Abstract
Our motivation is to enable non-specialists to use sophisticated biomechanical models in the clinic. To further this goal, in this study, we constructed a framework within 3D Slicer for automatically generating and solving patient-specific biomechanical models of the brain. This framework allows determining automatically patient-specific geometry from MRI data, generating patient-specific computational grid, defining boundary conditions and external loads, assigning material properties to intracranial constituents and solving the resulting set of differential equations. We used the Meshless Total Lagrangian Explicit Dynamics (MTLED) algorithm to solve these equations. We demonstrated the effectiveness and appropriateness of our framework on a case study of brain tissue deformations caused by placement of electrodes on the brain surface in intracranial electroencephalography (iEEG).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Patient-specific Modelling (PSM)
- Nonlinear computational biomechanics
- Brain
- Brain shift
- Automated computations
- Electrodes
- Deformations
1 Introduction
We are at the verge of a new exciting era of personalized medicine based on patient-specific scientific computations. These computations usually involve solving models described by boundary value problems of partial differential equations (PDEs). The most common and useful are models of biomechanics, bioheat transfer and bioelectricity.
In this paper, we are especially interested in patient-specific biomechanics as a tool to compute soft tissue deformations for operation planning and intraoperative guidance. While the methods for patient-specific biomechanical model generation [33] and solution [9, 10, 39] exist, they are very sophisticated and require very high level of specialist expertise from the users. Therefore, the objective of the work described here is to create an automatic framework so that these sophisticated computations can be conducted in the clinic by a non-specialist.
We integrated the framework [9, 39] to automate the process of generating and solving patient-specific biomechanical models into 3D Slicer (http://www.slicer.org/), an open-source software for visualization, registration, segmentation and quantification of medical data developed by Artificial Intelligence Laboratory of Massachusetts Institute of Technology and Surgical Planning Laboratory at Brigham and Women’s Hospital and Harvard Medical School [5].
We demonstrated the application of our framework using a case study of brain deformations caused by placement of electrodes on the brain surface in intracranial electroencephalography (iEEG). The case study is obtained from database of Computational Radiology Lab, Harvard Medical School. The paper is organized as follows: In Sect. 2, we presented the proposed framework. In Sect. 3, we showed our results based on the case study. Section 4 contains discussion and conclusion.
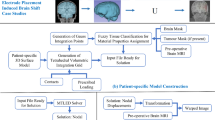
2 Proposed Framework
The four main steps of the proposed framework workflow (see Fig. 1) are as follows:
-
1.
Image Pre-processing
-
Determining patient-specific geometry from medical images
-
-
2.
Model Construction
-
Patient-specific computational grid generation
-
Defining boundary conditions and external load (displacements of the boundary)
-
Assigning patient-specific material properties to brain tissues
-
-
3.
Model Solution
-
Computation of tissue deformations using Meshless Total Lagrangian Explicit Dynamics Algorithm (MTLED)
-
-
4.
Image Warping, using the computed deformation field
The details of each step are given in Sects. 2.1–2.3. Image warping is done with example case study under Sect. 3.
2.1 Image Pre-processing
2.1.1 Determining Patient-Specific Geometry from Medical Images
To obtain the geometry of the brain, the skull needs to be removed from the preoperative MRI. We remove the skull and extract the brain volume using FreeSurfer software (http://surfer.nmr.mgh.harvard.edu/) (see Fig. 2). It is an open source software suite for processing and analyzing human brain medical resonance images (MRIs) [3]. Watershed algorithm is used by FreeSurfer to extract the brain portion from T1-weighted MRI [27]. We wrote python-based scripted modules within 3D Slicer to execute all the remaining steps.
After extracting brain volume, we use threshold filter [23] of 3D Slicer to select the brain parenchyma (see Fig. 3). We created a three dimensional surface model based on the selected region using model maker module of 3D Slicer, see Fig. 3 (3D view) [32]. We applied 10% Laplacian smoothing to control the smoothing on model [28]. We have selected 10% smoothing to avoid volume reduction.
Visualization of results of threshold filter along with visualization of surface geometry produced by surface model maker of 3D Slicer with 10% value of Laplacian filter. Visualization performed with 3D Slicer (www.slicer.org) [5]
2.1.2 Extracting Location of Electrodes
Information about the electrode locations is necessary for biomechanical modeling of neurosurgery [39]. We extracted locations (co-ordinates) of electrodes from the segmented electrode volume (image set containing electrode segmentations) using our procedure implemented as 3D Slicer extension, “electrodes-to-markups”. The procedure (see Fig. 5) consists of the following steps: (1) creating a binary label volume from segmented electrode volume, (2) splitting the binary label volume to segments corresponding to each electrode and (3) adding a point (3D space) at the centroid of segments defining each electrode. The conversion from segmented electrode volume to binary label volume (step (1)) is performed using PolySeg [24], a software library that provides automatic conversions between different representations (e.g. label map, surface) [24]. Splitting the binary label volume (step (2)) is performed using split island into segments and then segment statistics (step (3)) is used to get the centroid (center of mass of the segment) [24].
The intraoperative (post-implantation) positions of the electrodes were extracted (through segmentation) from the CT image (see Fig. 4) rigidly registered to the preoperative MRI. We projected the extracted electrodes (see Fig. 5c) onto the brain surface segmented from the preoperative MRI to determine the points corresponding to location of electrodes (referred to as projected electrodes) in the undeformed (preoperative) brain geometry [39]. We used these points to create an electrode sheet model by means of PolyData algorithm [29] from PyVista (www.pyvista.org), Python programming language library [29] (see Fig. 6). We then selected the nodes of brain model located under electrode sheet model (see Fig. 9). We define displacements on these selected surface brain nodes (see Sect. 3 for an example on calculating prescribed displacements) and we applied contacts (see section “Boundary Conditions”) on the remaining boundary nodes.
2.2 Model Construction
2.2.1 Patient-Specific Computational Grid Generation
In our method, we use unstructured cloud of nodes to discretize the geometry of interest instead of elements. In this study, we use a geometry conforming tetrahedral background grid [9, 39]. The displacements are calculated over the cloud of points formed by the vertices of the tetrahedra. Volumetric integration (a step in the MTLED solution method, see Sect. 3) is performed over background integration cells with four Gauss points per tetrahedral cell. Creating such background grids is fully automatic (i.e. does not require any manual correction) (see Fig. 7). It is very important to note that our tetrahedral integration grid is NOT a finite element mesh and does not need to conform to strict quality requirements demanded by the finite element method. MTLED incorporates Modified Moving Least Squares (MMLS) shape functions [11] which increases the set of admissible nodal distributions and allows very rapid generation of patient-specific discretization of acceptable quality [9]. Whereas in FEM, the linear tetrahedral elements exhibits volumetric locking, especially in case of soft tissues such as the brain, which are modelled as almost incompressible materials [36].
Our framework uses ACVD (Surface Mesh Coarsening and Resampling) [30] to construct a patient-specific triangulated brain surface (see Fig. 8b) which is then used for generating a 3D integration grid filled with tetrahedral integration cells (see Fig. 8c) using Gmsh [6]. The triangulated surface is also used for defining contacts. We automated all these steps and implemented them in 3D Slicer.
a Patient-specific tetrahedral integration grid with triangular surface mesh, b Example of triangulated patient specific brain surface mesh model, c Example of patient specific brain volumetric integration grid filled with tetrahedral cells (geometry conforming tetrahedral cells based biomechanical model)
2.2.2 Defining Boundary Conditions and External Load
2.2.2.1 Boundary Conditions
The stiffness of the skull is several orders of magnitude higher than that of the brain. Therefore, to define the boundary conditions for nodes other than displaced nodes on the exposed surface of the brain, a contact interface is defined between the rigid interface model of the skull and the deformable brain model. Nodes on the brain surface could not penetrate the skull, but could slide without friction or separate from the skull as described in [13].
We created a skull interface using the triangulated surface cells generated as described in Sect. 2.1.2 to define contacts automatically on the surface of patient specific brain biomechanical model.
2.2.2.2 External Load
Load can be defined either through forces (prescribing natural BCs) or displacements on the boundary (prescribing essential BCs). It is rather difficult to make patient-specific measurements of forces acting on the brain during surgery but there are well-established methods for determining the displacements on the boundaries from images. Furthermore, if we use forces, to accurately compute intraoperative deformations, we need accurate information about patient-specific material properties of the brain tissues. As there is no commonly established method to accurately determine patient-specific material properties of soft tissues from radiographic (MR, CT) images, we define the load through imposed motion (essential BCs). This makes the computed deformations only very weakly dependent on uncertainty in patient specific information about tissue material properties [20, 21, 34].
To define intraoperative loading, intraoperative information is required such as measurement of the current position of the exposed surface of the brain. This can be done through cameras [17] and a pointing tool of a neurosurgical station [25].
In this study, we defined the load by prescribing displacements (essential boundary conditions) on the brain surface deformed due to EEG electrode implantation [39]. The displacements were applied to the nodes located on the brain surface model directly under the electrode sheet (see Fig. 9). We applied displacements using smooth loading curve (3–4-5 polynomial) [12, 31].
2.2.3 Assignment of Patient-Specific Material Properties: Fuzzy Tissue Classification
Material properties of the intracranial constituents are assigned to integration points within the problem geometry through fuzzy tissue classification [1] algorithm. Hard segmentation of brain tissues is difficult to automate [4] and therefore it is incompatible with clinical workflows. Therefore, we integrated a fuzzy tissue classification algorithm [15, 16, 33, 38] into our framework to automatically assign material properties to brain tissues (see results in Fig. 10). Slight inaccuracies of tissue properties assignment do not affect the precision of intraoperative displacement prediction because the external load is defined though prescribed essential boundary condition motion rendering the problem Dirichlet-type [20, 38].
Result of automatic material property assignment (Young’s modulus of 3000 Pa for the brain and 100 Pa for CSF) using fuzzy tissue classification [33, 38]. Brain tissue and CSF (white) were used as cluster centers. Visualization performed with 3D Slicer www.slicer.org [5]
In this framework, a neo-Hookean constitutive model (see Table 1) was used for brain tissues with Poisson’s ratio of 0.49, whereas 0.1 was used for the ventricles [34, 35]. This simple model is used as the simulation belongs to the special class called displacement-zero traction problems (or Dirichlet-type problems) whose solutions are known to be weakly dependent on the unknown patient-specific material properties of the tissues [2, 22, 34].
2.3 Model Solution
2.3.1 Computation of Tissue Deformations: Meshless Total Lagrangian Explicit Dynamics Algorithm
MTLED is a numerically robust and accurate meshless algorithm [7, 9]. The method computes deformations at an unstructured cloud of nodes used to discretize the geometry instead of elements as in finite element methods, which requires a high quality mesh of problem geometry [33]. The algorithm uses explicit time integration based on the central difference method. Unlike implicit time integration, this does not require solving systems of equations at every time-step making the method robust in performing calculations [7].
MTLED was evaluated extensively in computing brain deformations on problem geometry based on patient specific MRI data. The simulation results presented were within limits of neurosurgical and imaging equipment accuracy (~1 mm) [9, 19]. The method is also capable of handling very large deformations as well as cutting [8].
Meshless methods are preferred to finite element methods, due to excessive element distortion, are unreliable in scenarios where human soft tissues undergo very large strains in the vicinity of contact with a surgical tool while MTLED gives reliable results for compressive strains exceeding 70% [9].
The MTLED solver uses three input files automatically generated using our framework within 3D Slicer, which are: (1) computational grid information file, (2) material properties and (3) external load information file. All remaining parameters of MTLED are set by default (see Table 2) and are based on the experience obtained through numerous applications in computing soft continua and soft tissue deformations. The end user can change these parameters as per requirements but we recommend that a non-specialist user leave them unaltered.
3 Case Study: Intracranial Electrodes Induced Brain Shift
For the purpose of this case study, we determined prescribed displacements using a BSpline transform obtained using scattered transform module [14]. The input to scattered transform is the original electrode positions derived from CT (see Fig. 11a) and the projected electrode positions (see Fig. 11b). The output is a BSpline transform. We applied the obtained BSpline transform to the undeformed (initial) brain surface nodes located under the electrode sheet to determine position of the corresponding nodes in the deformed (due to electrode implantation) brain geometry (see Fig. 11c) (yellow transformed brain nodes)). We computed the prescribed displacements as a difference between the locations (coordinates) of the corresponding brain surface nodes under electrode sheet in undeformed and deformed brain geometry.
External Load a Visualization of 3D intracranial electrodes (yellow) on a preoperative MRI rigidly aligned with CT image, b Visualization of projected electrodes (red) on MRI, c Visualization of brain nodes pre-transformed (red) and transformed (yellow) along with 3D brain model (blue with reduced opacity) on preoperative MRI rigidly aligned with CT image. Visualization performed with 3D Slicer www.slicer.org [5]
We used nodal displacements computed by MTLED (see Fig. 12) in the scattered transform module [14] in 3D Slicer to obtain a BSpline transform to warp the preoperative MRI image so that it corresponds to the brain configuration with electrodes implanted (see Fig. 12).
Visualization of the transformed preoperative MRI using deformation field computed by MTLED registered onto a CT with implanted intracranial electrodes. Visualization performed with 3D Slicer www.slicer.org [5]
The simulation presented in this study was performed on a HP ProBook with Intel Core i7 2.7 GHz processor and 8 GB of physical memory. The calculation time for generating automatically a patient-specific computational model with all details, including patient-specific geometry construction, craniotomy region selection, external loading and defining contacts was 180.87 s. The execution time of the MTLED solution algorithm (i.e. obtaining the deformed model) was 762 s. The time for warping the preoperative image with the deformation field extracted from the model was 0.9 s.
4 Discussion and Conclusion
In this paper we described the framework for automated solution of computational biomechanics problems described by partial differential equations of solid mechanics. We also demonstrated the effectiveness of this framework using a case study of brain deformation caused by the placement of electrodes on brain surface in intracranial electroencephalography (iEEG).
In this study, electrodes implanted brain shift is simulated with 21,788 nodes (i.e. ~65,364 differential equations are solved) and 55,470 integration cells. The model has 221,880 integration points. The patient specific biomechanical model construction, which involves defining the patient specific brain geometry from a preoperative MRI, patient-specific tetrahedral integration grid generation, defining boundary conditions and external loads, and assigning material properties to brain tissues, took 180.87 s of computer processing time. The solution of the model using our MTLED algorithm took 762 s and finally the image warping took 0.9 s.
We are interested in computation of the deformation field within the brain which is completely determined by the description of the displacement on the boundary and equations of solid mechanics. We are creating an automatic framework that facilitates such computation for wide range of neurosurgical procedures. We previously successfully validated through application in computation of the brain shift induced by craniotomy and comparison of the predicted deformations with the intraoperative MRI [26]. In this study we present an extension of this framework to even more challenging problems of computing brain deformations induced by implantation of intracranial electroencephalography (iEEG) electrodes [39]. We showcase it through application in the analysis of an epilepsy patient undergoing such implantation. The results are very promising. Nevertheless application in computer simulation of a selected surgical procedure conducted on one patient can be regarded only as a preliminary evaluation of our framework. Therefore, we plan to apply our framework in the analysis of more patients undergoing craniotomy induced brain shift, electrode implantation for epilepsy treatment, and brain tumour resection.
References
Bezdek, J. C., Ehrlich, R., & Full, W. (1984). FCM: The fuzzy c-means clustering algorithm. Computers & Geosciences, 10(2–3), 191–203.
Ciarlet, P. G. (1988). Mathematical elasticity. North Hollad.
Dale, A. M., Fischl, B., & Sereno, M. I. (1999). Cortical surface-based analysis: I. Segmentation and surface reconstruction. NeuroImage, 9(2), 179–194.
Dora, L., Agrawal, S., Panda, R., & Abraham, A. (2017). State-of-the-art methods for brain tissue segmentation: A review. IEEE Reviews in Biomedical Engineering, 10, 235–249.
Fedorov, A., Beichel, R., Kalpathy Cramer, J., Finet, J., Fillion Robin, J.-C., Pujol, S., Bauer, C., Jennings, D., Fennessy, F., Sonka, M., Buatti, J., Aylward, S., Miller, J., Pieper, S., & Kikinis, R. (2012). 3D Slicer as an image computing platform for the quantitative imaging network. Magnetic Resonance Imaging, 30(9), 1323–1341.
Geuzaine, C., & Remacle, J. F. (2009). Gmsh: A 3-D finite element mesh generator with built-in pre- and post-processing facilities. International Journal for Numerical Methods in Engineering, 79(11), 1309–1331.
Horton, A., Wittek, A., Joldes, G. R., & Miller, K. (2010). A meshless total Lagrangian explicit dynamics algorithm for surgical simulation. International Journal for Numerical Methods in Biomedical Engineering, 26, 977–998.
Jin, X., Joldes, G. R., Miller, K., Yang, K. H., & Wittek, A. (2014). Meshless algorithm for soft tissue cutting in surgical simulation. Computer Methods in Biomechanics and Biomedical Engineering, 17, 800–817.
Joldes, G., Bourantas, G., Zwick, B., Chowdhury, H., Wittek, A., Agrawal, S., Mountris, K., Hyde, D., Warfield, S. K., & Miller, K. (2019). Suite of meshless algorithms for accurate computation of soft tissue deformation for surgical simulation. Medical Image Analysis, 56, 152–171.
Joldes, G., Wittek, A., & Miller, K. (2009). Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation. Medical Image Analysis, 13(6), 912–919.
Joldes, G. R., Chowdhury, H. A., Wittek, A., Doyle, B., & Miller, K. (2015). Modified moving least squares with polynomial bases for scattered data approximation. Applied Mathematics and Computation, 266, 893–902.
Joldes, G. R., Wittek, A., & Miller, K. (2011). An adaptive dynamic relaxation method for solving nonlinear finite element problems. Application to brain shift estimation. International Journal for Numerical Methods in Biomedical Engineering, 27(2), 173–185.
Joldes, G. R., Wittek, A., Miller, K., & Morriss, L. (2008). Realistic and efficient brain-skull interaction model for brain shift computation. In Computational Biomechanics for Medicine III Workshop, MICCAI.
Joldes, G. R., Wittek, A., Warfield, S. K., & Miller, K. (2012). Performing brain image warping using the deformation field predicted by a biomechanical model (pp. 89–96). Springer.
Li, M., Miller, K., Joldes, G. R., Kikinis, R., & Wittek, A. (2016). Biomechanical model for computing deformations for whole-body image registration: A meshless approach. International Journal for Numerical Methods in Biomedical Engineering, 32(12).
Li, M., Wittek, A., Joldes, G. R., & Miller, K. (2016). Fuzzy tissue classification for non-linear patient-specific biomechanical models for whole-body image registration. In G. R. Joldes, B. Doyle, A. Wittek, P. M. F. Nielsen & K. Miller (Eds.), Computational biomechanics for medicine: Imaging, modeling and computing (pp. 85–96). Springer International Publishing.
Miga, M. I., Sun, K., Chen, I., Clements, L. W., Pheiffer, T. S., Simpson, A. L., & Thompson, R. C. (2016). Clinical evaluation of a model-updated image-guidance approach to brain shift compensation: Experience in 16 cases. International Journal of Computer Assisted Radiology and Surgery, 11(8), 1467–1474.
Miller, K., Chinzei, K., Orssengo, G., & Bednarz, P. (2000). Mechanical properties of brain tissue in-vivo: Experiment and computer simulation. Journal of Biomechanics, 33, 1369–1376.
Miller, K., Horton, A., Joldes, G. R., & Wittek, A. (2012). Beyond finite elements: A comprehensive, patient-specific neurosurgical simulation utilizing a meshless method. Journal of biomechanics, 45(15), 2698–2701.
Miller, K., & Lu, J. (2013). On the prospect of patient-specific biomechanics without patient-specific properties of tissues. Journal of the Mechanical Behavior of Biomedical Materials, 27, 154–166.
Miller, K., Wittek, A., & Joldes, G. (2011). Biomechanical modeling of the brain for computer-assisted neurosurgery. In Biomechanics of the brain (pp. 111–136). Springer.
Neal, M. L., & Kerckhoffs, R. (2010). Current progress in patient-specific modeling. Briefings in Bioinformatics, 11, 15.
Otsu, N. (1979). A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics, 9(1), 62–66.
Pinter, C., Lasso, A., & Fichtinger, G. (2019). Polymorph segmentation representation for medical image computing. Computer Methods and Programs in Biomedicine, 171, 19–26.
Pruthi, S., Dawant, B., & Parker, S. L. (2020). Initial experience with using a structured light 3D scanner and image registration to plan bedside subdural evacuating port system placement.
Safdar, S., Joldes, G., Zwick, B., Bourantas, G., Kikinis, R., Wittek, A., & Miller, K. (2021). Automatic framework for patient-specific biomechanical computations of organ deformation (pp. 3–16). Springer.
Ségonne, F., Dale, A. M., Busa, E., Glessner, M., Salat, D., Hahn, H. K., & Fischl, B. (2004). A hybrid approach to the skull stripping problem in MRI. NeuroImage, 22(3), 1060–1075.
Sorkine, O., Cohen-Or, D., Lipman, Y., Alexa, M., Rössl, C., & Seidel, H. P. (2004). Laplacian surface editing. In Proceedings of the 2004 Eurographics/ACM SIGGRAPH Symposium on Geometry Processing.
Sullivan, C., & Kaszynski, A. (2019). PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). Journal of Open Source Software, 4(37), 1450.
Valette, S., Chassery, J. M., & Prost, R. (2008). Generic remeshing of 3D triangular meshes with metric-dependent discrete Voronoi diagrams. IEEE Transactions on Visualization and Computer Graphics, 14(2), 369–381.
Waldron, K. J., & Kinzel, G. L. (2004). Kinematics, dynamics, and design of machinery. Wiley.
Lorensen, W. E., & Cline, H. E. (1987). Marching cubes: A high resolution 3D surface construction algorithm. In SIGGRAPH Computer Graphics, (Vol. 21, pp. 163–169). Association for Computing Machinery.
Wittek, A., Grosland, N., Joldes, G., Magnotta, V., & Miller, K. (2016). From finite element meshes to clouds of points: A review of methods for generation of computational biomechanics models for patient-specific applications. Annals of Biomedical Engineering, 44(1), 3–15.
Wittek, A., Hawkins, T., & Miller, K. (2009). On the unimportance of constitutive models in computing brain deformation for image-guided surgery. Biomechanics and Modeling in Mechanobiology, 8, 77–84.
Wittek, A., Joldes, G., Couton, M., Warfield, S. K., & Miller, K. (2010). Patient-specific non-linear finite element modelling for predicting soft organ deformation in real-time; Application to non-rigid neuroimage registration. Progress in Biophysics and Molecular Biology, 103, 292–303.
Wittek, A., Joldes, G. R., & Miller, K. (2019). Finite element algorithms for computational biomechanics of the brain (pp. 243–272). Springer.
Wittek, A., Miller, K., Kikinis, R., & Warfield, S. K. (2007). Patient-specific model of brain deformation: Application to medical image registration. Journal of Biomechanics, 40, 919–929.
Zhang, J. Y., Joldes, G. R., Wittek, A., & Miller, K. (2013). Patient-specific computational biomechanics of the brain without segmentation and meshing. International Journal for Numerical Methods in Biomedical Engineering, 29(2), 293–308.
Zwick, B.F., Bourantas, G.C., Safdar, S., Joldes, G.R., Hyde, D.E., Warfield, S.K., Wittek, A., & Miller, K. (2022). Patient-specific solution of the electrocorticography forward problem in deforming brain (preprint on ArXiv:2109.07164, submitted to Neuroimage).
Acknowledgements
The funding from NHMRC grants APP1162030; APP1144519 is gratefully acknowledged. The first author acknowledges scholarship funding from University Postgraduate Award. We also wish to thank 3D Slicer on-line community https://discourse.slicer.org/ whose members have made many valuable contributions. Our special thanks go to Dr. Andras Lasso of Laboratory for Percutaneous Surgery (PerkLab).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Safdar, S. et al. (2022). Automatic Framework for Patient-Specific Biomechanical Computations of Organ Deformation: An Epilepsy (EEG) Case Study. In: Nielsen, P.M., Nash, M.P., Li, X., Miller, K., Wittek, A. (eds) Computational Biomechanics for Medicine. MICCAI 2021. Springer, Cham. https://doi.org/10.1007/978-3-031-09327-2_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-09327-2_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-09326-5
Online ISBN: 978-3-031-09327-2
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)