencoder (TCAE) based on the Transformer model [14] and self-attention mechanism. The major theoretical and practical advantage of Transformer is that its multi-head self-attention can explicitly capture the correlation between time points, especially for those far away from each other and jointly attending to information from different representation subspaces, which is naturally suitable for our objective of holistic embedding of human brain function. We evaluate the proposed brain function embedding framework on the publicly available Human Connectome Project (HCP) task based fMRI dataset and it achieves state-of-the-art performance on brain state prediction downstream task. The interpretability of the learning embedding is also studied by exploring its relationship with the block-design paradigm. Overall, our method provides a novel and generic framework for representing the human brain’s functional architecture.
2 Methods
2.1 Overview
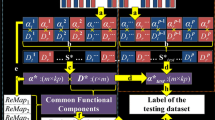
The proposed framework is shown in Fig. 1. We first illustrate the TCAE embedding framework used in this study in Sect. 2.2. Then, the learned embeddings are evaluated on a downstream task of brain state prediction which is introduced in Sect. 2.3.
Illustration of the proposed TCAE embedding framework. The 4D spatiotemporal fMRI data are firstly rearranged into 2D signal matrix and then input into the encoder. The output of the encoder is recognized as the learned embedding, which is used for the brain state prediction downstream task and reconstructing the input signal matrix.
2.2 Temporal-Correlated Autoencoder
In this section, we introduce the TCAE embedding framework based on the Transformer model and multi-head self-attention [14]. As illustrated in Fig. 1, TCAE has an encoder-decoder architecture. For both encoder and decoder, they consist of one embedding layer and one multi-head self-attention module. Specifically, in the encoder, the rearranged 2D fMRI signal matrix \({\textbf {S}}\in \mathbb {R}^{t\times n}\), where t is the number of time points and n is the number of voxels, is firstly embedded into a new feature matrix \({\textbf {S}}_f\in \mathbb {R}^{t\times m}\) through a fully-connected (FC) layer, where m is the reduced feature dimension \((m \ll n)\). Then, for attention head i, the self-attention map that captures the temporal correlations of different time points is computed as:
where \({\textbf {W}}_i^Q \in \mathbb {R}^{m\times k}\) and \({\textbf {W}}_i^K \in \mathbb {R}^{m\times k}\) are projection matrices and k is the feature dimension of the self-attention operation. With the self-attention maps, the output of attention head i can be computed as:
where \({\textbf {W}}_i^V \in \mathbb {R}^{m\times v}\) and v is the feature dimension for the output of attention heads. We then concatenate \({\textbf {S}}_{attn_i}\) along the feature dimension and transform the concatenated matrix into a new feature matrix \({\textbf {S}}_{multi} \in \mathbb {R}^{t\times m}\) as:
where \(Concat(\cdot )\) represents the concatenation operation. h is the number of heads and \({\textbf {W}}^O \in \mathbb {R}^{hv\times m}\) is the projection matrix. \({\textbf {S}}_{multi}\) is further fed into the feed forward layer to obtain the encoder output \({\textbf {E}}\in \mathbb {R}^{t\times m}\):
where \({\textbf {W}}_1\in \mathbb {R}^{m\times d_{ff}}\), \({\textbf {W}}_2\in \mathbb {R}^{d_{ff}\times m}\), \(b_1\) and \(b_2\) are biases. \(d_{ff}\) denotes the feature dimension of the inner layer. The encoder output \({\textbf {E}}\) is recognized as the learned embedding from our model. For the decoder, it fetches the encoder output \({\textbf {E}}\) to reconstruct the input fMRI signal matrix. The multi-head self-attention module in the decoder is same as that in the encoder except that the FC layer increases the feature dimension from m to n to match the input signal matrix.
The TCAE embedding framework is optimized in an unsupervised manner by minimizing the Mean Square Error (MSE) between the original fMRI signals \({\textbf {S}}\in \mathbb {R}^{t\times n}\) and their corresponding reconstruction \({\textbf {S}}'\in \mathbb {R}^{t\times n}\).
2.3 Prediction of Brain State
In this subsection, we introduce a brain state prediction task to evaluate the learned embedding. Specifically, each time point can be classified into a specific brain state according to the task that the subject participated in, e.g., math calculation or listening to a story. The prediction of brain state is performed by a two-stage manner. In the first stage, we pre-train a TCAE model to learn the embedding as described in Sect. 2.2. In the second stage, we fix the pre-trained model and obtain the embedding for each time point. The embedding is then input into a classifier consisting of two fully-connected (FC) layers with tanh/softmax as activation function, respectively. The classifier is optimized by minimizing the cross-entropy between predictions and labels. In this way, the learned embedding is not task-specific and the effectiveness and generalizability can be fairly evaluated.
3 Experiments
3.1 Dataset and Pre-Processing
We adopt the publicly available HCP task fMRI (tfMRI) dataset of S1200 release (https://www.humanconnectome.org/) [2]. In this paper, among 7 different tasks in HCP tfMRI dataset, Emotion and Language tasks are used as testbeds for our framework due to the space limit. The acquisition parameters of HCP tfMRI data are as follows: 90 \(\times \) 104 matrix, 72 slices, TR = 0.72 s, TE = 33.1 ms, 220 mm FOV, flip angle = 52\(^{\circ }\), BW = 2290 Hz/Px, in-plane FOV = 208 \(\times \) 180 mm, 2.0 mm isotropic voxels. The preprocessing pipelines of tfMRI data are implemented by FSL FEAT [16], including skull removal, motion correction, slice time correction, spatial smoothing, global drift removal (high-pass filtering) and registration to the standard MNI 152 4 mm space for reducing the computational overhead. Besides, time series from the voxels of 4D tfMRI data are rearranged into a 2D array with zero mean and standard deviation one. For a total of more than 1000 subjects in HCP S1200 release, we randomly select 600 subjects as training set, 200 subjects as validation set, and another 200 subjects as testing set for both two tasks, respectively. All the experimental results in our study are reported based on the testing set.
3.2 Implementation Details
In our experiments, we uniformly set the embedding dimension as 64, i.e., the embedding has 64 digits. For TCAE model, the m,k,v are set to 64 and \(d_{ff}\) is set to 128. For the predictor, the size of two FC layers are 64/32, respectively. The framework is implemented with PyTorch (https://pytorch.org/) deep learning library. We use the Adam optimizer [7] with \(\beta _1=0.9\) and \(\beta _2=0.999\). The batch size is 16 and the model is trained for 100 epochs with a learning rate 0.01 for both tasks on a single GTX 1080Ti GPU. It is noted that all experiments were performed with testing set based on model with the lowest loss on validation dataset.
 and
and  denotes the best and the second-best results, respectively.
denotes the best and the second-best results, respectively.3.3 Brain State Prediction Results
In this subsection, we evaluate the learned embedding on a brain state prediction downstream task. Here, we introduce several baseline methods for comparison: Autoencoder (AE), deep sparse recurrent autoencoder (DSRAE) [8], deep recurrent variational autoencoder (DRVAE) [13], spatiotemporal attention autoencoder (STAAE) [6]. AE denotes an autoencoder model consisting of one FC layer with tanh activation function in both encoder and decoder, which can be considered as a baseline without the multi-head self-attention module of our model. It is noted that DSRAE was designed for decomposing the spatial and temporal patterns as SDL; DRVAE model aimed at the augmentation of fMRI data; STAAE was proposed for the classification of Attention Deficit Hyperactivity Disorder (ADHD). We implement their network architectures and take the encoder’s output as embedding. The details about the configuration of those baselines can be found in supplemental materials. In Table 1, we report the brain state classification accuracy as well as the number of parameters (Params) and the number of multiply-accumulate operations (MACs) of those baselines and the proposed method with an embedding size of 64. The results for other embedding size and the hyper-parameter sensitivity analysis can be found in supplemental materials.
It is observed that all baselines have an accuracy over 0.6. The baseline AE outperforms all the other baselines in terms of prediction accuracy with much less parameters and MACs. This is probably because other baselines such as DRVAE are designed for a specific task which requires a specially designed architecture with more parameters. On the other hand, an architecture with more parameters may not be generalizable for our embedding task and thus degenerate the performance. Our proposed TCAE embedding framework introduces an additional self-attention module with slight increases in model parameters and computational operations compared with the baseline AE. But the performance gain is significant with the highest accuracy (two-sample one-tailed un-pair-wise t-test (p < 0.025, corrected)). This observation indicates that our model is compact and can learn a more generalizable embedding compared with other baselines.
3.4 Interpretation of the Learned Embedding
To explore the interpretation of the learned embedding, similar to [8], we assume that each digit of embedding corresponds to a temporal brain activity pattern, which can be obtained by extracting the digit value over time. With the extracted temporal pattern of each digit, we compute the Pearson Correlation Coefficient (PCC) between each pattern and the Hemodynamic Response Function (HRF) responses of task stimulus. From all digits, we select the one with the highest PCC value as the task-relevant digit which is an indicator of the embedding’ relevance to task stimulus. Here, we randomly select four subjects as examples to show the temporal pattern of task-relevant digit from the TCAE model as well as the corresponding HRF responses in Fig. 2.
 and
and  denotes the highest and the second-highest PCC, respectively.
denotes the highest and the second-highest PCC, respectively.Generally, the temporal pattern of task-relevant digit matches the corresponding HRF response well with PCC value larger than 0.45, indicating that the digits of the learned embedding are to some extent correlated to task stimulus. To quantitatively measure such correlation, we average the PCC of all subjects and compare it with those from baseline models in Table 2. It is observed that in Emotion task, the averaged PCC of TCAE’s task-relevant digit is larger than that of all compared baselines. However, in the Language task, AE and STAAE have the highest PCC for two task designs, respectively. A possible reason is that in Emotion task, the response of task stimulus is more complex and hard to be decoded and decomposed from the raw fMRI data. Our embeddings from TCAE model can better characterize such responses, which is in alignment with the highest brain state prediction accuracy in Table 1. While in the Language task, the responses are quite straightforward and can be easily captured by other deep learning models. It is consistent with overall higher brain state prediction accuracy in Table 1 than Emotion task. The TCAE embedding model may focus on more intrinsic responses and patterns which are still task-relevant but discriminative, resulting in a higher accuracy in brain state prediction but relatively lower PCC than baselines. Overall, the embedding from TCAE framework provides rich and meaningful information which is relevant to the brain’s response to task stimulus.
4 Conclusion
In this paper, we proposed a novel transformer-based framework that embeds the human brain function into a general, comparable, and stereotyped space where the brain activities measured by fMRI data in different brains and at different time points can be meaningfully and compactly represented. We evaluated the proposed framework in brain state prediction downstream task, and the results indicated that the learned embedding is generalizable and meaningful. It was also found that the embedding is relevant to the response of task stimulus. Our future works include evaluating the framework with more cognitive tasks in tfMRI and applying the embedding for disease diagnosis such as ADHD and Alzheimer’s disease.
References
Andersen, A.H., Gash, D.M., Avison, M.J.: Principal component analysis of the dynamic response measured by fMRI: a generalized linear systems framework. Magn. Reson. Imaging 17(6), 795–815 (1999)
Barch, D.M., et al.: Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189 (2013)
Bellman, R.E.: Adaptive Control Processes. Princeton University Press, Princeton (2015)
Calhoun, V.D., Adali, T.: Unmixing fMRI with independent component analysis. IEEE Eng. Med. Biol. Mag. 25(2), 79–90 (2006)
Dong, Q., et al.: Modeling hierarchical brain networks via volumetric sparse deep belief network. IEEE Trans. Biomed. Eng. 67(6), 1739–1748 (2019)
Dong, Q., Qiang, N., Lv, J., Li, X., Liu, T., Li, Q.: Spatiotemporal attention autoencoder (STAAE) for ADHD classification. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12267, pp. 508–517. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59728-3_50
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Li, Q., Dong, Q., Ge, F., Qiang, N., Wu, X., Liu, T.: Simultaneous spatial-temporal decomposition for connectome-scale brain networks by deep sparse recurrent auto-encoder. Brain Imaging Behav. 15(5), 2646–2660 (2021). https://doi.org/10.1007/s11682-021-00469-w
Liu, H., et al.: The cerebral cortex is bisectionally segregated into two fundamentally different functional units of gyri and sulci. Cereb. Cortex 29(10), 4238–4252 (2019)
Logothetis, N.K.: What we can do and what we cannot do with fMRI. Nature 453(7197), 869–878 (2008)
Lv, J., et al.: Holistic atlases of functional networks and interactions reveal reciprocal organizational architecture of cortical function. IEEE Trans. Biomed. Eng. 62(4), 1120–1131 (2014)
Mwangi, B., Tian, T.S., Soares, J.C.: A review of feature reduction techniques in neuroimaging. Neuroinformatics 12(2), 229–244 (2014)
Qiang, N., et al.: Modeling and augmenting of fMRI data using deep recurrent variational auto-encoder. J. Neural Eng. 18(4), 0460b6 (2021)
Vaswani, A., et al.: Attention is all you need. Adv. Neural Inf. Process. Syst. 30 (2017)
Wang, H., et al.: Recognizing brain states using deep sparse recurrent neural network. IEEE Trans. Med. Imaging 38(4), 1058–1068 (2018)
Woolrich, M.W., Ripley, B.D., Brady, M., Smith, S.M.: Temporal autocorrelation in univariate linear modeling of fMRI data. Neuroimage 14(6), 1370–1386 (2001)
Zhao, L., Dai, H., Jiang, X., Zhang, T., Zhu, D., Liu, T.: Exploring the functional difference of Gyri/Sulci via hierarchical interpretable autoencoder. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12907, pp. 701–709. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87234-2_66
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Zhao, L. et al. (2022). Embedding Human Brain Function via Transformer. In: Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2022. MICCAI 2022. Lecture Notes in Computer Science, vol 13431. Springer, Cham. https://doi.org/10.1007/978-3-031-16431-6_35
Download citation
DOI: https://doi.org/10.1007/978-3-031-16431-6_35
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16430-9
Online ISBN: 978-3-031-16431-6
eBook Packages: Computer ScienceComputer Science (R0)