Abstract
Hospital readmissions are frequent and costly events. Early risk prediction can lead to more effective resource planning and utilization. This paper presents a deep learning framework for predicting the risk of 30-day all-cause readmission given a patient journey dataset. The problem is posed as a binary classification. A novel personalized self-adaptive feature learning and embedding strategy is applied to learn the representations of patient journeys. We first introduce a Variable Attention module to capture the interdependencies of clinical features and generate attention feature representations. We then place a convolutional neural network (CNN) on the generated feature representations to estimate outcome probabilities. Demographic features, including sex and age, are then incorporated into a personalized representation used for adaptively fixing the output of CNN by modifying the network loss function. We successfully predict 30-day all-cause risk-of-readmission with area-under-receiver-operating-curve (AUROC) ranging between 0.838 to 0.858 and overall maximum accuracy of 77.34%.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Hospital readmission is a costly event, which imposes a tremendous burden on a nation’s healthcare system. In the USA, there were 7.8 million (19.6%) of hospital-discharged patients readmitted from 2003 to 2004, which accounted for $17.4 billion of hospital payments [1]. A recent study focused on the readmission rate of atherothrombotic disease in Western Australia reported that the cost of readmissions (A$30 million) accounted for 42% of the original admissions cost (A$71 million) [2]. Moreover, high readmission rates cause a disruption to the normality of hospital management, particularly in critical resources allocation such as inpatient beds. Thus, predicting readmission is critically important for more effective healthcare resource planning and utilization.
Employing a predictive model is one of the useful strategies to reduce the hospital readmission rate [3]. Specifically, machine learning (ML) or deep learning (DL) algorithms can be adopted to identify high-risk patients from electronic health records (EHRs) data so that corresponding preventive approach may be developed to minimize their risk-of-readmission. To this end, this paper investigates the issues and challenges associated with the prediction task, and addresses these by developing a DL-based predictive framework to predict the 30-day all-cause risk-of-readmission given a patient journey dataset.
In practice, it is challenging to learn the representations of patient journeys. Specifically, each patient journey includes two aspects: patient visit and feature levels. Further, the feature level consists of demographic and clinical features. As shown in Fig. 1, two anonymous patients visited the emergency department (ED). They were admitted to different care units. The diagnoses and procedures performed at each visit were recorded as the documented content for a single patient. Each clinical feature records an independent observation, while a set of features can represent the medical conditions of a patient at a given time point.
Patient journey samples. A patient journey is often a consequence of clinical patterns that are associated with specific sequences of clinical events. Usually, a patient walks in or takes an ambulance to the ED for medical treatment. The ED has a dedicated triage area, where nurses and doctors follow a specialized triage process and triages a patient on the basis of how critical the illness or injury is at the time of presentation to the ED. As a result, a patient is either admitted to the corresponding unit or discharged after medical treatments.
When predicting the risk-of-readmission, we aim to automatically model the contexts of patient journeys and generate new patient journey representations. The benefit of modeling patient journey contexts is that such a consideration can help us capture patient medical conditions for effective risk-of-readmission prediction. However, the patient visit process is often patient-specific, which is a consequence of a range of health problems or the environment of clinical (the capacity of the hospital to admit patients). As a result, a part of clinical features within a patient journey is irrelevant to the target prediction, and should be treated as noise for the risk-of-readmission. This issue has been largely disregarded in the existing studies on patient journey learning, which leads to significantly reduced accuracy of the prediction results. Thus, in order to capture patient medical conditions correctly, a good risk prediction method should be able to learn patient journey contexts by distinguishing the importance of features in each patient journey.
Another noticeable challenge stems from the heterogeneity in the disease and demographic features [4]. In practice, it is difficult to predict risk-of-readmission because of the heterogeneity in the disease and demographic features. Specifically, demographic features are usually regarded as a health context before admission. To fully model a patient journey, researchers usually examine the demographic features and combine them with the clinical features. However, if two patients were admitted with heart failure, one is 20-year-old, another 80, then the corresponding health status may have significant differences, resulting in different risk-of readmission. Additionally, due to the complexity and diversity of all cause inpatient data, a certain amount of diseases are patient-specific.
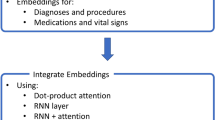
To jointly tackle the above issues, in this paper, we propose a novel DL predictive framework, which can automatically learn the representations of the patient journey and effectively perform risk-of-readmission prediction. Firstly, it proposes a Variable Attention module, which is composed of a 1D-CNNs and the self-attention mechanism [5]. The 1D-CNN is developed to capture the interdependencies of clinical features to model each patient journey context and generate feature representations, which are then used as query and key vectors in the self-attention mechanism. The key-query pairs are used to compute the inner dependency weights, then used to update the values. A series of attention feature representations are generated for each patient journey. We place a customized CNN on attention feature representations to perform the predictive modeling. Secondly, it introduces a personalized characterization representation to fix the output of the neural network adaptively, which is achieved by adding two additional terms into the network loss function. The personalized characterization representation is formed by exploiting a standard logistic function to automatically and adaptively learn demographic feature distribution and importance and then embed the newly generated feature representations into one overall representation. This framework enables learning and embedding of demographic and clinical features self-adaptively take place so that their respective contributions to final outputs are captured.
Our major contributions are as follows:
-
1.
We propose a novel deep learning framework for predicting 30-day all-cause risk-of-readmission by fully learning patient journey representations.
-
2.
We designed a personalized feature learning and embedding strategy to incorporate demographic and clinical features. Meanwhile, we modify the network loss function to adjust their contributions in the framework.
-
3.
We conduct extensive experiments on a real patient journey dataset to validate our proposed framework. The empirical results demonstrated significant prediction performance improvement across the task.
2 Basic Notation and Problem Definition
Our patient journey dataset consists of patients’ time-ordered visiting records, which is denoted by V, i.e., \(V =\{V_{1}, V_{2}, ..., V_{\mathcal {T}}\}\). Each visiting record \(V_{t}\) consists of demographic and clinical features. Demographic features contains age and sex. For each \(V_{t}\), we use \(D_{t} \in R^{1 \times M}\) and \(C_{t} \in R^{1 \times N}\) separately to denote demographic and clinical features at time t. For each \(V_{t}\), we provided the visit event-level label for binary task, \(Y_{t}\) = 1 denotes a patient is readmitted within 30-day, otherwise \(Y_{t}\) = 0. The goal of this task is to predict \(Y_{t}\) by learning \(V =\{V_{1}, V_{2}, ..., V_{\mathcal {T}}\}\) from the given dataset.
3 The Proposed Framework
To tackle the aforementioned challenges, in this work, we propose a novel deep learning framework. Figure 2 provides an overview of the proposed framework. In the rest of this section, we introduce the modules of the framework separately.
3.1 Personalized Feature Learning and Embedding
Clinical Features. Learning new feature representations for diagnosis is at the heart of healthcare analytics [6]. In particular, our diagnostic information contains a medical diagnosis, syndrome, or symptom. To model its impact, we used a logistic function as follows:
where \(C_{Diag}\) denotes diagnosis and \(W_{Diag}\) \(\in \) R is the specific parameter to model the corresponding impact for the prediction task. \(\varphi \) is a predefined scalar and set \(\varphi \) = 90 (see Sect. 4.2, there are 181 types of diagnosis).
Capturing the Interdependencies of Clinical Features and Generating Attention Feature Representations. In this subsection, we propose Variable Attention module, which consists of a 1D-CNN and the self-attention mechanism [5]. Specifically, we first apply a 1D convolution operation on clinical features to learn the interdependencies of clinical features and generate the new feature representations. Then, the max-pooling operation [7] is used to extract the most important feature representations. Last, the pooling results are used as query and key vectors in the attention mechanism. Note that 1D convolution and max-pooling operations separately work in the horizontal and vertical directions.
With the help of convolution operation, the attention mechanism can learn the interdependencies of clinical features at a larger nonlinear space. Moreover, the obtained max-pooling results provide the most important feature representations used for the attention mechanism. Furthermore, one notable advantage of the module is that an interpretable attention map is given after the training, which gives valuable information about the target variables on how much they are correlated to each other.
Mathematically, in the 1D-CNN, the shape of output matrix corresponds to the shape of input matrix as follows: \(L_{out} = (L_{in} - filter\_size + 2 \times padding)/stride + 1\). We define the stride is 1, and \(filter\_size = 2 \times padding + 1\). For clinical features C, we have the following 1D convolution operation:
where \(Conv1D(\cdot )\) denotes the 1D convolution operation and \(H \in R^{\mathcal {T} \times N}\) denotes the new feature representations. Both have the same shape. The max-pooling operation extracts the most important feature representations from H. The pooling result is used as the query vector \(q \in R^{1 \times N}\) and key vector \(k \in R^{1 \times N}\) for the attention mechanism.
The query-key pairs are used to compute the inner dependency weights and then update the values. Mathematically, the formula is defined as below:
where \(\alpha \in R^{N \times N}\) generated for a patient journey can explain the causative and associative relationships between diagnoses and procedures performed at each patient journey. For simplicity, we use AttC to denote Attention(C) in the following sections.
Demographics Features. In this work, we mainly consider patient age and gender when predicting risk-of-readmission [8, 9]. Moreover, age was categorized by the Australian Institute of Health and Welfare (AIHWFootnote 1) into below 1 year, 1–4 years, 5–14 years, 15–24 years, 25–34 years, 35–44 years, 45–54 years, 55–64 years, 65–74 years, 75–84 years, over 85 years. In the same vein, we used logistic function again to model age and sex in order to learn their distributions and importance as follows:
where \(D_{AGE}\) denotes age and \(W_{AGE}\) \(\in \) R is the specific parameter to model the impact of age for prediction task. The parameter is used to regularize variable outputs in order to achieve its optimization. \(\varphi \) is a predefined scalar. We used age groups instead of patients’ actual ages and set \(\varphi \) = 5 (based on above 11 subgroups of age).
where \(D_{SEX}\) denotes sex and \(W_{SEX}\) \(\in \) R is the specific parameter to model the impact of sex for prediction task. \(\varphi \) is a predefined scalar and set \(\varphi \) = 1.
To better characterize age and sex, we embed them into a personalized characterization representation.
where \(W_{base}^{emb}\) is an embedding matrix and base consists of \(f_{AGE}\) and \(f_{SEX}\).
3.2 Personalized Prediction
We customize a CNN with a 1D convolutional layer and a max-pooling layer. The convolutional layer is placed on the attention feature representations. We use a combination of m filters with s different window sizes. We use l to denote the size of feature window and \(AttC_{j: j + l -1}\) denote the concatenation of l clinical features from \(AttC_{j}\) to \(AttC_{j + l -1}\). A filter \(W_{f} \in R^{1 \times l}\) is applied on the window of l clinical features to produce a new feature \(f_{j} \in R\) with the ReLU activation function as follows:
where \(b_{f} \in R\) is a bias term and ReLU(f) = max(f, 0).
This filter is applied to each possible window of clinical features in the whole \(\{AttC_{1: l}, AttC_{2: l + 1}, ..., AttC_{N - l + 1: N}\}\) to generate a feature map \(f \in R^{N - l + 1}\) as follows:
The max-pooling layer is placed on the feature map f. Each filter produces a feature. Since we have m filters with s different window sizes, the final vector representation of \(AttC_{t}\) can be obtained by concatenating all the extracted features, e.g., \(z_{t} \in R^{ms}\). A fully connected softmax layer is used to estimate outcome probabilities as follows:
where \(W_{Y} \in R^{n \times ms}\) and \(b_{Y} \in R^{n}\) are the learnable parameters and n is the number of target labels (e.g., 0 or 1 in binary classification).
Let \(\theta \) be the set of all the parameters in CNN. \(\hat{Y}_{t}\) and \(Y_{t}\) are separately prediction probability vector and ground truth. The cross-entropy between ground truth \(Y_{t}\) and outcome probabilities \(\hat{Y}_{t}\) is used to estimate the loss. The objective function can be defined as follows:
Given the input data \(_{t}\) to predict its true label vector \(Y_{t}\), we can obtain outcome probability vector \(\hat{Y}_{t} = p(Y_{t}|C_{t}; \theta ))\). Now we use the proposed personalized characterization representation \(f^{t}_{base}\) (demographic features) to fix the output of CNN adaptively. The objective function can be rewritten as follows:
where \(\alpha \), \(\beta \) are the hyper-parameters and \(\mathcal {L^{'}(W)}\) is the average cross entropy between \(f^{t}_{base}\) and ground truth \(Y_{t}\). The \(\mathcal {L^{'}(W)}\) is defined as follows:
In summary, Eq. (12) incorporates two additional loss terms, both of which are relevant with demographic features.
\(\alpha \frac{1}{\mathcal {T}} \sum ^{\mathcal {T}}_{t=1} KL (f^{t}_{base}|P(Y_{t}|C_{t}; \theta ))\) is the KL loss between personalized characterization representation and prediction distributions, which is used to fix the prediction results achieved by CNN.
Another loss function of \(\mathcal {L^{'}(W)}\) represents the self-adaptive process for demographic features. It provides a bridge between distributions of demographic features and ground truth, where each demographic feature can achieve its optimization by updating its values with the learning process.
4 Experimental Setup
4.1 Dataset Description
We presented one case study on the risk-of-readmission prediction, using a patient journey dataset from a metropolitan hospital in Australia. The data set being used is the administrative part of the EHRs from the hospital for the whole of 2018 and 2019. It contains demographic, admission (emergency or elective, location of care and treating clinical team) and discharge data, and detailed diagnostic information. It also contains time and location stamp information that records every occasion a patient is moved between locations within the hospital. Ethical approval was obtained for access to the dataset. Table 1 presents an overview of all the selected features.
4.2 Data Preprocessing
We selected the patient data that was readmitted within 30-day from the discharge based on the standardized HRRP readmission measure [10]. Each patient has a unique patient URN (ID) for distinguishing multiple admissions such as readmission. We examined the number of days between the current admission and him/her previous discharge for a given patient and checked if it is less than 30-day. Moreover, to accurately learn the common patient behaviors, we selected a diagnosis with more than 100 visits from the dataset. The final dataset includes 181 diseases and 61264 records (roughly 83% of the total data). Besides, we also used the undersampling approach [11] to address the label imbalance problem.
4.3 Baseline Approaches
To validate the performance of the proposed framework for our prediction task, we compare the proposed framework with logistic regression (LR), Gaussian Naive Bayes (GNB), Support Vector Machine (SVM), Random Forest (RF), AdaBoost, Explainable Boosting Machine (EBM) [12], and two CNN models. EBM is a strong baseline that has been applied in 30-day risk-of-readmission prediction and reported state-of-the-art accuracy. Besides, in ablation experiments, we present one variant of the proposed framework (CNN+). It only incorporates the attention mechanism [5].
4.4 Implementation Details and Evaluation Strategies
We implement the proposed framework with PyTorch 0.2.0. We implement ML approaches with scikit-learnFootnote 2 and EBMFootnote 3. We use the same settings for the proposed framework and other CNN models. Specifically, we set the size of filter windows (l) from 2 to 5 with s = 100 filter maps. Moreover, we propose using 1 as the size of filter windows that can introduce more nonlinearities without modifying the size of the input and thus enhance the expression ability of the neural network [13]. We also use regularization (\(l_{2}\) norm with the coefficient 0.001), and drop-out strategies (drop-out rate is 0.5) for all DL approaches. For training models, we employ a standard train/test/validation split. The validation set is used to select the best values of parameters. To evaluate binary outcomes, we calculate AUROC, Accuracy, F1 Score, and Brier Score Loss (BSL). We perform 100 repeats for all used approaches and report the average performance.
5 Results and Discussion
5.1 Performance Evaluation
Table 2 shows the performance of all approaches on the dataset. The results indicate that the proposed framework can outperform other baseline methods. Specifically, the risk-of-readmission was predicted with an AUROC of 0.8480 and one standard deviation of 0.010. Moreover, we find that the proposed framework can outperform CNN. Therefore, it indicates that the proposed personalized feature learning and embedding can improve the accuracy of risk-of-readmission prediction. Another important finding was that the proposed framework could outperform the CNN+. Therefore, it indicates that incorporating the self-attention mechanism and 1D-CNN is better for improving predictive accuracy than using the self-attention mechanism alone.
5.2 Clinical Feature Interdependencies
One aspect of the proposed method is that it explicitly captures the interdependencies of clinical features and generates attention feature representations. This is achieved by applying Variable Attention module to each patient journey. The module provides an interpretable attention map after the training, which gives valuable information about the target variables on how much they are correlated to each other. Therefore, this makes the proposed method explainable. To showcase this feature, we visualized four patient journeys, which correspond to patients A-D. These examples come from the test dataset. Patients A and B were readmitted with Heart Failure and Shock but with significant differences in other clinical features. Patients C and D were readmitted with Chronic Obstructive Airways Disease but with minor differences in other clinical features.
Figure 3 shows the clinical feature interdependencies of patients A–D. The attention scores calculated by the proposed Variable Attention module are shown. Note that for each attention score, we round up to 2 decimal places. The ordinates of the figure are the Query features, and the abscissas are the Key features. The boxes in the figures show how much each Key feature responds to the Query when a Query feature makes a query. We find that Variable Attention module can figure out clinical feature interdependencies for four patients. These feature interdependencies can be explained in part by attention scores. e.g., Variable Attention module figures out there are relatively high interdependencies between most clinical features and Diag. Additionally, Variable Attention module figures out that the part of the clinical features is also likely to respond strongly to themselves, which is denoted by the diagonal of four matrices.
6 Conclusion
In this work, we have presented a novel DL framework of personalized learning and embedding features with the aim of predicting risk-of-readmission. Experiments on a real patient journey dataset show that our framework demonstrated significant prediction performance improvement. The findings from this study make several contributions to the current literature. Firstly, we have proven that prediction of 30-day all-cause risk-of-readmission in hospitals is possible using patient journeys. Secondly, personalized feature learning and embedding contribute in several ways to our understanding of the importance of clinical and demographic features and provide a basis for further research. Lastly, it is a scalable framework, which can readily be of broad use to the scientific and health care communities. A number of possible future studies using the same experimental setup are apparent such as admission prediction at the time of triage and hospital admission location prediction.
References
Jencks, S.F., Williams, M.V., Coleman, E.A.: Rehospitalizations among patients in the medicare fee-for-service program. N. Engl. J. Med. 360(14), 1418–1428 (2009)
Atkins, E.R., Geelhoed, E.A., Knuiman, M., Briffa, T.G.: One third of hospital costs for atherothrombotic disease are attributable to readmissions: a linked data analysis. BMC Health Serv. Res. 14(1), 1–9 (2014)
Zhou, H., Della, P.R., Roberts, P., Goh, L., Dhaliwal, S.S.: Utility of models to predict 28-day or 30-day unplanned hospital readmissions: an updated systematic review. BMJ Open 6(6), e011060 (2016)
Reddy, B.K., Delen, D.: Predicting hospital readmission for lupus patients: an RNN-LSTM-based deep-learning methodology. Comput. Biol. Med. 101, 199–209 (2018)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, pp. 5998–6008 (2017)
Cai, X., Gao, J., Ngiam, K.Y., Ooi, B.C., Zhang, Y., Yuan, X.: Medical concept embedding with time-aware attention. In: Proceedings of the 27th International Joint Conference on Artificial Intelligence, pp. 3984–3990 (2018)
Collobert, R., Weston, J., Bottou, L., Karlen, M., Kavukcuoglu, K., Kuksa, P.: Natural language processing (almost) from scratch. J. Mach. Learn. Res. 12(ARTICLE), 2493–2537 (2011)
Berry, J.G., et al.: Age trends in 30 day hospital readmissions: us national retrospective analysis. BMJ 360 (2018)
Maali, Y., Perez-Concha, O., Coiera, E., Roffe, D., Day, R.O., Gallego, B.: Predicting 7-day, 30-day and 60-day all-cause unplanned readmission: a case study of a Sydney hospital. BMC Med. Inform. Decis. Mak. 18(1), 1–11 (2018)
McIlvennan, C.K., Eapen, Z.J., Allen, L.A.: Hospital readmissions reduction program. Circulation 131(20), 1796–1803 (2015)
Lemaître, G., Nogueira, F., Aridas, C.K.: Imbalanced-learn: a Python toolbox to tackle the curse of imbalanced datasets in machine learning. J. Mach. Learn. Res. 18(1), 559–563 (2017)
Nori, H., Jenkins, S., Koch, P., Caruana, R.: InterpretML: a unified framework for machine learning interpretability. arXiv preprint arXiv:1909.09223 (2019)
Lin, M., Chen, Q., Yan, S.: Network in network. arXiv preprint arXiv:1312.4400 (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, Y., Qin, S. (2022). Hospital Readmission Prediction via Personalized Feature Learning and Embedding: A Novel Deep Learning Framework. In: Fujita, H., Fournier-Viger, P., Ali, M., Wang, Y. (eds) Advances and Trends in Artificial Intelligence. Theory and Practices in Artificial Intelligence. IEA/AIE 2022. Lecture Notes in Computer Science(), vol 13343. Springer, Cham. https://doi.org/10.1007/978-3-031-08530-7_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-08530-7_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08529-1
Online ISBN: 978-3-031-08530-7
eBook Packages: Computer ScienceComputer Science (R0)