Abstract
Since the beginning of the COVID-19 pandemic, more than 350 million cases and 5 million deaths have occurred. Since day one, multiple methods have been provided to diagnose patients who have been infected. Alongside the gold standard of laboratory analyses, deep learning algorithms on chest X-rays (CXR) have been developed to support the COVID-19 diagnosis. The literature reports that convolutional neural networks (CNNs) have obtained excellent results on image datasets when the tests are performed in cross-validation, but such models fail to generalize to unseen data. To overcome this limitation, we exploit the strength of multiple CNNs by building an ensemble of classifiers via an optimized late fusion approach. To demonstrate the system’s robustness, we present different experiments on open source CXR datasets to simulate a real-world scenario, where scans of patients affected by various lung pathologies and coming from external datasets are tested. Promising performances are obtained both in cross-validation and in external validation, obtaining an average accuracy of 93.02% and 91.02%, respectively.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Since the start of the pandemic, the [1] recorded more than 350 million cases and 5 million deaths caused by the acute respiratory syndrome COVID-19. To control and reduce the spread of the pandemic, different testing modalities, like the reverse transcriptase-polymerase chain reaction (RT-PCR), have been introduced to validate the presence of the virus in patients.
Further to laboratory tests, there have also been efforts to use medical images as a means to diagnose COVID-19 pneumonia [2], mainly using computed tomography (CT) and chest X-ray (CXR) scans. The choice of the imaging modality carries pros and cons. Thanks to its high specificity and its facility of recognizing the different stages of the pathology, CT is the key modality for diagnosing lung pathologies. However, on CT scans it is hard to differentiate between COVID-19 positive patients and those affected by other lung pathologies [2]. Moreover, with CT scanning there is a high risk of contamination for both patients and clinicians, since the cleaning procedure of the scanners is not trivial. Conversely, although CXR has less sensitivity than CT, it is more used for its cost-effectiveness, compactness and limited cross-infection. With the CXR modality there is also the possibility of using portable scanners, useful in emergency care units or at the patients’ house, facilitating the control of the virus also in underdeveloped countries.
Over the last decade, deep-learning (DL) has demonstrated to be one of the best solutions to overcome challenges coming from multiple fields of study since it can extrapolate information from data useful for the task at hand [7, 22]. Therefore, during the COVID-19 pandemic, researchers have developed DL models able to diagnose COVID-19 on CXR. The state-of-the-art has focused mainly on two classification tasks. The first detects COVID-19 pneumonia in a binary classification task distinguishing between images of patients suffering from COVID-19 and those not affected by this disease, including healthy subjects and those affected by other pneumonia. This task is shortly referred to as COVID-19 vs. non-COVID-19 in the following. The second aims to discriminate images of patients affected by COVID-19 pneumonia, other types of pneumonia and healthy subjects shortly named COVID-19 vs. Pneumonia vs. Healthy hereinafter. Providing a survey of the work on these tasks is out of the scope of this contribution, but the interested readers can refer to [31] for further details. However, it is worth noting that the analysis of the literature reveals a major limitation: such models do not reflect a real-world where patients affected by different lung diseases, further to pneumonia, arrive at hospitals and are scanned for diagnosis. For instance, a model trained on the non-COVID-19 vs. COVID-19 task, where the non-COVID-19 class includes only healthy patients, is not useful in this scenario since the model is not specific to the COVID-19 diseases. Similarly, in the Healthy vs. Pneumonia vs. COVID-19 task, the algorithm learns how to classify between patients affected by COVID-19, healthy patients and pneumonia, but it is not able to detect other lung diseases. These motivations go hand-in-hand with clinical motivations, which state that it is important to detect if a patient is healthy or is affected by pulmonary disease, discriminating between a generic pulmonary disease, COVID-19 pneumonia and other types of pneumonia [5] since each therapy is different [21].
In the literature, the few papers which extend the 2-class and the 3-class classification tasks to other pulmonary diseases are few in number. In [21] the authors used a pre-trained CNN which uses texture descriptors of CXR images [8, 30] to recognize different types of pneumonia: COVID-19, SARS, MERS, Pneumocystis, Streptococcus, Varicella and healthy cases. In [9] the authors proposed a CNN model similar to the InceptionNetV3 that screens COVID-19 positive cases from other types of Pnuemonia, Tubercolosis patients and healthy cases, using a CXR dataset [8, 18, 25, 30]. Finally, in [3] the authors presented a transfer learning approach working with a pre-trained CNN on a CXR dataset [8, 30] to discriminate between four classes: COVID-19 pneumonia, other pneumonia, other diseases and healthy patients.
A general limitation of most approaches processing CXR scans for classification goals is that they do not externally validate the models on never-seen data because only a simple hold-out or a cross-validation (CV) scheme are usually used to compute the performance. This favorably biases the performance concerning a real-world scenario where CXR scans come from different scanners and hospitals, which makes non-trivial the generalization of the model. Such issue is also confirmed by our results reported in the next sections: we find that state-of-the-art CNNs have high performance when tested in CV, but they drop when tested on an external data source. To overcome this limitation, and also to deal with the need of extending the 2- and 3-class task to more classes, in this paper we present a method to algorithmically build an ensemble of pre-trained CNNs that performs a 4-class classification task on CXR scans, where we have patients affected by COVID-19, other pneumonia, other lung diseases and healthy subjects, shortly referred as COVID-19 vs. Pneumonia vs. Other vs. Healthy. Figure 1 shows examples of images belonging to those four classes. The extension to a 4-class scenario may seem straightforward, but given that many lung diseases are collected in the new class, it extends the capabilities of the system, which now can work with a vast number of lung conditions. To make the DL model usable in clinical practice, we present an approach that performs well not only in CV but is also robust to external validation.
The manuscript is organized as follows: Sect. 2 presents the datasets used for training and testing, Sect. 3 shows the proposed method and explains the experiments followed, and Sect. 4 shows and discusses the results obtained, and finally Sect. 5 provides the concluding remarks.
2 Materials
The scientific community has focused on gathering various COVID-19 open-access datasets. Among them, here we focus on those containing CXR images that we put together to reflect as much as possible a real-world scenario where different lung diseases are studied and where the scans are collected from multiple centers, augmenting the variance in the data. As also discussed in [21], inter-center variability is a crucial step [9] to make the algorithms more robust.
We collected images of patients affected by COVID-19 by exploiting two COVID-19 multi-centric datasets, namely AIforCOVID [27] and COVIDX [29]. The former is used for training, whilst the latter is for external validation. Furthermore, images for the other three classes (i.e. pneumonia, other pulmonary diseases and healthy cases) were retrieved from the NIH CXR dataset [30] and they are used to set up the training and validation datasets.
The AIforCOVID dataset [27] is composed of anteroposterior and posteroanterior views of 1100 COVID-19 positive patients, with a mild or severe outcome collected from six different Italian hospitals.
The well-known COVIDX dataset [29] is composed of both COVID-19 positive and negative CXR scans: here we retrieved 16690 scans of the positive class since the non-COVID-19 cases came from the NIH CXR dataset [30].
The NIH CXR dataset [30] contains 112120 CXR images in anteroposterior view collected from NIH clinical center’s internal PACS systems: it includes 60361 scans of healthy cases, 1431 scans of patients affected by pneumonia and 50328 scans of cases affected by other lung pathologies, which are atelectasis, cardiomegaly, effusion, infiltration, mass, nodule, and pneumothorax. To have a balanced dataset for training, we randomly selected 1100 images for each of the three classes. The remaining 108820 scans were used for external validation.
To sum up, the dataset used for experiments performed in CV is composed of 1100 scans for each of the four classes, whereas the one used in external validation accounts for 16690 scans for the COVID-19 class, 331 scans for the pneumonia class, 49228 scans for the other lung pathologies class, and 59261 scans for the healthy class.
3 Methods
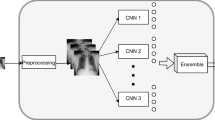
Our DL approach works with CXR images to perform a 4-class classification task, which discriminates between COVID-19 cases, pneumonia cases, healthy patients and patients affected by other lung diseases, and it algorithmically builds an optimized late fusion ensemble of multiple pre-trained CNNs. The idea stems from observing that today many pre-trained CNNs are available, permitting researchers and practitioners to explore many different deep architectures by exploiting transfer learning even when the available dataset would not permit training from scratch. Furthermore, once several CNNs have been trained, a question arises: is it better to pick the CNN with the best performance on a validation set or to explore the possibility to build an ensemble of CNNs? Indeed, it is well known that in many cases, ensembles of classifiers combined in late fusion have provided better performance than single learners [14]. This happens since fusing multiple models provides complementary and more powerful data representation, and the success of such a mixture relies on having diverse classifiers [14] offering different and complementary points of view to the ensemble. Moreover, in the case we opt for the ensemble, there is another question: which are the CNNs to be included in the ensemble? To this end, denoting with n and k the number of available CNNs and the number of CNNs to included in the ensemble, a researcher should explore \(\gamma = \sum _{k=2}^{n} \left( {\begin{array}{c}n\\ k\end{array}}\right) \) combinations to find which is the best one. But putting together the CNNs with the largest performance not always retrieves the best ensemble. This happens because the CNNs should provide wrong classifications on the same samples: this phenomenon can be measured by the diversity score, which measures how much the classifications returned by a mixture of classifiers vary on a set of data. In this respect, here we present a multi-objective solution to this search that returns the ensemble and therefore the set of CNNs, maximizing accuracy and diversity scores on a validation set. Figure 2 shows the whole pipeline that is further described in the next subsections.
3.1 Pre-processing
As a first step, there is the need to align the CXR images because they are collected from different centers: the goal is to obtain a cropped image of the bounding box containing the patient’s lung, excluding unnecessary regions of the scan. This is performed using a U-Net [23] trained on the Montgomery County CXR collection [18] and the Japanese Society of Radiological Technology repository [25] with a total of 7717 CXR scans of non-COVID-19 patients, which extrapolates the mask of the lung pixels. Given the mask, the cropped image is the minimum squared bounding box containing both lungs.
The U-Net was trained for 100 epochs using a binary cross-entropy loss function and an Adam optimizer after resizing the images to a 3\(\,\times \,\)256\(\,\times \,\)256 normalized tensor. To prevent overfitting, we applied a random augmentation, which consists of a random rotation (\(\pm {25}^\circ \)), random horizontal and vertical shift (\(\pm 25\) pixels), and random zoom (0–0.2%). To remove any artifact, we selected the top two biggest segmented regions representing the lungs. We also assessed the U-Net performance by running a 5-fold CV on the two aforementioned datasets that returned an average Dice score equal to 96.32%.
3.2 Training of Single CNNs
We individually trained and tested 20 different CNNs with a stratified 10-fold CV where the train-validation-test split is 70-20-10%. They are well-known state-of-the-art CNNs [20] pre-trained on the ImageNet dataset [10]: AlexNet [19], VGG11, VGG13, VGG16, VGG19 [26], GoogLeNet [28], ResNet18, ResNet34, ResNet50, ResNet101, ResNet152 [15], WideResNet50 [33], ResNeXt50 [32], SqeezeNet1(0), SqeezeNet1(1) [17], DenseNet121, DenseNet161, DenseNet169, DenseNet201 [16], and MobileNetV2 [24].
After the alignment phase, the images are resized to a 3\(\,\times \,\)224\(\,\times \,\)224 tensor and normalized. To prevent overfitting, during training a random augmentation is performed: random horizontal and vertical random shift (\(\pm 7\) pixels), flip along the vertical axis, random rotation (\(\pm {45}^\circ \)) and elastic transformation (\(\alpha = 20-40\), \(\sigma =7\)). All the CNNs are trained using the cross-entropy as loss function, with a maximum of 300 epochs and an early stopping of 25 epochs on the validation set. We used a batch size of 32 and used stochastic gradient descent as optimizer with an initial learning rate of 0.001 and a momentum of 0.9, a learning rate scheduler with a step size of 7, and \(\gamma =0.1\).
3.3 Ensemble Optimization
As already mentioned, the composition of the ensemble is determined by maximizing both the accuracy and the diversity scores provided by the ensemble itself on a validation set. While the accuracy ACC is uniquely defined, the diversity can be measured using different scores heuristically set, which are divided into pairwise and non-pairwise measures, although the former usually perform better than the latter [6]. In this work, the used the pairwise double-fault score \(DF_{i,j}\), which is the proportion of samples miss-classified by the classifiers i and j. For a team of c classifiers, the averaged double-fault \(\overline{DF}\) over all pairs of classifiers is given by \(\overline{DF} = \frac{2}{c(c-1)}\sum _{i=1}^{c-1} \sum _{j=i+1}^{c} DF_{i,j}\). Both ACC and \(\overline{DF}\) range in [0, 1], and the higher the values, the more accurate and diverse the models. In practice, given c classifiers collected in the set \(\mathbf {C} = \{C_i\}_{i=1}^c\) our method looks for the combination of \(k \le c\) models maximizing both the accuracy and the double-fault score (\(\overline{DF}\)) on a validation set among all the \(\theta \) possible combinations which are collected in the set \(\mathbf {\Theta } = \{\varTheta _j \}_{j=1}^{\theta }\), where \(\varTheta _j\) denotes one of the possible mixture of classifiers. The method returns \(\hat{\varTheta }\) containing the set of k classifiers from \(\mathbf {C}\) that constitute the best ensemble so that \(\hat{\varTheta } = \{ C_i \in \varTheta _j | j = \hbox {arg min}_{\varTheta _j \in \mathbf {\Theta }} F \}\), where F is the objective function defined as \(F=(1-ACC(\varTheta _j))^2 + (1-\overline{DF}(\varTheta _j))^2\). Let us also notice that, as proofed in [12, 13], solving this two-objective minimization problem corresponds to finding the Pareto optimum of the optimization problem that has a unique solution.
Furthermore, the method can work with any aggregation rule combining the outputs provided by the single classifier in the ensemble. In this respect, here we use the majority voting rule, which assigns the most common label among the classifications since it has demonstrated to be the most performing in many applications [4]. To prevent any tie, we considered only odd values of k in [3, 20], resulting in 9 combinations.
Notice also that, to prevent any bias, the optimization is performed on a validation set without intersections with the test and external validation sets.
4 Results and Discussions
Tables 1, 2, 3 and 4 show all the results achieved in terms of accuracy and recall for each of the four classes. On the one hand, Tables 1 and 2 present the performance attained by each of the 20 CNNs when the experiments were performed in 10 fold CV and on the external dataset, respectively. On the other hand, in the case of CV, each row of Table 3 reports the scores achieved by the ensemble returning the minimum F among all the possible mixture of classifiers in \(\mathbf {\Theta }\); note that such ensembles were built considering only odd values of k to avoid any ties in the final decision. Furthermore, for any k, we fixed the ensemble in Table 3 and applied it to the external dataset: the corresponding results are shown in Table 4. All such four tables also show in the first column the rank of each row.
In Table 1 we notice that the models provide satisfactory performance in CV, but Table 2 reveals that they do not generalize well to images belonging from a cohort different from the one used for training, as the drop of 5–8% in accuracy suggests. Similar behavior occurs for the recalls of each class. This observation is strengthened by the fact that the results in external validation are attained on a set of thousands of images, much larger than the set used for training. This finding also confirms previous work showing that DL suffers from this limitation in several bio-medicine applications [11]. This behavior also occurs for the models used by the authors in [3], which worked with the AlexNet, VGG16, and ResNet50.
Let us now focus on the results attained by the optimized ensembles (Tables 3 and 4). We notice in all the cases in Table 3 that the accuracy is larger than 90% and the ensembles, whatever the number of single CNNs used, always outperform the results provided by the single deep networks. This suggests that the ensemble of classifiers successfully exploits the diversity introduced by the different CNNs. Furthermore, the ensemble is robust to external validation since its performance drops to a lesser extent, i.e. around 2–3%, for any k. Among all the ensembles, the combination with the highest accuracy in both the experiments has \(k=3\), showing that the best choice of k can be obtained prior to the external validation. This combination is composed of the VGG11, ResNet34, and DenseNet161 CNNs, which do not correspond to the top 3 single models.
Furthermore, we assess if the performance of the CNNs and of the ensembles are different. To this goal, rather than focusing on the best models, we run the pairwise t-test between the distributions of each performance score. In other words, both in CV and external validation, we compare the performance scores between the single CNNs and the ensembles, finding that they are statistically different (p-value \(\le \) 0.05). In particular, this result holds not only for the global accuracy but also for the recall of each class. The statistical significance of the performance differences is also confirmed when comparing the best CNN and the best ensemble.
Finally, we deepen how the optimization works on the validation set. To this goal, the left panel in Fig. 3 shows the values of accuracy and diversity computed for any of the \(\theta \) ensembles we tested. Straightforwardly, the best ensemble is the one lying in the top right corner of this plot. Furthermore, we notice that the lowest diversity values correspond to the lowest accuracy, confirming the empirical observation that any mixture of classifiers should include learners making errors on different samples. Observing the colors, we notice that there is a concentric scheme as k rises, showing that ensembles of lower values of k have a higher range of accuracy and diversity. This suggests that randomly picking three or even more CNNs and including them in an ensemble does not guarantee to get larger performance than using one of the CNNs. To further prove the importance of having the diversity in the objective function F we also performed an ablation experiment where its contribution is neglected: this means that the composition of the best ensemble is determined only by maximizing the accuracy. The results, not shown here for space reason, reveal that for any k the performance of an ensemble built using our F is better than those attained maximizing only the accuracy. Let us now focus on the right panel, which zooms the plot close to the top right corner, showing level curves that correspond to points where F is constant. As already reported, the two-objective optimization problem is solved by an ensemble of 3 classifiers. We also notice that sub-optimal performances are attained not by other ensembles with other three classifiers but, rather, by ensembles with more classifiers. Nevertheless, the positions of the colored circles confirm that maximizing one of the two scores is not enough to get the best performance. Indeed we note that the diversity drops as the accuracy increases. Furthermore, as the number of classifiers in the mixture increases, the multi-objective function F drops, and in some cases also the diversity and the accuracy drop. This empirically suggests that ensuring diversity while keeping large accuracy becomes more difficult as the number of classifiers in the ensemble increases.
5 Conclusions
In this manuscript we presented an approach to build an optimized ensemble combining several CNNs via a late fusion approach. The goal is to obtain a classifier robust to CXR scans of multiple pulmonary diseases coming from multiple data sources, as it happens in the real world. In an effort to deploy the solution in practice, the results on the one side show that our approach is able to generalize to unseen data, overcoming the limits of single classifiers. On the other side, the rankings shown in Tables 3 and 4 reveal that the best ensemble in CV is also the best in the external validation, an observation that does not hold in the case of single CNNs, confirming the robustness of the method. Future works are directed towards the external validation on other public, as well as to extend the number of classes.
References
Worldmeters coronavirus. https://www.worldometers.info/coronavirus/. Accessed 01 Feb 2022
Aljondi, R., et al.: Diagnostic value of imaging modalities for COVID-19: scoping review. J. Med. Internet Res. 22(8), e19673 (2020)
Basu, S., et al.: Deep learning for screening COVID-19 using chest x-ray images. In: 2020 IEEE Symposium Series on Computational Intelligence (SSCI), pp. 2521–2527. IEEE (2020)
Brown, G., et al.: Diversity creation methods: a survey and categorisation. Inf. Fusion 6(1), 5–20 (2005)
Brunese, L., et al.: Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from x-rays. Comput. Methods Programs Biomed. 196, 105608 (2020)
Cavalcanti, G.D., et al.: Combining diversity measures for ensemble pruning. Pattern Recogn. Lett. 74, 38–45 (2016)
Cipollari, S., et al.: Convolutional neural networks for automated classification of prostate multiparametric magnetic resonance imaging based on image quality. J. Magn. Reson. Imaging 55(2), 480–490 (2022)
Cohen, J.P., et al.: COVID-19 image data collection: prospective predictions are the future. arXiv preprint arXiv:2006.11988 (2020)
Das, D., Santosh, K.C., Pal, U.: Truncated inception net: COVID-19 outbreak screening using chest x-rays. Phys. Eng. Sci. Med. 43(3), 915–925 (2020). https://doi.org/10.1007/s13246-020-00888-x
Deng, J., et al.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. IEEE (2009)
Futoma, J., et al.: The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Health 2(9), e489–e492 (2020)
Guarrasi, V., et al.: A multi-expert system to detect COVID-19 cases in x-ray images. In: 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), pp. 395–400. IEEE (2021)
Guarrasi, V., et al.: Pareto optimization of deep networks for COVID-19 diagnosis from chest x-rays. Pattern Recogn. 121, 108242 (2022)
Hansen, L.K., et al.: Neural network ensembles. IEEE Trans. Pattern Anal. Mach. Intell. 12(10), 993–1001 (1990)
He, K., et al.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Huang, G., et al.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Iandola, F.N., et al.: SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and \(<\)0.5MB model size. arXiv preprint arXiv:1602.07360 (2016)
Jaeger, S., et al.: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 4(6), 475 (2014)
Krizhevsky, A.: One weird trick for parallelizing convolutional neural networks. arXiv preprint arXiv:1404.5997 (2014)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Pereira, R.M., et al.: COVID-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 194, 105532 (2020)
Pouyanfar, S., et al.: A survey on deep learning: algorithms, techniques, and applications. ACM Comput. Surv. (CSUR) 51(5), 1–36 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sandler, M., et al.: MobileNetV2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018)
Shiraishi, J., et al.: Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 174(1), 71–74 (2000)
Simonyan, K., et al.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Soda, P., et al.: AIforCOVID: predicting the clinical outcomes in patients with COVID-19 applying AI to chest-x-rays. An Italian multicentre study. Med. Image Anal. 74, 102216 (2021)
Szegedy, C., et al.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
Wang, L., et al.: Covid-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 10(1), 1–12 (2020)
Wang, X., et al.: ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2097–2106 (2017)
Wynants, L., et al.: Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. Brit. Med. J. 369 (2020)
Xie, S., et al.: Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1492–1500 (2017)
Zagoruyko, S., et al.: Wide residual networks. arXiv preprint arXiv:1605.07146 (2016)
Acknowledgements
This work is partially funded by: POR CAMPANIA FESR 2014–2020, AP1-OS1.3 project “Protocolli TC del torace a bassissima dose e tecniche di intelligenza artificiale per la diagnosi precoce e quantificazione della malattia da COVID-19” CUP D54I20001410002; EU project “University-Industrial Educational Centre in Advanced Biomedical and Medical Informatics (CeBMI) No. 612462-EPP-1-2019-1-SK-EPPKA2-KA”; “AI against COVID-19 Competition”, organized by IEEE SIGHT Montreal, Vision and Image Processing Research Group of the University of Waterloo, and DarwinAI Corp., and sponsored by Microsoft.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Guarrasi, V., Soda, P. (2022). Optimized Fusion of CNNs to Diagnose Pulmonary Diseases on Chest X-Rays. In: Sclaroff, S., Distante, C., Leo, M., Farinella, G.M., Tombari, F. (eds) Image Analysis and Processing – ICIAP 2022. ICIAP 2022. Lecture Notes in Computer Science, vol 13231. Springer, Cham. https://doi.org/10.1007/978-3-031-06427-2_17
Download citation
DOI: https://doi.org/10.1007/978-3-031-06427-2_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-06426-5
Online ISBN: 978-3-031-06427-2
eBook Packages: Computer ScienceComputer Science (R0)