Abstract
Nowadays, liquid biopsy represents one of the most promising techniques for early diagnosis, monitoring, and therapy screening of cancer. This novel methodology includes, among other techniques, the isolation, capture, and analysis of circulating tumor cells (CTCs). Nonetheless, the identification of CTC from whole blood is challenging due to their extremely low concentration (1–100 per ml of whole blood), and traditional methods result insufficient in terms of purity, recovery, throughput and/or viability of the processed sample. In this context, the development of microfluidic devices for detecting and isolating CTCs offers a wide range of new opportunities due to their excellent properties for cell manipulation and the advantages to integrate and bring different laboratory processes into the microscale improving the sensitivity, portability, reducing cost and time. This chapter explores current and recent microfluidic approaches that have been developed for the analysis and detection of CTCs, which involve cell capture methods based on affinity binding and label-free methods and detection based on electrical, chemical, and optical sensors. All the exposed technologies seek to overcome the limitations of commercial systems for the analysis and isolation of CTCs, as well as to provide extended analysis that will allow the development of novel and more efficient diagnostic tools.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
One of the big challenges in cancer is achieving an early-stage diagnosis. Currently tumors are not found until symptoms appear or by chance when the patient undergoes a medical test which in both situations can be too late for the patient. Many analysts now argue that the development of efficient methods for screening, early diagnosis, and monitoring, are promising technologies to achieve a high-efficiency therapy and to reduce cancer mortality [1]. As has been reported, several biological body fluids like blood, urine, and saliva contain biomarkers, which could be DNA, RNA, proteins, or whole cells [1, 2]. In recent years, interest has arisen in the development of new technologies to detect those biomarkers from body fluids, such as liquid biopsies [1]. The emergence of liquid biopsies has been useful for the diagnosis of physiological conditions, inflammatory processes and specially represents a good alternative tool for non-invasive analysis of tumor-derived materials.
Currently, tissue biopsies represent the gold standard for tumor profiling. Nonetheless, this method displays many limitations that include the invasiveness, risk and depending on some anatomical locations is not easy (or even impossible) to obtain [3]. In fact, it provides a limited vision of the tumor profile, considering that tumors are heterogeneous composed of different subpopulations of cells, which display a variability of genetic and epigenetic changes. In addition to differences between primary and metastatic lesions [3]. Therefore, the tissue biopsies fail to represent the overall tumor profile, capture the alterations in different parts, and monitor the diseases progression [4].
On the other hand, liquid biopsies could be done through routine blood extraction and analysis, a procedure that is much easier and less invasive than a tumor biopsy. In this context, liquid biopsies are a cheaper, faster, non-invasive alternative to conventional biopsies, that can be used for personalized cancer therapy [1, 3]. In a broad sense, liquid biopsy is based on the isolation of biomarkers from blood that can be used for cancer diagnostic and monitoring. This definition englobes circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosomes [1, 2, 5].
1.1 What Are the Circulating Tumor Cells?
Since 1869, Ashworth discovered the tumor cells in the peripherical blood and proposed the concept of a CTC [6]. These CTCs are cancer cells, which leave the primary tumor and enter the bloodstream initiating a process called metastasis that is responsible for almost 90% of cancer deaths [7]. From the biological point of view, these cells do not bind to the extracellular matrix (ECM) and survive in the bloodstream because of their resistance to apoptotic process known as “anoikis” [8], as well as factors associated with epithelial and mesenchymal plasticity or stem cell-like properties.
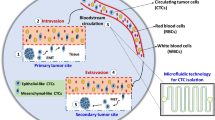
Metastasis is a biological complex process, which involves cell migration, invasion, arrest at secondary and primary parts, intrusion of tumor cells in bloodstream, dissemination, extravasation at distant parts, colonization and finally, the formation of a metastatic secondary tumor clinically detectable (Fig. 16.1) [7, 10]. Recent studies indicate the high heterogeneity of CTC population, including CTCs clusters, individual CTCs, epithelial CTCs, hybrid epithelial-mesenchymal CTCs, mesenchymal CTCs and stem-like CTCs [11]. In fact, it has been found that CTCs clusters show distinct features regarding to individual CTCs, such as phenotype, sign of gene expression and nature [12, 13]. Moreover, other studies have revealed that CTCs clusters may have 100 times more metastatic potential than single CTCs [14]. The presence of CTCs in blood has been recognized in many types of cancer, such as breast cancer, colon cancer, lung, prostate among others [14]. In fact, previous clinical studies in patients with breast cancer have correlated the presence of CTCs with an increase in tumor burden, aggressiveness, as well as decreased time to relapse [10].
Liquid biopsy and CTCs origin and progression under therapeutic application. Circulating proteins (CPs), circulating tumor DNA (ctDNA), circulating tumor cells (CTCs) and extracellular vesicles (EVs) enter to the circulation and can be used to detect minimal tumor generation and monitor tumor heterogeneity. CTCs will be generated by the primary tumor and cooperate with TEPs (tumor-educated blood platelets) to survive and enter in circulation as single CTCs or CTCs clusters. Furthermore, EVs represent pre-metastatic scavengers that resist to immune damage and allow metastasis in secondary areas. After target therapy is applied, the drug-resistant cancer cells will proliferate by adaptive evolution. Thus, liquid biopsy allows to predict metastasis or relapse. Natural cancer development is represented by the red arrows, meanwhile the development of tumors after therapy is shown by the black arrows. Licensed under CC-BY 4.0 International License. The Final, Published Version of This Article Is Available at [9]
In these circumstances, isolation and detection of CTCs that are circulating into the bloodstream could be used for early cancer metastasis detection as well as for monitoring tumor’s responsiveness to radio/chemotherapy and developing personalized patient treatments. Some of the diagnostic possibilities that CTCs detection would offer can be summarized as follows [15].
-
The presence of CTCs in blood is highly associated with metastatic risk but also indicates primary tumor existence. As it is known that CTCs are present in blood from very early stages of the disease and prior to the appearance of symptoms, a device that allows the detection of CTCs could be used for both determining metastatic risk and early cancer detection.
-
It is also known that the number of CTCs present in the blood sample is related to the stage of the patient’s disease. Thus, another advantage could be to monitor tumor’s responsiveness to therapy. This would be done by running routine blood analysis and comparing the number of CTCs prior and after the treatment.
-
Another important issue is tumor heterogeneity, both within a tumor and between the primary tumor and its metastases, which cannot be captured by a simple tissue biopsy. This heterogeneity accounts for the genotypic differences between different regions of the tumor or between the primary and the secondary tumors. It is known that almost all tumors treated with any therapy acquire resistance because of tumor heterogeneity, clonal evolution, and selection [16]. Therefore, CTCs isolation and analysis would enable to understand better the phenomena of metastatic drug resistance.
-
Tumors are very prone to suffer mutations in their genome, so their molecular profile changes frequently, and so does the effective treatment required. Thus, liquid biopsy would allow monitoring the molecular profile of the tumor just by taking periodic blood samples from the patient. Therefore, according to the results obtained the clinicians can find out if the treatment is correct or if it must be adjusted due to mutations.
It is therefore clear that the development of a platform allowing CTCs isolation and detection from blood samples would be a major improvement in the study and treatment of cancer. Nevertheless, the identification of CTC from whole blood is challenging, as the number of CTCs in 1 mL of blood from a cancer patient is only 1–100, in comparison with 109 hematologic cells [17]. Hence, the traditional methods to isolate cells, like flow cytometry, density gradient centrifugation, and immunocapture by magnetic beads do not have enough sensitivity to detect rare cells like CTCs [18].
Despite the high number of scientific publications related to CTCs detection, there is only one CTC test approved by the FDA, the CellSearch® system by Menarini Silicon Biosystems [19]. This system uses magnetic particles coated with antibodies that bind to the protein EpCAM (epithelial cell adhesion molecule) for quantifying the CTCs in metastatic breast, prostate, and colon [20]. In the last few decades, EpCAM was considered as universal tumor biomarker for epithelial-derived cancer types [21]. Nevertheless, several studies indicate that some CTCs can be EpCAM-negative [22] such as those which had an epithelial-to-mesenchymal transition and mesenchymal origin. Moreover, CTCs isolated from patients display a wide range of EpCAM expression, where a part of these cells are EpCAM-negative [23, 24] for instance in some advanced lung cancer [23, 25, 26]. This represents one of the main drawbacks of this platform as well as, it requires expensive equipment and allows neither 100% purity nor isolation of viable cells for further culture and studies.
To overcome these limitations, several microfluidic platforms have also been developed. Indeed, Lab-on-a-Chip (LOC) technologies have been exploited in the recent decades for several biomedical applications such as diagnostics, biochemical assays, and drug discovery among others. Microfluidics and LOC technologies are directly correlated terms since LOC technologies aim to integrate and bring different laboratory processes into the microscale to exploit the advantages that working at this length scale provides. Therefore, these devices are frequently composed of microfluidic elements like microchannels, micropumps, microvalves, etc. to enable processing small (micro scaled) amounts of liquid.
Nowadays, most of the analytical and diagnostic assays are done with benchtop equipment in hospitals and/or centralized laboratories, which are either operated by trained personnel or composed of chains of automated pipetting robots with the associated increment of power consumption and space demand. Microfluidic LOC platforms arise as an alternative to the present model of diagnostics since these offer a wide range of new opportunities that can be summarized as: [27, 28].
-
Portability due to its reduced dimensions.
-
Higher sensitivity.
-
Faster results obtaining.
-
Reduced laboratory space.
-
Lower cost per test due to less quantity of reagent required.
In addition, apart from those advantages stated above, working at the microscale allows taking advantage of the unique phenomena taking place at such scale:
-
Gravitational force loses importance.
-
Well-defined laminar flow.
-
Controllable diffusion.
-
High degree of parallelization.
Therefore, all the characteristics mentioned before, together with the fact that microfluidic LOC devices have the perfect size for cell manipulation and that they have the possibility to play with well-defined particle forces related to inertial effects [29, 30], make them ideal for building CTCs isolation devices that can be used for liquid biopsies.
2 Microfluidic-Based Isolation of CTCs
The application of microfluidic devices to the detection and isolation of circulating tumor cells is quite recent, the firsts scientific works arising towards 2005. The first scientist to create a considerable impact in the field was Prof. Toner and his group at Harvard Medical School with the publishing in 2007 of a research paper [31] in which they obtained promising results concerning the isolation of CTCs on a microchip capable of efficient and selective separation of viable CTCs from peripheral whole blood samples, mediated by the interaction of target CTCs with antibody (EpCAM)-coated microposts under precisely controlled laminar flow conditions, and without requisite pre-labeling or processing of samples. At that moment, this was revolutionary and encouraged a lot of scientists, all with the same purpose of developing a lab-on-chip device to detect and isolate CTCs. If we now look back on that paper, it has become the most cited paper regarding CTCs isolation using microfluidics with almost 3000 citations up to date.
Since then, many other microfluidic devices have been proposed for the separation of CTC from blood. They can be based in microfiltration, deterministic lateral displacement, centrifugation, inertial focusing, affinity-based methods, among others [29, 32,33,34,35]. They separate according to cell properties like size, density, shape, deformability, or biomarkers expression [27, 36, 37]. The parameters used to evaluate the performance of this technology include purity, throughput, recovery rate and cell viability. The purity is associated to the blood cells depletion, which indicates the number of CTCs compared to blood cells. Meanwhile, recovery rate reflects the ratio of targeted cells collected in the CTCs outlet to the total number of CTCs, throughput refers to the amount of blood sample (usually in milliliters) that can be processed in the device per unit of time and thus the number of cells that can be captured and, finally, cell viability indicates if the cells recovered are alive [29, 36]. Considering the low concentration of CTCs on the blood, it is important to guarantee that all the CTCs are being recovered by the device and that they are viable. Thus, the cells recovered could be used in personalized drug screening [38, 39]. Overall, with the emergence of personalized medicine the latest developed devices for CTCs separation are focused on the detection of CTCs via label-independent methods, low pre-treatment, large volume of sample processing in a short time, high purity, and possibility to propagate the isolated CTCs [40, 41].
2.1 Affinity-Based Methods
The affinity-based microfluidic devices are based on the use of specific antibodies attached on the channels walls to capture target cells. These devices have been developed as positive enrichment techniques when the antibodies are used to directly capture CTCs and negative enrichment techniques if antibodies are used to target blood cells. The first positive enrichment platforms were devices composed of series of posts functionalized with EpCAM antibodies as explained before [31]. Nevertheless, the cells follow the fluid streamlines of the laminar flow, which limit the interaction between the CTCs and the antibody modified surface. To overcome this, herringbone-like structures were proposed [35, 37, 42,43,44] that promotes fluid mixing in the channel, thus increasing particle-surface interaction. Figure 16.2 presents a representation of particles trajectories in a rectangular channel and in a channel with a herringbone structure. In Fig. 16.2a, the particles follow the fluid streamlines in a rectangular straight channel, while in Fig. 16.2b, the herringbone structure promotes a mixing, so the particles move transversally on the channel with more probability to reach the functionalized channel walls.
Graphic representation of the particles trajectories in (a) traditional rectangular channel and in a (b) herringbone device. Figure from [42] Copyright 2010 National Academy of Sciences
Different herringbone chips were studied for positive enrichment, including the surface functionalization with different antibodies (Fig. 16.4b) [46], specific for some cell lines. Some of them have two opposed surfaces with herringbone structures to increase the mixing [35, 37, 42, 44]. Moreover, to decrease the non-specific cell adherence, Wang et al. proposed a wavy herringbone structure [35]. However, one of the major disadvantages of this system is associated with the difficulty of eluding CTCs after being captured, this process could have an impact on their viability [34].
Recently, new methods have been developed for improving positive enrichment methods such as hydrogel microparticles (MP) functionalized with EpCAM antibodies, which have the advantages of water-like reactivity, biologically compatible materials, and synergy with various analysis platforms (Fig. 16.3) [35]. In this method the hydrogel particles are synthesized using degassed mold lithography (DML), as a result, the porosity and functionality of the MPs increase achieving an effective conjugation with antibodies [48]. In addition, the MPs functionalization is based on carbodiimide cross-link chemistry conjugated to antibodies through carboxyl groups. Also, NHS and EDC chemistry was used to covalently attach Neutravidin protein to the carboxyl groups. Finally, biotinylated anti-EpCAM antibodies were used for avidin–biotin reaction [47]. Moreover, other methods based on Immunofunctionalized hydrogels have been successfully developed for capturing CTCs (Fig. 16.4c).
Positive enrichment methods based on hydrogel microparticles (MP) and EpCAM antibodies. (a) Hydrogel microparticles synthesized by degassed mold Lithography (DML). Thus, UV-induced radical polymerization with prepolymers (i) acrylic acid (ii), polyethylene glycol diacrylate (PEGDA) and (iii) polyethylene glycol (PEG), hydrogel microparticles containing carboxyl groups are synthesized. (b) Image of polymerized hydrogel microparticles (MP). Scale bar 200μm. (c) Interaction between avidin protein and biotin allows the anti-EpCAM–biotin conjugation. This principle is based on the reaction between carboxyl groups in the particles and primary amines in NeutraAvidin with the help of N-hydroxysuccinimide (NHS) and N-(3-dimethylaminopropyl)-N′-ethycarbodiimide hydrochloride (EDC). (d) Circulating tumor cells (CTCs) captured by functionalized hydrogel microparticles with EpCAM antibody. Licensed under CC-BY 4.0 International License. The final, published version of this article is available at [47]
(a) NanoVelcro CTC Chip is composed of a patterned silicon nanowire (SiNW) substrate and herringbone features which promote the helical flow in the microchannel improving the interaction between CTCs and anti-EpCAM coated SiNW substrate. Licensed under Creative Commons Attribution 3.0 License (CC BY 3.0). The final, published version of this article is available at [45]. (b) Herringbone microfluidic device composed of 16 parallel microchannels for cells capture (left image). The center image is a Schematic representation for capture and analysis of plasma cells in microfluidic device. In this technology, the microchannels are coated with biotinylated CD138 antibodies, which capture the cells from the flow. Finally, capture cells are staining with anti-κ immunoglobulin for their identification. Fluorescence image shows the cells captured from a clinical sample. Red represents the CD138 and green anti-κ immunoglobulin (Right image). Licensed under Creative Commons Attribution 3.0 License (CC BY 3.0). The final, published version of this article is available at [46]. (c) Schematic representation of fabrication and degradation of patterned photodegradable hydrogel films. Thus, a PDMS master is covered with hydrogel and irradiated at 405 nm light. Subsequently, a microfluidic herringbone channel is bonded on the top of the hydrogel film. The hydrogel can be degraded under flow condition using irradiation of 365 nm light. Finally, the CTCs can be collected for further processing [38]. Copyright (2018), Elsevier
Other studies were focused on increasing the capture efficiency of CTCs, such as the GEDI chip [49]. In this case, the optimization was based on the displacement, size, and shape of microposts, as well as the use of a specific prostate antigen (PSMA). On the other hand, among the commercial methods available, CEE™ microfluidic chip is characterized by randomly located microposts functionalized with streptavidin, which allow to capture targeted CTCs with biotinylated antibodies [50]. Meanwhile, The NanoVelcro CTC chip is composed of silicon (Fig. 16.4a) nanowire substrates (SiNWs) functionalized with EpCAM for CTCs capture [35, 51].
All the previous methods have a limitation, though. As mentioned before, not all CTC express a specific membrane marker. Potentially important CTCs subpopulations like mesenchymal and stem cell like CTCs would be missed [52]. Besides, the CTCs would need to be eluted after being captured, and this process might have an impact on the CTC viability [14, 24]. So that, affinity-based techniques based on negative enrichment were proposed.
Negative enrichment of CTCs is an affinity-based method that has the purpose of removing hematopoietic cells by targeting specific antigens that are not expressed by the CTCs as, for instance, CD45 (leukocyte common antigen) [53]. For example, CTC-iChip was developed by Ozkumur et al., which eliminate the blood cells based on physical properties and CD45/CD15 expression [54]. Hyun et al., proposed a herringbone device for negative enrichment by targeting of leukocytes with the surface immobilized with CD45 after the blood lysis or centrifugation for eliminating the erythrocytes [17]. Moreover, other technology known as the geometrically activated surface interaction (GASI) chip was fabricated, which was similar to the Herringbone chip but with microvortexing features aimed at increasing the number of captured leukocytes [54]. On the other hand, a novel work was reported by Fatih Sarioglu and colleagues, they developed a method for negative enrichment of CTCs using whole blood. This method is composed by 3D microfluidic device which captures the leukocytes based on immuno-enhanced microfiltration, also allows the depletion of erythrocytes, and remains the CTCs in suspension [55]. Overall, negative enrichment allows the capture of CTCs with low or no expression of EpCAM and the CTCs can be collected intact and viable for subsequently clinical analysis [56]. However, as the population of leukocytes in the sample is usually very high, even large depletion rates are not always ensuring good results concerning the purity of the sample. In addition, negative enrichment is used to capture the leukocytes after the blood sample has been already processed to eliminate the erythrocytes, so that, this methodology has the potential to be used to complement other techniques by enriching their results in terms of purity.
Recent evidence suggests, however, that the entire CD45+ population should not be considered as a discriminant for isolating CTCs from a population of leukocytes, due to some CTCs displaying expression levels of CD45 and some leukocytes subpopulation such as non-lymphocytes, which show low expression of CD45. To overcome the limitations of employing antibodies, aptamers have emerged as a potential alternative for the isolation of CTCs. These are single-stranded oligonucleotides such as RNA, DNA, or peptides that bind to targets such as proteins with a high specificity and sensitivity [50]. Moreover, it was also discovered that an in vitro process called “systematic evolution of ligands by exponential enrichment” (SELEX) allows the synthesis and selection of aptamers in a straightforward manner. In fact, the potential use of aptamers for isolating CTCs has been successfully evaluated with samples spiked with different cancer cell lines and in patients’ samples [57]. Based on the previous findings, the key advantage of using aptamers for the isolation of CTCs is that they can be prepared in different panels targeting several proteins expressed on available cancer cells, for which it is not necessary to know the precise targets [58, 59].
Finally, both negative and positive enrichment methods can be combined with magnetic-activated cell sorting. In this case, magnetic microbeads are coated with the antibodies and a magnetic field is used to attract the target cells bounded to the microbeads [29, 60]. However, these devices have limited throughput due to the time needed for the force to act on the particles [30].
2.2 Label-Free Methods
The devices discussed within the previous section have, as a main drawback, the fact that they rely on the expression of a certain biomarker present on the cell membrane. However, this condition is not always satisfied, and this might lead to a loss of CTCs and affect the performance of the device in terms of the recovery rate. As an alternative, several microfluidic devices have been proposed for the enrichment of CTCs from blood based exclusively on physical properties of the CTCs, such as size, density, mechanical plasticity, and dielectric properties [61]. Among these methods, the ones that have been more exploited are size-based methods [62, 63].
2.2.1 Size-Based Methods
This methodology considers the difference on size between CTCs, erythrocytes, and leukocytes. As was previously indicated, the diameter of CTCs usually ranges from 15μm to 20μm [18], erythrocytes are between 6μm and 8μm, meanwhile, the leukocytes go from 6μm to 20μm, where neutrophils represent between 40% and 75% with diameters that go from 10μm to 12μm [64]. Among size-based methods, one can basically find filters, in which the sample flow through an array of microscale constrictions and inertial microfluidic sorting devices, in which the cells are separated due to size-dependent inertial fluid forces.
2.2.1.1 Filtering Microfluidic Sorting Devices
Normally, these kinds of devices capture the CTCs as they are bigger in size, allowing to pass the other cellular components. Depending on the filtering principle, they can use either chromatography columns, pillars, or pores [65].
Chromatography is a classical technique for separating components of a mixture based on their ability to pass through a column with a porous material. This method is used for separating molecules in a label-free manner [65, 66]. In 2011, Hongshen Ma et al. used this principle, but in an opposite behavior for CTCs separation. Thus, dynamic microstructures have the advantages of filtration and hydrodynamic manipulation, wherewith is possible to discriminate cells based in size and deformability, meanwhile the cells are in a continuous flow [65].
On the other hand, an outstanding device designed in the low Reynolds number regime is known as deterministic lateral displacement (DLD) (Fig. 16.5a and b). In this technology, the smaller cells follow the streamlines and pass through a series of posts without net lateral displacement. Meanwhile, the bigger cells change to a different streamline when enter in contact with the pillars and are laterally moved from the original streamline [69]. The evidence from previous studies has shown the potential of this technology in separating cancer cells from blood with a performance of 80% [67]. In general, devices based on pillars use an array of microposts that form constrictions. Mohamed et al. developed a microfluidic device composed by pillars of four successively decreasing clearances from 20μm to 5μm (Fig. 16.5c) [65, 68].
(a) Microfluidic chip composed of a deterministic lateral displacement (DLD) channel with mirrored triangular micropost array. Thus, bigger cells like cancer cells and some leukocytes were concentrated in the middle of the channel, meanwhile the smaller cell such as the erythrocytes and most of the leukocytes follow the streamlines flow direction. Finally, the capture channel is a PDMS layer with herringbone structures modified with EpCAM. These structures promoting the capture of CTCs. Copyright (2013), Elsevier [67]. (b) Fluorescent images of cancer cells captured on the chip surface. Cells were stained with Vybrant® DyeCycle™ Green. Copyright (2013), Elsevier [67]. (c) Microfluidic devices for cancer isolation based on cell size and deformability. The images show blood samples spiked with MDA231 cells, where all the blood cells flow freely through the device, but the MDA231 cells were retained between the gaps. Copyright (2009), Elsevier [68]
On the other hand, technology based on pores consist of a membrane with holes which demonstrated a recovery rate higher than 85%. In fact, there are some commercial devices based on this principle, such as Rarecells®, Screencell®, and Clearcells® [32]. The principle of this technology is based on the force applied to the cells, which depends on the flow rate and is related with the deformability of the cells. Thus, the flow rates and the cross-section of the membrane constrictions are the key parameters that define the efficiency of this kind of devices. Nonetheless, the major disadvantage of this method is the clogging, when it is used with whole blood sample and as a result, the flow rate changes. The flow rate modification triggers a low throughput and change in the limit separation size. Moreover, these microfluidic technologies do not allow recover the CTCs from the membrane for further clinical analysis [65].
2.2.1.1.1 Inertial Microfluidic Devices
Among the systems proposed in the literature, microfluidic devices based on the inertial focusing are promising, which could overcome the limitations of other methods that use pillars, pores, or labeling approaches with the advantage of achieving a high throughput [67]. Several studies have revealed that in a Reynolds number between stokes flow (Re < <1) and inviscid flow, namely, in a range from 1 to approximately 100, forces from inertial effects appear such as drag forces from Dean flows, shear gradient lift forces, and wall effect lift forces, which are balanced for achieving the size-based separation [30, 70].
Overall, the system can be described by the channel Reynolds number (Re) and the particles Reynolds number (Rep) by the following equations, where ρf and 𝜇 are the fluid density and dynamic viscosity, Um is the maximum fluid velocity in the channel, Dh represents the hydraulic diameter and a the particle diameter. In a rectangular channel Dh = 2WH/(W + H), in which W is the channel width and H the channel height.
In 2007, Di Carlo et al. published a pioneer work, in which the inertial focusing was used to control the particles position in microfluidics devices with curved channels according to the particles size. They demonstrated that particles did not follow the fluid streamlines but migrated across them as the inertial forces became significant. The particle’s position in the microchannel was related to the particle’s diameter [30].
Traditionally, the inertial lift forces and drag forces are orthogonally on a particle [71], but in a curved channel a secondary cross-sectional flow start to appear (Dean flows). Then, the Dean flow triggers the particles experience a drag force on the same axis as the shear gradient and the wall effect lift forces [30]. Hence, the balance between these forces cause different equilibrium positions of particles depending on their size. Thus, small particles follow the Dean flow while big particles are under a stronger lift force.
The Dean flow can be described by the Dean number through the Eq. (16.3), in which r is the radius of channel curvature [30, 72]. Moreover, inertial lift forces and drag forces can be calculated as indicate the Eqs. (16.4 and 16.5).
In Eq. (16.4) Re represents the channel Reynolds number, μ is the fluid dynamic viscosity, ρ is the fluid density, UF is the average velocity of the fluid and ap is the particles diameter. Meanwhile, fL is the lift coefficient that corresponds to a complex function of the Re and the cross-sectional positions of particles xp.
As previously mentioned, Dean Drag Force can be expressed as Eq. (16.5). Where ρ is the fluid density, UF is the average velocity of the fluid and ap is the particles diameter. Meanwhile, Dh is the cross-sectional hydraulic diameter and R is the radius of the microchannel [71, 72].
Up to now, several spiral devices have been reported for isolating CTCs from the blood applying inertial forces [36]. As explained before, the separation is based on the difference of size. Thus, the lift force is important for bigger cells like CTCs, meanwhile is not significant for smaller cells such as erythrocytes and small leukocytes. The blood cells mainly follow the Dean flow due to the Drag force, and do not focus on certain positions. Finally, by carefully designing the device geometry, the blood cells will leave the system using one device outlet, according to the Dean cycle and the CTCs are focused on another outlet [51], thus obtaining separation.
Some devices developed by previous researchers included a sheath flow inlet to initially confine the blood cells in one wall, so all of them can follow the Dean cycle [29, 36, 51, 55, 73]. Sun et al. proposed a double spiral device with 6 loops and alternation of the flow direction due to an S-turn. The device has 20 a low aspect ratio compared to other devices in the literature (H/W = 0.167) and present one inlet and three outlets [18]. Moreover, spiral design with a trapezoidal cross-section was also proposed to generate stronger Dean flow than in a traditional rectangular cross-section; this Device is composed of 3 loops and a recovery rate of 80% was obtained [29, 36]. Some years ago, devices with multiplexed setup were proposed to increase the throughput [29, 73], which are composed of 4 spirals in parallel that were stacked forming a multiplexed device with 40 spirals, reaching a throughput of ~500 mL/min. The system was tested for the separation of Chinese hamster ovary cells (CHO) and yeast [29]. Also, cascade microfluidic devices were proposed including the integration of more than one spiral [70, 74]. These technologies demonstrated a recovery rate between 80% and 90% and a throughput up to 2 mL/min [70]. Figure 16.6 represents different spiral microfluidic technologies.
(a) Multiplexed spiral device. The device is composed by four inputs in the middle of the sorting unit. The figure shows the CTCs outlet and waste outlets are indicated. Licensed under CC BY 4.0 International License. The final, published version of this article is available at [75] (b) Spiral microfluidic device with trapezoidal cross-section microchannels. The device has two modes: filtration and fractionation. In filtration mode, the particles suspended are trapped and focused near the outer wall under strong vortices. Meanwhile, in the fractionation mode, smaller particles (red) are trapped in Dean vortices and keep near the outer wall, but the bigger particles (blue) are focused near the inner wall. Thus, the particle separation by size is achieved. Basically, changing from one mode to other depends on the magnitude of the hydrodynamic forces inside the microchannels. Licensed under CC BY 4.0 International License. The final, published version of this article is available at [76] (c) Representation of triple-microchannel spiral microfluidic chip composed by slits for isolation of CTCs. The three microchannels are interconnected by arrays of slits, which allow that the cells pass from one main microchannel to other through the flow direction. Licensed under CC BY 4.0 International License. The final, published version of this article is available at [77]
2.2.2 Dielectrophoresis
The dielectrophoresis (DEP) represents a label-free, precise, and low-cost diagnostic method [78]. This method is based on the dielectric cell properties, due to the cells are electrically neutral but they can be polarized, which depends on their polarity and conductivity. Thus, when the cells are subjected to a non-uniform electric field, a dielectrophoretic force (FDEP) starts to appear, whose magnitude and direction depend on the dielectric properties of the cells, the medium, their size and shape, as well as the frequency of the electric field [78]. It has been reported two types of FDEP, a positive dielectrophoresis (PDEP) which appears when the cell polarization is bigger than the medium. Thus, the cells move towards the strong electric field region. On the other hand, there is a negative dielectrophoresis (nDEP) that appears if the polarization of the cells is smaller than the medium and therefore the cells move in the opposite direction [79]. This method allows that cells can be differentiated depending on the polarizability [80].
Currently, microfluidic devices with electrodes embedded that produce the AC electric field have been developed (Fig. 16.7a and b). The benchtop device in the literature presents a recovery rate between 70% and 90% [83]. Alazzam et al. [84] demonstrated a yield of 95% by applying this methodology. The main drawback is the low throughput. Therefore, this methodology is used as a complement to the other ones, as the sized based methods. This has been explored by Moon et al. [85] who combines hydrodynamic focusing with dielectrophoresis and obtained 162-fold enrichment of the MCF-7 cells—CTCs cells model over RBCs at a 7.6 mL/h flow rate [86].
(a) Schematic representation of ApoStream flow chamber. This device applies an AC electric field to the sample and is composed of electroplated copper and gold electrodes on the bottom part of the flow chamber. The sample was introduced into the flow chamber, and the cancer cells are collected in the other rectangular port. The principle of separation is based on the DEP forces, which pull the cancer cells through the bottom chamber and repel the other cells [80]. Reprinted from, with the permission of AIP Publishing. (b) Microfluidic device composed of electrodes, which allow the isolation of cancer cells due to the DEP effect and the difference in cell size. Thus the target cells were trapped onto the sensing electrodes. Finally, the impedance measurements allow to identify the presence of cancer cells. Copyright (2018), Elsevier [81]. (c) Microfluidic pH Sensor for detection of cancer cells based on the measurements between a silver–silver chloride and zinc oxide electrodes for CTCs recognition in blood. The device detects the cancer cells based on changes of pH in the extracellular environment [82]. Copyright © 2017, American Chemical Society
In contrast with traditional technologies reported, microfluidics-integrated separation method combined with ODEP (Optically Induced Dielectrophoresis) represent a novel strategy for complex cell manipulation, which involve suspension, transportation, collection, and purification of cancer cell. This method was validated with 8 mL of blood samples with H209 cancer cell clusters, as result an excellent recovery rate up to 91.5% ± 5.6% was achieved [56].
3 Microfluidic-Based Detection of CTCs
In general, the CTC separation methods described so far have in common the absence of integrated detection of the isolated tumor cells into the microdevice. Most of the systems, to elucidate the presence of the CTCs, use fluorescent-labeled antibodies specifically attached to the CTC, and the fluorescence label is detected with an external microscope. However, some authors have gone one step further and have combined microfluidic isolation techniques with integrated sensors for CTCs analysis in situ. We have summarized here some representative examples using either electrical, optical, or chemical sensors.
Field effect transistors (FET) are semiconductor components with three terminals (gate (G), source (S) and drain (D)) [87]. One of the major challenges of using FETs as biosensors is achieving the immobilization of affinity reagents such as antibodies or aptamers on the open gates [88]. Furthermore, Yi-Hong Chen and their colleagues developed a microfluidic device composed by CTC-specific aptamers functionalized on a FET surface and it is composed of a dual-layer with two inlets and 14 individual trapping chambers. The chip was tested with human colon cancer cell lines (HCT-8), as a CTC model and blood samples were spiked with HCT-8 cells. As result, the device was able to capture a maximum of 42 from a total of 1000 cancer cells [52].
Some years ago, Pulikkathodia and colleagues developed a high electron mobility transistor (HEMT), which is composed of a multiplexed sensor integrated into a microfluidic channel to detect colorectal cancer cells (HTC-8) [42]. Besides, the rise of impedance spectroscopy represents an excellent tool for label-free characterization of cells, which provides information about electrical cell parameters [89]. Some devices were developed based on this principle, such as the microfluidic device with circular electrodes designed by Nguyen and Jen. This Device was validated with A549 lung CTCs and blood, which was able to discern between the two cell populations based on their different resistivity [87]. It is worth also mentioning that in 2014, Hywel Morgan and colleagues used a single-cell microfluidic impedance cytometry to determine the dielectric features of MCF-7 cells and discriminate them from leukocytes. Moreover, it was demonstrated that the combination of the impedance cytometry with magnetic beads conjugated with antibodies enables to detect very low amount of MCF-7 (~100 in 1 ml blood) [90].
To date, some optically read-out methods and their integration into microfluidic devices have been designed, which include reflectance spectroscopy, surface plasmon resonance, and evanescent wave sensing, among others. In 2012, Kumeria and colleagues reported a microfluidic nanopore reflectometric interference spectroscopy (RIfS) device composed of microchannels and Anodic Aluminum Oxide (AAO) substrate modified with anti-EpCAM for detecting CTCs [91]. Thus, when the CTCs binding to the EpCAM antibody on AAO Surface, a wavelength shift in the Fabry-Perot interference fringe appears [91].
With respect to the chemical-based sensing devices, according to the study provided by Tzu-Keng Chiu et al., the metabolic performance of cancer cells as, for example, the production of lactic acid by CTCs represents a promising approach. This technology can count the cells by the formation of a micro-droplet and optical transduction of lactic acid for cell single detection. Unfortunately, the device cannot detect the presence of similar cells like leukocytes [92].
Finally, the metabolic change produced by the CTC that produces a reduction in the surrounding pH was also used as a method for differentiating cancer cell lines. The PH studies were performed by potentiometric methods with Ag/AgCl reference electrode and a ZnO working electrode. Nonetheless, the proposed device was not tested with blood samples (Fig. 16.7c) [82].
4 Conclusions
Microfluidic technologies have emerged as high-impact technologies in the field of circulating tumor cells isolation and detection, which represent a novel method for early cancer diagnosis, as well as an excellent tool for monitoring the disease evolution and treatment efficiency. There has been a great advance in the isolation and detection of circulating tumor cells using microfluidic devices within the past 15 years as they offer a high-throughput, compact, and economic alternative to the presently established methods. Many of the developed devices have indeed tried to overcome the limitations of commercial technologies such as CellSearch.
This chapter provided a compilation of literature related to the developed microfluidic technologies and the principles behind them for detection and separation of CTCs from others blood components. Also, it was defined the potential of introducing sensors, physical, chemical, and biological principles in the separation, detection, and analysis of CTCs through microfluidic devices.
With respect to the isolation of CTCs, many different methods have been proposed up to date, which we can classify into affinity-based methods, when a target molecule is used to capture the cells and label-free methods if the cells are sorted based on their physical characteristics without the need for labeling.
The affinity-based methods have the main drawback that they rely on a specific interaction between the microfluidic system walls and the cells membrane. However, this condition is not always satisfied and might lead to the miss of cancer cells and then affect the recovery rate. In addition, positive enrichment methods directly capture CTCs and so this result in not trustable cells for further analysis. Besides, affinity-based methods work better at low cell concentration, which is not the case for blood samples. Nevertheless, they have proved to be a good alternative to use as a complementary method, after a previous purification to remove high cell contamination.
On the other hand, with respect to label-free methods, the devices based on lateral displacement and microfiltration are more prone to clogging, so they cannot process high volume of samples. The devices based on dielectrophoretic forces present very good selectivity, but they have low throughput due to the weak electrical forces compared to the hydrodynamic drag. Inertial microfluidic devices offer a good alternative that solves some of the previous limitations as they can deal with high cell concentration without clogging, have proven an excellent cell recovery and offer very high throughput. However, high sample concentration leads to increased cell-cell interaction, which lower the purity and the inherent overlapping in size between leukocytes and CTCs also lowers purity.
As an overall conclusion, we can therefore state that microfluidic lab-on-a-chip devices have the potential to make a breakthrough for the isolation and detection of CTCs from blood samples. Nevertheless, after a careful reviewing of the available literature, there is no device with the necessary standard for parameters like purity, recovery rate, throughput, and cell viability as it is needed for its use in clinical settings. A convenient strategy would probably be to prioritize a high (or full) cell recovery and a good quality of the isolated cells (viability) to have a representative population for further studies. Indeed, CTCs are very scarce, and we cannot afford to lose any information about the tumor.
References
Martins I et al (2021, Feb) Liquid biopsies: applications for cancer diagnosis and monitoring. Genes 12(3):349. https://doi.org/10.3390/genes12030349
Mattox AK, Bettegowda C, Zhou S, Papadopoulos N, Kinzler KW, Vogelstein B (2019, Aug) Applications of liquid biopsies for cancer. Sci Transl Med 11(507):eaay1984. https://doi.org/10.1126/scitranslmed.aay1984
Heitzer E, Ulz P, Geigl JB (2015, Jan) Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 61(1):112–123. https://doi.org/10.1373/clinchem.2014.222679
Xie F, Li P, Gong J, Tan H, Ma J (2018, May) Urinary cell-free DNA as a prognostic marker for KRAS-positive advanced-stage NSCLC. Clin Transl Oncol 20(5):591–598. https://doi.org/10.1007/s12094-017-1754-7
Michela B (2021, Jul) Liquid biopsy: a family of possible diagnostic tools. Diagnostics 11(8):1391. https://doi.org/10.3390/diagnostics11081391
“25806217”
Guan X (2015, Sep) Cancer metastases: challenges and opportunities. Acta Pharm Sin B 5(5):402–418. https://doi.org/10.1016/j.apsb.2015.07.005
Li S et al (2019, Apr) Shear stress promotes anoikis resistance of cancer cells via caveolin-1-dependent extrinsic and intrinsic apoptotic pathways. J Cell Physiol 234(4):3730–3743. https://doi.org/10.1002/jcp.27149
Qiu J et al (2020) Refining cancer management using integrated liquid biopsy. Theranostics 10(5):2374–2384. https://doi.org/10.7150/thno.40677
Chaffer CL, Weinberg RA (2011, Mar) A perspective on cancer cell metastasis. Science 331(6024):1559–1564. https://doi.org/10.1126/science.1203543
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S (2009, Aug) Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 11(4):R46. https://doi.org/10.1186/bcr2333
Fabisiewicz A, Grzybowska E (2017, Jan) CTC clusters in cancer progression and metastasis. Med Oncol 34(1):12. https://doi.org/10.1007/s12032-016-0875-0
Geethadevi A, Parashar D, Bishop E, Pradeep S, Chaluvally-Raghavan P (2017, Dec) ERBB signaling in CTCs of ovarian cancer and glioblastoma. Genes Cancer 8(11–12):746–751. https://doi.org/10.18632/genesandcancer.162
Plaks V, Koopman CD, Werb Z (2013, Sep) Circulating tumor cells. Science 341(6151):1186–1188. https://doi.org/10.1126/science.1235226
Woestemeier A et al (2020, Mar) Clinical relevance of circulating tumor cells in esophageal cancer detected by a combined MACS enrichment method. Cancers 12(3):718. https://doi.org/10.3390/cancers12030718
Gerlinger M et al (2012, Mar) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366(10):883–892. https://doi.org/10.1056/NEJMoa1113205
Hyun K-A, Jung H-I (2014) Advances and critical concerns with the microfluidic enrichments of circulating tumor cells. Lab Chip 14(1):45–56. https://doi.org/10.1039/C3LC50582K
Sun J et al (2012) Double spiral microchannel for label-free tumor cell separation and enrichment. Lab Chip 12(20):3952. https://doi.org/10.1039/c2lc40679a
Menairini Silicon Biosystems. CellSearch - circulating tumor cell test. https://www.cellsearchctc.com/, 2022
Miller MC, van Doyle G, Terstappen LWMM (2010) Significance of circulating tumor cells detected by the cellsearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol 2010:1–8. https://doi.org/10.1155/2010/617421
Eslami-S Z, Cortés-Hernández LE, Alix-Panabières C (2020, Aug) Epithelial cell adhesion molecule: an anchor to isolate clinically relevant circulating tumor cells. Cell 9(8):1836. https://doi.org/10.3390/cells9081836
Woo D, Yu M (2018, Nov) Circulating tumor cells as ‘liquid biopsies’ to understand cancer metastasis. Transl Res 201:128–135. https://doi.org/10.1016/j.trsl.2018.07.003
Mikolajczyk SD et al (2011) Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J Oncol 2011:1–10. https://doi.org/10.1155/2011/252361
de Wit S et al (2015, Dec) The detection of EpCAM+ and EpCAM– circulating tumor cells. Sci Rep 5(1):12270. https://doi.org/10.1038/srep12270
Gabriel MT, Calleja LR, Chalopin A, Ory B, Heymann D (2016, Apr) Circulating tumor cells: a review of non–EpCAM-based approaches for cell enrichment and isolation. Clin Chem 62(4):571–581. https://doi.org/10.1373/clinchem.2015.249706
Krebs MG et al (2012, Feb) Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 7(2):306–315. https://doi.org/10.1097/JTO.0b013e31823c5c16
Sajeesh P, Sen AK (2014, Jul) Particle separation and sorting in microfluidic devices: a review. Microfluid Nanofluid 17(1):1–52. https://doi.org/10.1007/s10404-013-1291-9
Mark D, Haeberle S, Roth G, von Stetten F, Zengerle R (2010) Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. Chem Soc Rev 39(3):1153. https://doi.org/10.1039/b820557b
Warkiani ME, Wu L, Tay AKP, Han J (2015, Dec) Large-volume microfluidic cell sorting for biomedical applications. Annu Rev Biomed Eng 17(1):1–34. https://doi.org/10.1146/annurev-bioeng-071114-040818
di Carlo D, Irimia D, Tompkins RG, Toner M (2007, Nov) Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci 104(48):18892–18897. https://doi.org/10.1073/pnas.0704958104
Nagrath S et al (2007, Dec) Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450(7173):1235–1239. https://doi.org/10.1038/nature06385
Vona G et al (2000, Jan) Isolation by size of epithelial tumor cells. Am J Pathol 156(1):57–63. https://doi.org/10.1016/S0002-9440(10)64706-2
Jiang X et al (2017) Microfluidic isolation of platelet-covered circulating tumor cells. Lab Chip 17(20):3498–3503. https://doi.org/10.1039/C7LC00654C
Lin Z, Luo G, Du W, Kong T, Liu C, Liu Z (2020, Mar) Recent advances in microfluidic platforms applied in cancer metastasis: circulating tumor cells’ (CTCs) isolation and tumor-on-A-Chip. Small 16(9):1903899. https://doi.org/10.1002/smll.201903899
Wang S, Thomas A, Lee E, Yang S, Cheng X, Liu Y (2016) Highly efficient and selective isolation of rare tumor cells using a microfluidic chip with wavy-herringbone micro-patterned surfaces. Analyst 141(7):2228–2237. https://doi.org/10.1039/C6AN00236F
Al-Faqheri W, Thio THG, Qasaimeh MA, Dietzel A, Madou M, Al-Halhouli A (2017, Jun) Particle/cell separation on microfluidic platforms based on centrifugation effect: a review. Microfluid Nanofluid 21(6):102. https://doi.org/10.1007/s10404-017-1933-4
Xue P, Zhang L, Guo J, Xu Z, Kang Y (2016, Dec) Isolation and retrieval of circulating tumor cells on a microchip with double parallel layers of herringbone structure. Microfluid Nanofluid 20(12):169. https://doi.org/10.1007/s10404-016-1834-y
LeValley PJ et al (2019, Feb) Immunofunctional photodegradable poly(ethylene glycol) hydrogel surfaces for the capture and release of rare cells. Colloids Surf B: Biointerfaces 174:483–492. https://doi.org/10.1016/j.colsurfb.2018.11.049
Zhang X, Ju S, Wang X, Cong H (2019, Aug) Advances in liquid biopsy using circulating tumor cells and circulating cell-free tumor DNA for detection and monitoring of breast cancer. Clin Exp Med 19(3):271–279. https://doi.org/10.1007/s10238-019-00563-w
Zhou J, Kulasinghe A, Bogseth A, O’Byrne K, Punyadeera C, Papautsky I (2019, Dec) Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Microsyst Nanoeng 5(1):8. https://doi.org/10.1038/s41378-019-0045-6
Kulasinghe A et al (2017, Mar) Enrichment of circulating head and neck tumour cells using spiral microfluidic technology. Sci Rep 7(1):42517. https://doi.org/10.1038/srep42517
Stott SL et al (2010, Oct) Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci 107(43):18392–18397. https://doi.org/10.1073/pnas.1012539107
Xue P et al (2014, Mar) Isolation and elution of Hep3B circulating tumor cells using a dual-functional herringbone chip. Microfluid Nanofluid 16(3):605–612. https://doi.org/10.1007/s10404-013-1250-5
Xue P, Wu Y, Guo J, Kang Y (2015, Apr) Highly efficient capture and harvest of circulating tumor cells on a microfluidic chip integrated with herringbone and micropost arrays. Biomed Microdevices 17(2):39. https://doi.org/10.1007/s10544-015-9945-x
He W et al (2016, Mar) Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget. https://doi.org/10.18632/oncotarget.8305
Qasaimeh MA et al (2017, May) Isolation of circulating plasma cells in multiple myeloma using CD138 antibody-based capture in a microfluidic device. Sci Rep 7(1):45681. https://doi.org/10.1038/srep45681
Lee NJ et al (2020, Jan) Affinity-enhanced CTC-capturing hydrogel microparticles fabricated by degassed Mold lithography. J Clin Med 9(2):301. https://doi.org/10.3390/jcm9020301
Kim HU, Lim YJ, Lee HJ, Lee NJ, Bong KW (2020) Degassed micromolding lithography for rapid fabrication of anisotropic hydrogel microparticles with high-resolution and high uniformity. Lab Chip 20(1):74–83. https://doi.org/10.1039/C9LC00828D
Gleghorn JP et al (2010) Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip 10(1):27–29. https://doi.org/10.1039/B917959C
Turetta M et al (2018, Dec) Emerging Technologies for Cancer Research: towards personalized medicine with microfluidic platforms and 3D tumor models. Curr Med Chem 25(35):4616–4637. https://doi.org/10.2174/0929867325666180605122633
Hou S et al (2013, Mar) Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Ed 52(12):3379–3383. https://doi.org/10.1002/anie.201208452
Yu M et al (2013, Feb) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339(6119):580–584. https://doi.org/10.1126/science.1228522
Ferreira MM, Ramani VC, Jeffrey SS (2016, Mar) Circulating tumor cell technologies. Mol Oncol 10(3):374–394. https://doi.org/10.1016/j.molonc.2016.01.007
Ozkumur E et al (2013, Apr) Inertial focusing for tumor antigen–dependent and –independent sorting of rare circulating tumor cells. Sci Transl Med 5(179):179ra147. https://doi.org/10.1126/scitranslmed.3005616
Guglielmi R et al (2020, Dec) Technical validation of a new microfluidic device for enrichment of CTCs from large volumes of blood by using buffy coats to mimic diagnostic leukapheresis products. Sci Rep 10(1):20312. https://doi.org/10.1038/s41598-020-77227-3
Hyun K-A, Lee TY, Jung H-I (2013, May) Negative enrichment of circulating tumor cells using a geometrically activated surface interaction Chip. Anal Chem 85(9):4439–4445. https://doi.org/10.1021/ac3037766
Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D (2009) Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst 134(9):1808. https://doi.org/10.1039/b904476k
Song Y et al (2013, Apr) Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem 85(8):4141–4149. https://doi.org/10.1021/ac400366b
Racila E et al (1998, Apr) Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci 95(8):4589–4594. https://doi.org/10.1073/pnas.95.8.4589
Mishima Y et al (2017, Apr) The mutational landscape of circulating tumor cells in multiple myeloma. Cell Rep 19(1):218–224. https://doi.org/10.1016/j.celrep.2017.03.025
Esmaeilsabzali H, Beischlag TV, Cox ME, Parameswaran AM, Park EJ (2013, Nov) Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv 31(7):1063–1084. https://doi.org/10.1016/j.biotechadv.2013.08.016
Burger R, Ducrée J (2012, May) Handling and analysis of cells and bioparticles on centrifugal microfluidic platforms. Expert Rev Mol Diagn 12(4):407–421. https://doi.org/10.1586/erm.12.28
Warkiani ME et al (2016, Jan) Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc 11(1):134–148. https://doi.org/10.1038/nprot.2016.003
Smerage JB, Hayes DF (2006, Jan) The measurement and therapeutic implications of circulating tumour cells in breast cancer. Br J Cancer 94(1):8–12. https://doi.org/10.1038/sj.bjc.6602871
Jin C et al (2014) Technologies for label-free separation of circulating tumor cells: from historical foundations to recent developments. Lab Chip 14(1):32–44. https://doi.org/10.1039/C3LC50625H
Gerhardt T, Woo S, Ma H (2011) Chromatographic behaviour of single cells in a microchannel with dynamic geometry. Lab Chip 11(16):2731. https://doi.org/10.1039/c1lc20092e
Liu Z et al (2013, Sep) High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosens Bioelectron 47:113–119. https://doi.org/10.1016/j.bios.2013.03.017
Mohamed H, Murray M, Turner JN, Caggana M (2009, Nov) Isolation of tumor cells using size and deformation. J Chromatogr A 1216(47):8289–8295. https://doi.org/10.1016/j.chroma.2009.05.036
Inglis DW (2009, Jan) Efficient microfluidic particle separation arrays. Appl Phys Lett 94(1):013510. https://doi.org/10.1063/1.3068750
Zhang Z, Ramnath N, Nagrath S (2015, Sep) Current status of CTCs as liquid biopsy in lung cancer and future directions. Front Oncol 5:209. https://doi.org/10.3389/fonc.2015.00209
Amini H, Lee W, di Carlo D (2014) Inertial microfluidic physics. Lab Chip 14(15):2739. https://doi.org/10.1039/c4lc00128a
di Carlo D (2009) Inertial microfluidics. Lab Chip 9(21):3038. https://doi.org/10.1039/b912547g
Warkiani ME et al (2014) Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip 14(1):128–137. https://doi.org/10.1039/C3LC50617G
Abdulla A, Liu W, Gholamipour-Shirazi A, Sun J, Ding X (2018, Apr) High-throughput isolation of circulating tumor cells using cascaded inertial focusing microfluidic channel. Anal Chem 90(7):4397–4405. https://doi.org/10.1021/acs.analchem.7b04210
Khoo BL, Shang M, Ng CH, Lim CT, Chng WJ, Han J (2019, Dec) Liquid biopsy for minimal residual disease detection in leukemia using a portable blast cell biochip. NPJ Precis Oncol 3(1):30. https://doi.org/10.1038/s41698-019-0102-5
Warkiani ME, Tay AKP, Guan G, Han J (2015, Sep) Membrane-less microfiltration using inertial microfluidics. Sci Rep 5(1):11018. https://doi.org/10.1038/srep11018
Chen H (2018, Dec) A triplet parallelizing spiral microfluidic Chip for continuous separation of tumor cells. Sci Rep 8(1):4042. https://doi.org/10.1038/s41598-018-22348-z
Gascoyne PRC, Wang X-B, Huang Y, Becker FF (1997, May) Dielectrophoretic separation of cancer cells from blood. IEEE Trans Ind Appl 33(3):670–678. https://doi.org/10.1109/28.585856
Lee D, Hwang B, Kim B (2016, Dec) The potential of a dielectrophoresis activated cell sorter (DACS) as a next generation cell sorter. Micro Nano Syst Lett 4(1):2. https://doi.org/10.1186/s40486-016-0028-4
Gupta V et al (2012, Jun) ApoStream™, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 6(2):024133. https://doi.org/10.1063/1.4731647
Nguyen N-V, Jen C-P (2018, Dec) Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens Bioelectron 121:10–18. https://doi.org/10.1016/j.bios.2018.08.059
Mani GK, Morohoshi M, Yasoda Y, Yokoyama S, Kimura H, Tsuchiya K (2017, Feb) ZnO-based microfluidic pH sensor: a versatile approach for quick recognition of circulating tumor cells in blood. ACS Appl Mater Interfaces 9(6):5193–5203. https://doi.org/10.1021/acsami.6b16261
Low WS, Wan Abas WAB (2015) Benchtop technologies for circulating tumor cells separation based on biophysical properties. Biomed Res Int 2015:1–22. https://doi.org/10.1155/2015/239362
Alazzam A, Stiharu I, Bhat R, Meguerditchian A-N (2011, Jun) Interdigitated comb-like electrodes for continuous separation of malignant cells from blood using dielectrophoresis. Electrophoresis 32(11):1327–1336. https://doi.org/10.1002/elps.201000625
Moon H-S et al (2011) Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 11(6):1118. https://doi.org/10.1039/c0lc00345j
Liao C-J et al (2018, Oct) An optically induced Dielectrophoresis (ODEP)-based microfluidic system for the isolation of high-purity CD45neg/EpCAMneg cells from the blood samples of cancer patients—demonstration and initial exploration of the clinical significance of these cells. Micromachines 9(11):563. https://doi.org/10.3390/mi9110563
Tietze U, Schenk C, Gamm E (2008) Field effect transistor. In: Electronic circuits. Springer, Berlin, Heidelberg, pp 169–268. https://doi.org/10.1007/978-3-540-78655-9_3
Han K-H, Han A, Frazier AB (2006, Apr) Microsystems for isolation and electrophysiological analysis of breast cancer cells from blood. Biosens Bioelectron 21(10):1907–1914. https://doi.org/10.1016/j.bios.2006.01.024
Gu W, Zhao Y (2010, Nov) Cellular electrical impedance spectroscopy: an emerging technology of microscale biosensors. Expert Rev Med Devices 7(6):767–779. https://doi.org/10.1586/erd.10.47
Spencer D, Hollis V, Morgan H (2014, Nov) Microfluidic impedance cytometry of tumour cells in blood. Biomicrofluidics 8(6):064124. https://doi.org/10.1063/1.4904405
Kumeria T, Kurkuri MD, Diener KR, Parkinson L, Losic D (2012, May) Label-free reflectometric interference microchip biosensor based on nanoporous alumina for detection of circulating tumour cells. Biosens Bioelectron 35(1):167–173. https://doi.org/10.1016/j.bios.2012.02.038
Chiu T-K, Lei K-F, Hsieh C-H, Hsiao H-B, Wang H-M, Wu M-H (2015, Mar) Development of a microfluidic-based optical sensing device for label-free detection of circulating tumor cells (CTCs) through their lactic acid metabolism. Sensors 15(3):6789–6806. https://doi.org/10.3390/s150306789
Acknowledgments
CIBER-BBN is an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. The Nanobioengineering group in the Institute of Bioengineering of Catalonia (IBEC) has support from the Commission for Universities and Research of the Department of Innovation, Universities, and Enterprise of the Generalitat de Catalunya (2017 SGR 1079) and is part of the CERCA Program/Generalitat de Catalunya. This work is partially supported by Obra Social “La Caixa” project “Understanding and measuring mechanical tumor properties to improve cancer diagnosis, treatment, and survival: Application to liquid biopsies”.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Sierra-Agudelo, J., Rodriguez-Trujillo, R., Samitier, J. (2022). Microfluidics for the Isolation and Detection of Circulating Tumor Cells. In: Caballero, D., Kundu, S.C., Reis, R.L. (eds) Microfluidics and Biosensors in Cancer Research. Advances in Experimental Medicine and Biology, vol 1379. Springer, Cham. https://doi.org/10.1007/978-3-031-04039-9_16
Download citation
DOI: https://doi.org/10.1007/978-3-031-04039-9_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-04038-2
Online ISBN: 978-3-031-04039-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)