Abstract
The members of the family Reoviridae (reoviruses) consist of 9–12 discrete double-stranded RNA (dsRNA) segments enclosed by single, double, or triple capsid layers. The outer capsid proteins of reoviruses exhibit the highest diversity in both sequence and structural organization. By contrast, the conserved RNA-dependent RNA polymerase (RdRp) structure in the conserved innermost shell in all reoviruses suggests that they share common transcriptional regulatory mechanisms. After reoviruses are delivered into the cytoplasm of a host cell, their inner capsid particles (ICPs) remain intact and serve as a stable nanoscale machine for RNA transcription and capping performed using enzymes in ICPs. Advances in cryo-electron microscopy have enabled the reconstruction at near-atomic resolution of not only the icosahedral capsid, including capping enzymes, but also the nonicosahedrally distributed complexes of RdRps within the capsid at different transcriptional stages. These near-atomic resolution structures allow us to visualize highly coordinated structural changes in the related enzymes, genomic RNA, and capsid protein during reovirus transcription. In addition, reoviruses encode their own enzymes for nascent RNA capping before RNA releasing from their ICPs.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Reoviridae
- Reoviruses
- Reovirus transcription
- Double-stranded RNA (dsRNA)
- Capsid
- Capsid proteins
- Capsid layers
- Inner Capsid Particle (ICP)
- Core
- RNA-dependent RNA polymerase (RdRp)
Introduction
Reoviridae is the largest family of double-stranded RNA (dsRNA) viruses and consists of 15 recognized genera. The members of the Reoviridae family (hereafter referred to as reoviruses) have been isolated from various mammals, birds, reptiles, fish, crustaceans, marine protists, insects, ticks, arachnids, plants, and fungi and include a total of 75 virus species with wide geographical distributions (King et al. 2011).
The virions of reoviruses consist of 9–12 discrete dsRNA segments enclosed by single, double, or triple capsid layers. The virions can be divided into two parts on the basis of their functions: a transcriptionally competent inner capsid particle (ICP, also known as the core) and an outer layer of capsid proteins that coats the ICP for protection and membrane penetration; some single-layered reoviruses do no possess the outer capsid layer. Because of the absence of a lipid bilayer that can fuse with the cell membrane, reoviruses deliver their ICPs into the cytoplasm through membrane disruption facilitated by capsid proteins present in the outer layer. The sequence and structural organization of the outer capsid proteins of reoviruses are highly diverse, thus enabling these proteins to penetrate different cell membrane barriers.
The members of the Reoviridae family are divided into two subfamilies on the basis of the structural organization of their capsid: Sedoreovirinae and Spinareovirinae (Carstens 2010). A common process adopted by reoviruses involves the uncoating of the outer capsid layer after their delivery into the cytoplasm of the host cell where ICPs remain intact. ICPs are a multienzyme machine for RNA synthesis and capping. Similar to most dsRNA viruses, the ICPs of reoviruses have a conserved T = 1 icosahedral innermost capsid shell that is composed of 60 copies of two conformers of a shell protein. Shell proteins present in reoviruses belonging to various genera have a similar structural topology despite lacking a detectable amino acid sequence identity (Fig. 17.1). In the cytoplasm, the virus-encoded RNA-dependent RNA polymerase (RdRp), which is located under the icosahedral five-fold vertex in the innermost capsid shell, transcribes nascent RNA by using the minus strand of the genomic dsRNA segment as a template within the inner capsid while avoiding the dsRNA-activated defense mechanisms of the host cell (Lawton et al. 1997; Mertens 2004; Nibert and Baker 2003).
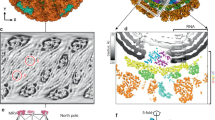
Innermost capsid shells and shell proteins of reoviruses. (a) Innermost shells. Two conformers of the shell protein of each reovirus are in red and green, respectively. The two conformers in the asymmetric unit are in dark red and forest green. (b) Superimpositions of the two conformers of the reoviruses
Reoviruses possess their own protein with capping enzymes and cap the nascent RNA before releasing it from transcribing ICPs. The 5′-end capping of nascent RNA is a conserved process in eukaryotes and viruses. The cap structure is involved in the utilization of the host machinery for the efficient translation of viral mRNA, the evasion of the host’s antiviral innate immune response, and the encapsidation and transcription of viral RNA. Capping enzymes found in the reoviruses of the Spinareovirinae subfamily are present in each turret-like structure located on the outer surface of the innermost capsid shell, whereas those in the reoviruses of the Sedoreovirinae subfamily are encapsidated within the innermost capsid shell.
The structures of reoviruses have been extensively studied through X-ray crystallography and cryo-electron microscopy (cryo-EM). Studies have identified the various structural characteristics of reoviruses and their functional roles in the virus life cycle. This chapter summarizes the structural studies of reoviruses published during the past years. Considerable technical progress and advancements have been made in high-resolution cryo-EM and single-particle analysis. In particular, this chapter focuses on recent in situ structural studies of RdRp within viral capsids that employed these advanced technologies. Together with the findings of the biochemical and genetic analyses of reovirus transcription, these in situ structural studies of RdRp within reovirus capsids have enhanced the understanding regarding the structure–function relationships of endogenous transcription.
Icosahedral Capsid Structures of Reoviruses
Reoviruses in the Subfamily Spinareovirinae
The virions of reoviruses belonging to the subfamily Spinareovirinae consist of an ICP coated by a layer of three-fold heterohexamers of two coat proteins (represented by the Orthoreovirus, Aquareovirus, and Oryzavirus genera) or only the ICP (represented by the Dinovernavirus and Cypovirus genera). The double-layered virions and their ICPs are of approximately 820 and 800 Å in diameter, respectively. The ICP consists of many nonicosahedral segmented genomic dsRNAs and an equal copy number of the RdRp complex enclosed by a T = 1 icosahedral capsid formed by the shell, clamp, and turret proteins.
The innermost shell of ICPs is formed by two conformers of a plate-like shell protein. The shell proteins in reoviruses with known structures are all topologically conserved. Five monomers of conformer A form a star-like complex at each five-fold vertex and five monomers of conformer B pack into the gap between two adjacent monomers of conformer A. Twelve of the decamers form the innermost capsid shell (Fig. 17.1a). Both conformers have a similar plate-like structure with no strongly defined domain boundaries. The plate-like shell protein can be divided into two subdomains on the basis of the major difference between two conformers: subdomains I and II; the subdomains undergo a minor rigid body tilt relative to one another (Reinisch et al. 2000) (Fig. 17.1b). Subdomain II (also known as the apical domain) is an insert into subdomain I. Five copies of the tip of the apical domain form a five-fold pore at the vertex of the icosahedral five-fold axis on the capsid shell for the exit of nascent mRNA. The N-terminal regions of both subunits are not resolved in icosahedral reconstruction because of the lack of icosahedral symmetry. The asymmetric structures and functions of the N-terminal regions are described below.
The outer surface of the innermost shell of reoviruses belonging to the subfamily Spinareovirinae is decorated with nodular “clamp” (also called “cement”) proteins (Fig. 17.2a–c). The copy number of clamp proteins varies among different genera of Spinareovirinae. For example, dinovernavirus has a total of 60 clamp proteins that are located around the 12 turrets (Auguste et al. 2015). Cypovirus, aquareovirus, and oryzavirus have 120 clamp proteins, with the additional 60 proteins located around the three-fold vertices (Zhou et al. 2003; Cheng et al. 2008; Miyazaki et al. 2008). In addition to the 120 clamp proteins present in the cypovirus, aquareovirus, and oryzavirus, orthoreovirus possesses an additional 30 proteins located on the two-fold vertices (Zhang et al. 2005a, b; Yan et al. 2011). All clamp proteins are located on the border of and interact with two or three copies of neighboring shell proteins, and they are indispensable for the stability of the innermost capsid shell (Reinisch et al. 2000).
Capsid structures of three turreted reoviruses. The colors of the capsid proteins are labeled. (a) and (b) Capsid structures of the single-layered dinovernavirus and cypovirus. The flexible fiber at each five-fold vertex is not shown. (c) Inner capsid structure of aquareovirus. (d) Capsid structure of double-layered aquareovirus. (e) Cut-open view of the capsid structure of aquareovirus
Reoviruses belonging to the subfamily Spinareovirinae are also known as “turreted” reoviruses because of the presence of a distinct pentameric turret on each five-fold vertex of the innermost shell (Fig. 17.2d). The turret is formed by the five copies of turret proteins and is involved in the catalysis of 5′ mRNA cap synthesis (Miyazaki et al. 2008; Zhang et al. 2003, 2005b; Cheng et al. 2010, 2011; Yan et al. 2011; Zhu et al. 2014). The capping role of the turret protein is described in the following section.
The turret proteins of reoviruses belonging to the genera Orthoreovirus and Aquareovirus have a C-terminal three-domain flap (Fig. 17.3a). The three domains in the flap have structures related to the immunoglobulin fold (Reinisch et al. 2000; Cheng et al. 2010). By contrast, reoviruses belonging to the genera Oryzavirus, Dinovernavirus, and Cypovirus lack the flap in the turret protein (Fig. 17.3b) (Miyazaki et al. 2008; Auguste et al. 2015; Cheng et al. 2011). The flap has been observed to swivel about a hinge between virions and the ICPs of mammalian orthoreovirus (MRV) (Chandran et al. 1999; Dryden et al. 1993), between an aquareovirus virion (Cheng et al. 2010) and an aquareovirus particle with a partially degraded outermost protein (Zhang et al. 2010; Wang et al. 2018), and also between two types of baboon reovirus (BRV) virions (Yan et al. 2011). The significance of flap swiveling to reovirus function or assembly remains to be deciphered. Some reoviruses have a fiber that binds to cell surface receptors during infection (Lee et al. 1981). The fiber at the icosahedral five-fold vertex is held by turret flaps in MRV and avian reovirus (ARV) (Dryden et al. 1993; Zhang et al. 2005b) or by a brace domain in cypovirus (Cheng et al. 2011). The presence or absence of the fiber is not dependent on genera. For example, MRV and ARV but not BRV have fibers, although they all belong to the genus Orthoreovirus.
The ICPs of orthoreovirus and aquareovirus are coated by an outer layer of 200 three-fold symmetrical heterohexamers arranged on an incomplete T = 13 icosahedral lattice, exposing turrets on icosahedral vertices (Fig. 17.2d and e). By contrast, oryzavirus has only 60 heterohexamers surrounding the turret (Miyazaki et al. 2008). Interactions between the ICP and heterohexamer are mediated by the clamp proteins (Fig. 17.2e). Each three-fold heterohexamer comprises three copies of the protector protein located on top of the three copies of the membrane penetration protein (Fig.17.4a). The trimer of the penetration protein plays crucial roles in the viral penetration of the cell membrane (Liemann et al. 2002). A myristoyl group (Fig. 17.4a) is covalently linked to the N terminus and embedded in a hydrophobic pocket of the trimer of the penetration protein (Liemann et al. 2002; Zhang et al. 2010). When the reovirus virion interacts with proteases in the intestinal lumen where reoviruses infect intestinal cells, the proteases strip off the protector protein, resulting in the formation of infectious subviral particles (ISVPs) (Chang and Zweerink 1971; Silverstein et al. 1970). The ISVPs are characterized by the loss of the protector protein through proteolytic degradation (Nibert and Fields 1992). Concomitantly, the Asn42-Pro43 bond in the penetration protein of the orthoreovirus or aquareovirus ISVPs is cleaved through autocleavage (Fig. 17.4b), dividing the protein into a myristoylated N terminus and C terminus (Liemann et al. 2002; Zhang et al. 2010). This autocleavage is a critical step in the cell entry of coated turreted reoviruses (Nibert et al. 1991a). The myristoylated N-terminus must be released from the hydrophobic pocket, forming a penetration pore or disrupting the membrane for the translocation of the reovirus ICP into the cytoplasm (Nibert et al. 1991b; Odegard et al. 2004).
The cell entry mechanism of the single-layered cypovirus and dinovernavirus remains unclear. The cypovirus virions are embedded in a micrometer-sized crystal polyhedron that is made of the trimers of the viral polyhedrin protein (Coulibaly et al. 2007), which has a similar central β-sandwich and an overall trimeric shape to the membrane penetration protein of orthoreovirus and aquareovirus (Liemann et al. 2002; Cheng et al. 2010). However, polyhedrin has a new fold and evolved to assemble in vivo into three-dimensional cubic crystals rather than icosahedral shells (Coulibaly et al. 2007). By contrast, dinovernavirus is not embedded and lacks any homolog to these trimeric capsid and polyhedron proteins (Auguste et al. 2015).
Reoviruses in Sedoreovirinae
Reoviruses belonging to the subfamily Sedoreovirinae, represented by the Rotavirus and Orbivirus genera, consist of an ICP coated by a layer of outer capsid proteins (Fig. 17.5). The ICP, which is a double-layered particle (DLP or core), consists of nonicasahedral segmented genomic dsRNAs and an equal copy number of RdRp enclosed by an icosahedral architecture formed by the two layers of capsid proteins.
The innermost shell is formed by 120 copies of the two conformers of the shell protein, which is topologically conserved with that of reoviruses belonging to the subfamily Spinareovirinae (Fig. 17.1). A T = 13 icosahedral layer of the 260 trimers of an intermediate protein surrounds the innermost shell (Grimes et al. 1998; McClain et al. 2010). Reoviruses belonging to the subfamily Sedoreovirinae are also known as “nonturreted” reoviruses because they do not have a distinct pentameric turret on each five-fold vertex of the innermost shell. In addition, no clamp proteins are present on the shell; therefore, the surface of the ICP is relatively smooth. The protein that functions as a viral RNA-capping enzyme is enclosed in the capsid shell of nonturreted reoviruses (Kumar et al. 2020; Sutton et al. 2007).
The infectious virions of nonturreted reoviruses are a triple-layered particle (TLP) of approximately 1000 Å in diameter. Compared with the conserved architecture and topology of ICP proteins, the outer layer coated on the ICP is considerably more diversified. Similar to turreted reoviruses, the outer-layer proteins of nonturreted reoviruses undergo substantial conformational changes to facilitate cell membrane penetration and strip off after the ICP is translocated into the cytoplasm during infection. The ICP of rotavirus (Fig. 17.5a) consists of an innermost shell formed by 120 copies of two VP2 conformers coated by a layer of 260 VP6 trimers (McClain et al. 2010). The outer layer of the rotavirus virion, which serves as the molecular machinery for host-cell binding and penetration, contains two proteins, namely VP4 and VP7 (Fig. 17.5b). Each of 260 VP7 trimers directly clamps onto the trimer of the intermediate protein VP6, forming a continuous perforated capsid (Settembre et al. 2011; Chen et al. 2009). A trimeric spike of VP4 emerges from each of the local six-fold channels formed by the six trimers of VP6 and VP7 around the five-fold vertex. Efficient infection requires the cleavage of VP4 by trypsin into two fragments: VP8* for receptor binding and VP5* for membrane penetration. The spike appears only in the structure of trypsin-treated particles, suggesting the rigidification of the spike upon proteolysis (Crawford et al. 2001; Trask et al. 2012). The ICP of bluetongue virus (BTV, Fig. 17.5c) belonging to the genus Orbivirus consists of an innermost shell formed by 120 copies of two VP3 conformers and an intermediate layer of 260 VP7 trimers (Grimes et al. 1998). The outer capsid has 60 triskelion-like VP2 spikes, surrounded by 120 globular trimers of VP5 (Fig. 17.5d). Each VP2 trimer binds atop four VP7 trimers; each VP5 trimer bridges the channel formed by six surrounding VP7 trimers (Zhang et al. 2016; Roy 2017). VP2 binds to surface glycoproteins and facilitates the endocytosis of the virion, whereas VP5 penetrates the host cell membrane and delivers the ICP into the host cytosol. Unlike the membrane penetration protein VP4 of rotavirus, the VP5 of BTV does not appear to be proteolytically processed and is likely to fold into a metastable conformation following translation (Roy 2017).
Reovirus RdRp
Reovirus possesses a segmented dsRNA genome consisting of each of a full set of RNA segments with conserved sequences at 5′ and 3′ ends, along with an equal number of RdRps. The number of genome segments in reoviruses varies from 9 to 12, depending on the genus to which the reovirus belongs. Each RNA segment is associated with a single RdRp under each of the five-fold vertices (Periz et al. 2013). RdRps are capable of both RNA replication and transcription within the reovirus ICP (Gillies et al. 1971; Shatkin and Sipe 1968; Skehel and Joklik 1969; Tao et al. 2002; Lu et al. 2008). During reovirus ICP assembly, each RdRp recognizes a capped viral mRNA segment based on the conserved sequence at 5′ and 3′ ends (Tao et al. 2002; Lu et al. 2008). To ensure the encapsidation of a full set of mRNAs into the ICP, higher order structures may form between different mRNA segments (Tao et al. 2002; Borodavka et al. 2017). The ICP assembly is accompanied by a single round of the RdRp-driven synthesis of minus-strand RNAs complementary to mRNAs (replication), eventually forming genomic dsRNA segments in mature virions. Upon reovirus infection, ICPs remain intact after the delivery of reoviruses into the host cell’s cytoplasm, and RdRps can repeatedly transcribe RNA from the minus-strand RNA genome within ICPs. Reoviruses have been used as models to study RNA transcription and capping processes (Borsa and Graham 1968; Levin et al. 1970; Furuichi and Miura 1975; Furuichi 1978; Smith and Furuichi 1980; Lawton et al. 1997; Yang et al. 2012; Yu et al. 2015).
The structures of recombinant orthoreovirus and rotavirus RdRps with various RNA substrates have been determined through X-ray crystallography (Tao et al. 2002; Lu et al. 2008). The two RdRp structures exhibit a typical overall architecture of RNA viruses. Both contain a polymerase domain surrounded by an N-terminal domain and a C-terminal bracelet domain. Both the N-terminal domain and C-terminal bracelet domain are crucial for catalytic activity (Wehrfritz et al. 2007). Similar to other polynucleotide polymerases (Ferrer-Orta et al. 2006), the right-handed polymerase domain can be further divided into three subdomains, namely, fingers, thumb, and palm. The three domains fold together into a cage-like structure. The hollow catalytic center in the polymerase domain is connected to the exterior through four well-defined channels that are used for RNA template entry, NTP entry, template exit (or dsRNA exit in the replication mode), and transcript exit. Compared with other RdRps with known structures, the template exit channel is unique in the reovirus RdRp structure, indicating that reovirus RdRps can internally separate nascent dsRNA duplexes (McDonald et al. 2009). The presence of an extended loop (priming loop) between the fingers and palm subdomains is a distinctive feature of the two reovirus RdRps. The structures of orthoreovirus RdRp soaked with the template Mg2+ and NTPs show that the priming loop (residues 558–565) can serve as a platform for stabilizing an NTP at the priming site during the initiation stage and retract to allow the passage of the nascent dsRNA duplex during the elongation stage (Tao et al. 2002). A pair of Mg2+ ions is crucial for the formation of the phosphodiester bond. Residues essential for anchoring the pair of Mg2+ ions are Asp585, Asp734, and Asp735. In the rotavirus RdRp structure, the priming loop bends away from the active site, leaving it in a retracted state incapable of supporting an NTP and initiating RNA synthesis, thus preventing the rotavirus RdRp from forming a functional initiation complex (Lu et al. 2008). A cap analog soaked into both orthoreovirus and rotavirus RdRp crystals sits in a groove near the tip of the thumb subdomain of the two RdRps. This cap-binding site may serve to anchor the capped plus strand of genomic dsRNA during transcription.
High-resolution in situ structures of RdRp at different transcriptional stages are necessary for fully understanding the highly coordinated RNA transcription process in the confined shell of the ICP. The structures of RdRp complexes and the genome within the cypovirus capsid have been determined for the first time by using cryo-EM and the capsid subtraction–based symmetry mismatch reconstruction method (Liu and Cheng 2015; Cheng 2015; Liu 2015). Subsequently, an increasing number of structures of RdRp and genomes within viral capsids have been resolved (Zhang et al. 2015; Dai et al. 2017; Wang et al. 2018; Ding et al. 2018; Ding et al. 2019; Ilca et al. 2019; He et al. 2019; Kaelber et al. 2020; Jenni et al. 2019). In the following sections, we summarize the in situ structural studies of various reovirus RdRps within reovirus particles at different stages. These in situ structural studies of RdRps and genomic RNA within nontranscribing and transcribing reovirus capsids that we summarize below have provided insights into dynamic molecular mechanisms through which RdRp interacts with the shell protein and dsRNA segment, initiates transcription, and ensures polymerase processivity.
Structure of the RdRp Complex and Genome in a Nontranscribing Mature Virion
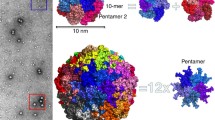
The in situ structures of genomes and RdRp complexes within the mature virions of cypovirus and aquareovirus have been determined at a near-atomic resolution (Liu and Cheng 2015; Wang et al. 2018; Zhang et al. 2015). The overall structures of genomes and RdRp complexes within the two turreted reoviruses are similar. The genome is composed of regularly distributed spherical layers that are formed by the structures of discontinuous dsRNA fragments running in parallel and associated with RdRp complexes (Fig. 17.6a). Each RdRp complex, which consists of an RdRp and a cofactor protein, is anchored at the inner surface of the capsid slightly off center from the icosahedral five-fold vertex and surrounded by multiple layers of dsRNA resembling a nest-shaped layered structure in a relatively crowded capsid shell. The distance between two adjacent dsRNA fragments within the same layer is approximately 25–28 Å, whereas the distance between two adjacent layers is approximately 28–30 Å. The inner layers are less well-organized compared with the outer layers of dsRNA fragments (Liu and Cheng 2015). The layers of the dsRNA fragment resemble the organization of the cholesteric liquid crystal (de Gennes and Prost 1993), an observation that is consistent with an earlier assumption that the dsRNA genome would form liquid crystalline arrays within the highly condensed capsid (Gouet et al. 1999). This liquid crystalline organization, which provides molecules with a certain degree of fluidity (de Gennes and Prost 1993), may provide mobility for dsRNA during transcription in the crowded capsid. RdRp complexes and the associated dsRNA fragments appear to be organized in a pseudo D3 symmetry. However, RdRp complexes at 6 of the 12 vertices exhibit stronger density, whereas those at the remaining six positions show weaker densities, implying partial (two-third) occupancy; therefore, only 10 RdRp complexes are located under the 12 vertices in the capsid (Liu and Cheng 2015). In the subsequent study, 10 RdRp complexes were resolved, leaving two vertices occupied only by dsRNA (Zhang et al. 2015). In aquareovirus with 11 RNA segments, only 11 of the 12 vertices are occupied by RdRp complexes (Wang et al. 2018; Ding et al. 2018).
Structures of the genome and RdRp complexes within the nontanscribing mature cypovirus and aquareovirus virions. The color scheme for the proteins and domains is labeled. (a) RdRp complex and dsRNA within cypovirus. Left: half of the icosahedral capsid (gray) is removed to show the structures of the genomic dsRNA (first layer in orange and second layer in yellow), RdRp VP2 (magenta), and cofactor protein VP4 (cyan). Middle: Zoom-in view of the dsRNA, RdRp, and cofactor protein in the capsid. Right: The cap of the plus strand of genomic dsRNA interacts with an A/G base binding site formed by the Tyr184 and Arg37 of the N-terminal domain at the entrance of the template entry channel. (b) RdRp complex and dsRNA within aquareovirus. The color scheme is identical to that of panel A
The structures of RdRps within the mature virions of cypovirus and aquareovirus share a similar architecture to the X-ray crystal structures of the recombinant RdRps of orthoreovirus and rotavirus (Tao et al. 2002; Lu et al. 2008). Both RdRps contain a polymerase domain surrounded by an N-terminal domain and a C-terminal bracelet domain. The catalytic centers of RdRps are connected to the exterior through four well-defined tunnels. In the mature virion of cypovirus, a dsRNA fragment traverses across the template exit channel and closely interacts with the bracelet and polymerase domains (Fig. 17.6a). This dsRNA fragment was previously misinterpreted as the capped 5′ end of the plus strand of a dsRNA (Liu and Cheng 2015). The catalytic center of the polymerase domain is occupied by a loop (residues 925–932) in the bracelet domain, resulting in the partial blockage of the transcript exit, template entry, and template exit channels (Liu and Cheng 2015; Zhang et al. 2015; Li et al. 2017). The cap of the plus strand of genomic dsRNA interacts with an A/G base binding site formed by the Tyr184 and Arg37 of the N-terminal domain at the entrance of the template entry channel (Fig. 17.6a) (Cui et al. 2019). This A/G base binding site is adjacent to the cap-binding site between the N-terminal domain and the tip of the thumb subdomain (Tao et al. 2002; Li et al. 2020; Liu and Cheng 2015; Zhang et al. 2015). The RdRp and dsRNA in the mature virion of aquareovirus (Fig. 17.6b) exhibit similar interactions with that of cypovirus (Fig. 17.6a). The differences are that dsRNA binding does not cause the partial blockage of the template entry and exit channels. In contrast, many elements are flexible in the aquareovirus RdRp structure, resulting in a hollow central catalytic cavity (Wang et al. 2018).
RdRps are in complex with a cofactor protein in cypovirus and aquareovirus (Liu and Cheng 2015; Wang et al. 2018; Ding et al. 2018; Zhang et al. 2015). The cofactor protein is located on the RdRp surface region between the two channels for the RNA template entry and NTP entry (Fig. 17.6). In cypovirus, the cofactor protein VP4 consists of an N-terminal and a C-terminal NTPase domain. The N-terminal domain is formed by two small β-sheets and several α-helices. The main body of the C-terminal NTPase is a Walker-A α/β motif and contains a GTP molecule at a predicted NTP binding site (Zhang et al. 2015). The RdRp cofactor protein VP4 in aquareovirus has the same interaction with RdRp as that observed in cypovirus. The structure of the aquareovirus VP4 can be divided into three domains: an N-terminal nodule domain, a plate domain, and a C-terminal domain (Wang et al. 2018; Ding et al. 2018). The N-terminal nodule domain, which resides in the corner between a copy of the shell protein VP3A and the RdRp protein VP2, interacts with the dsRNA fragment roughly parallel to the capsid shell (Wang et al. 2018). The plate domain is roughly rectangular and resides in the corner between the shell protein VP3A and the VP2 polymerase domain and interacts with a surface region of the VP2 polymerase domain between the two channels for template entry and NTP entry. The aquareovirus VP4 N-terminal nodule domain exhibits no similarity to its counterpart in cypovirus VP4; by contrast, the plate domain resembles the C-terminal NTPase domain in cypovirus. In addition, the aquareovirus VP4 plate domain contains fully conserved motifs (KxxxK and SDxxG) for NTP binding in the μ2 homology proteins of turreted reoviruses (Noble and Nibert 1997; Kim et al. 2004; Nibert and Kim 2004). An additional density, which is located at the conserved NTP binding site surrounded by conserved residues (Lys410, Lys414, and Ser439), can be assigned to the NTP or the single phosphate group corresponding to the ɣ-phosphate group of the bound NTP (Wang et al. 2018; Ding et al. 2018). The aquareovirus VP4 has an additional C-terminal domain, which does not exist in the cypovirus VP4. The C-terminal domain is located at the entrance of the template entry channel and interacts with the dsRNA fragment approaching the RdRp template entry channel. By contrast, no interaction was observed between the cypovirus cofactor protein VP4 and genomic RNA. The cofactor protein remains structurally unchanged during the transcription of turreted reoviruses.
The architecture and domains of the RdRp in nonturreted reoviruses are similar to those of the RdRp in turreted reoviruses. However, the RdRp in nonturreted reoviruses is not in complex with the cofactor protein. The RdRp monomer anchors to the inner capsid shell surrounding the icosahedral five-fold axis, a location similar to that of the RdRp complex in turreted reoviruses. Unlike the D3 symmetric distribution of RdRps found in turreted reoviruses, the position of an RdRp in nonturreted reoviruses at any particular vertex does not correlate with those of RdRps under other five-fold vertices. In other words, the distribution of the RdRp has none of the symmetries that would form a subgroup of the icosahedral point group (Jenni et al. 2019). The stochastic distribution of RdRps rules out any unique long-range order for RNA; therefore, the overall RNA structure in the capsid has not been resolved (Jenni et al. 2019; He et al. 2019; Ding et al. 2019). In the mature virion of BTV, the 5′ cap of the positive strand of a dsRNA genome binds to residue His40 at the entrance of the template entry channel. A dsRNA fragment closely interacts with the bracelet and polymerase domains (He et al. 2019). These interactions are similar to those observed in turreted reoviruses. In the mature virion of rotavirus, the RdRp appears to interact with dsRNA at the cap-binding site (Jenni et al. 2019).
RdRp in Transcribing Reoviruses
Transcribing reovirus particles can be obtained in vitro by treating reovirus virions or ICPs with a transcription reaction buffer including Mg2+, ATP, GTP, CTP, UTP, and S-adenosyl-L-methionine (SAM) (Bartlett et al. 1974; Smith and Furuichi 1980; Van Dijk and Huismans 1980; Lawton et al. 1997; Drayna and Fields 1982). To elucidate the transcriptional mechanisms of viruses, many structures of transcribing reoviruses have been resolved (Lawton et al. 1997; Pesavento et al. 2001; Mendez et al. 2008; Yang et al. 2012; Zhu et al. 2014; Yu et al. 2015). Recently, the in situ structures of the reovirus RdRps at different transcriptional stages have been determined using a combination of capsid subtraction, symmetry mismatch, classification, and local reconstruction methods (Liu and Cheng 2015; Zhang et al. 2015; Li et al. 2020; Cui et al. 2019; Ding et al. 2019; Jenni et al. 2019).
The transcription process in the reovirus capsid can be divided into two stages, namely initiation and elongation, in which the RdRps undergo stepwise conformational changes. The transcription process is activated by the removal of outer-layer proteins in nonturreted reoviruses (He et al. 2019; Jenni et al. 2019) and the binding of SAM in the single-layered turreted cypovirus (Yu et al. 2015; Cui et al. 2019). Transcriptional activation is reflected by the expansion of the capsid chamber resulting from the outward tilt of the apical domains of the five copies of shell proteins around the five-fold vertex. For double-layered turreted reoviruses, the removal of the outer layer of aquareovirus alone does not cause capsid expansion (Cheng et al. 2008), although the removal of the outer layer is required for elongation but not initiation in orthoreovirus (Farsetta et al. 2000).
Initiation
Genomic dsRNA binds to the RdRp through the binding of the capped 5′ end of the plus strand to the site adjacent to the entrance of the template entry channel of the RdRp in the nontranscribing mature reovirus. Because only the 3′ end of a single-stranded template can enter the catalytic center, dsRNA must unwind first. This RdRp state, in which two separate single-stranded RNAs bind to the entrance of the template entry channel, was observed in the rotavirus DLP (Ding et al. 2019; Jenni et al. 2019). The capped 5′ end of the genomic plus strand (nontemplate RNA) interacts with the RdRp cap-binding site, whereas the minus strand (template RNA) interacts with an RNA binding β-sheet in the finger subdomain at the entrance of the template entry channel (Ding et al. 2019; Jenni et al. 2019). The absence of any RNA density in the RdRp active site at this stage suggests that the RdRp represents a preinitiation state (Jenni et al. 2019). A helix–loop–helix subdomain of the rotavirus RdRp N-terminal domain, which extends toward the genomic plus strand in the preinitiation state and retracts in the transcriptional elongation state, was proposed to play a role in separating the genomic duplex (Ding et al. 2019).
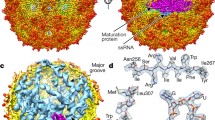
In the initiation state of the cypovirus RdRp, the dsRNA fragment, which traverses across the template exit channel and interacts with the bracelet and polymerase domains in the mature virion, is removed. Accordingly, blocked channels in the nontranscribing RdRp are unblocked through the conformational change of the bracelet domain. The path between the transcript exit channel of the RdRp and the five-fold pore on the shell is blocked by a three-helix bundle (residues 1091–1133) in the RdRp bracelet domain. The dsRNA unwinds at the entrance of the template entry channel, and the template RNA (minus strand) enters into the positively charged channel (Fig. 17.7a and b) (Li et al. 2020; Cui et al. 2019). The 3′ end of the template RNA enters the catalytic site of the transcribing cypovirus RdRp, making base pairs with a dinucleotide product strand and an incoming NTP (Fig. 17.7c) (Li et al. 2020). The end of the short RNA duplex, which consists of the RNA template and the product dinucleotide, is buttressed by a so-called switch loop (residues 1080–1088) from the bracelet domain (Li et al. 2020; Liu and Cheng 2015) and the priming loop (residues 516–524). The 5′ triphosphate of the first product nucleotide interacts with the surrounding positively charged residues of Lys521, Arg738, Arg770, His1084, Lys1085, and Arg1146. The triphosphate of the incoming NTP interacts with two Mg2+ ions, which are coordinated by Asp547, Asp680, and Asp681 (Fig. 17.7c). In the initiation stage of the cypovirus RdRp, Cui et al. (2019) modeled only one nucleotide product and an incoming NTP pairing with the template. However, they observed the dinucleotide product in their cryo-EM density map (EMDB ID: EMD-20582), although the density of the first base in the dinucleotide was slightly weaker. Except for conformational changes in the RdRp and RNA that directly interact with the RdRp, the remaining region of the dsRNA genome is well organized and identical to that observed in the nontranscribing mature cypovirus capsid. The plus strand (nontemplate RNA) is pushed toward a cleft formed by the N terminal and thumb in the initiation RdRp. This cleft, which becomes narrower than that of the RdRp in the nontranscribing mature cypovirus, functions as a cap-binding site to immobilize nontemplate RNA (Fig. 17.7d). The first base (A) of the plus strand interacts with the A/G base binding site, and the cap of the plus strand binds to the cap-binding site adjacent to the A/G base binding site (Cui et al. 2019).
RdRp and cofactor protein of cypovirus at initiation stage. (a) RdRp (magenta) and cofactor protein (cyan) and associated dsRNA (gray) under the turret (gray). (b) Cut-open view of the RdRP and associated dsRNA. The genomic dsRNA unwinds at the template entry channel, with the minus-strand RNA (template) inserting into the channel and the plus-strand RNA being tethered to the cap-binding site at the channel entrance. (c) The template RNA makes base pairs with a dinucleotide product strand and an incoming NTP in the RdRp catalytic site. (d) Zoom-in view of the cap-binding site
The RdRp transition from the initiation to the elongation state is suggested to be a rate-limiting step in RNA viruses (Harrus et al. 2010; Appleby et al. 2015; Scrima et al. 2012). The capture of the RdRp structure at the initiation stage in the transcribing cypovirus suggests that most RdRps in transcribing viral particles remain at this stage, thus providing the evidence for the existence of this slow step.
Elongation
The initiation is followed by the elongation step. A study reconstructed the cypovirus RdRp structure at early- and later-elongation stages. The cypovirus sample used for the reconstruction was treated with the transcription reaction buffer in the absence of CTP; therefore, the transcription is halted at the position where the first C appears (Cui et al. 2019). In the RdRp at the early-elongation stage, the template RNA inserts into the polymerase domain. In the initiation state, the conformations of the priming loop and switch loop block the pathway of the template and transcript. By contrast, in the early-elongation state, the switch loop moves away and forms part of the transcript exit channel. Moreover, the priming loop retracts, allowing the movement of the template and transcript (Cui et al. 2019). An 8-base-pair template–transcript complex was observed in the polymerase domain (Fig. 17.8). The thumb remains closed and immobilizes nontemplate RNA at the early-elongation stage, which is identical to that of the RdRp at the initiation stage. In addition, the priming loop in the rotavirus RdRp at the elongation stage was observed to adopt a retracted conformation to make room for the growing template and product (Jenni et al. 2019). Therefore, conformational changes in the switch loop and priming loop ensure polymerase processivity and begin the transition from the initiation to elongation stage. For the rotavirus RdRp, a unique C-terminal plug, which is located in the template exit channel in the nontranscribing rotavirus RdRp, is displaced in the RdRp at the elongation stage to allow template RNA to exit (Jenni et al. 2019; Ding et al. 2019).
RdRp and cofactor protein of cypovirus at later-elongation stage. The newly synthesized transcript is in purple, and the color scheme of dsRNA genome, RdRp, and cofactor is identical to that in Fig. 17.6. (a) RdRp and cofactor protein under the capsid shell (gray). The transcript (purple) is released through a five-fold pore in the capsid shell. The turret is not shown for clarity. (b) Cut-open view of the RdRp catalytic site. The transcript and template RNA are separated by two negatively charged residues Asp1089 and Asp1090
In the cypovirus RdRp structure at the later-elongation stage, the newly synthesized transcript and template RNA form eight base pairs and then are separated at the ninth base pair by two negatively charged residues, namely Asp1089 and Asp1090, of the bracelet domain (Cui et al. 2019). The Asp1089 guides the template RNA through the template RNA exit channel. The three-helix bundle in the bracelet domain shifts at the elongation stage, resulting in a tunnel connecting the transcript exit channel and the five-fold pore on the capsid shell (Fig. 17.8). The Asp1090 guides the newly synthesized transcript to pass through the transcript exit channel and release through the five-fold pore. A similar template RNA–transcript separation mechanism is observed in rotavirus. A five-helix bundle (residues 923–996), which is the counterpart of the three-helix bundle in the cypovirus, splits the template–transcript and mediates the exit of the transcript in the transcribing rotavirus RdRp (Jenni et al. 2019; Ding et al. 2019). Compared with the nontranscribing rotavirus RdRp, the five-helix bundle shifts. Simultaneously, a tip loop (resides 346–373) of a copy of the shell protein, which blocks the transcript–exit site in the nontranscribing DLP structure, also shifts. These conformational changes result in the formation of a continuous tunnel between the RdRp transcript exit channel and the five-fold pore on the capsid shell. The transcript extends from the five-fold pore through the tunnel (Jenni et al. 2019; Ding et al. 2019). Therefore, the bracelet domain of the reovirus RdRp not only functions as a helicase but also plays a role in directing the template and transcript to exit through their respective channels.
The nontemplate RNA binding cleft in the RdRp at the later-elongation stage adopts the open conformation in contrast to the closed conformation in the initiation and early-elongation stages. The nontemplate RNA leaves from the nontemplate RNA binding cleft and slides through a positively charged nontemplate RNA track located outside the RdRp. Outside the polymerase core, the nontemplate RNA reanneals with the template RNA. The reannealed dsRNA of the transcription bubble is located at the same position as the dsRNA that traverses across the template exit channel in the nontranscribing RdRp. The traverse dsRNA, which binds to the template exit channel of each RdRp in the nontranscribing cypovirus, is suggested to be the tail-terminal end of each genomic dsRNA segment, namely the other end relative to the capped-terminal end of each genomic RNA (Cui et al. 2019).
A previous study suggested that the 5′ end of the plus-strand RNA always binds to the cap-binding site at the entrance of the template entry channel during the whole transcription process (Tao et al. 2002). The negative-strand RNA reanneals with the plus-strand RNA after emerging from the template exit channel. Thus, the 3′ end of the negative-strand RNA easily reinserts into the template entry channel and reinitiates after the completion of each round of mRNA synthesis. The in situ structures of the cypovirus RdRp in the initiation stage with an RNA fork and the elongation stage with a complete transcription bubble suggest that after reannealing, the capped dsRNA end may follow the tail-terminal end of genomic dsRNA, a so-called “ouroboros” model of conservative transcription. The capped dsRNA end and tail-terminal dsRNA were proposed to belong to the same segment of genomic RNA (Cui et al. 2019). In this model, the cap is not always bound to the cap-binding site; thus, no undesirable kinks or sharp U-turns are present on the dsRNA genome during elongation. However, the capped dsRNA end following the tail-terminal end has not been directly observed, and the mechanism through which the two dsRNA ends interact remains unclear. In addition, the transcription bubble was not observed in the rotavirus RdRp in the elongation state (the plus-strand RNA was not resolved) (Jenni et al. 2019; Ding et al. 2019).
Roles of the Shell Protein in the RdRp Transcription
In the capsid, RdRp anchors to the inner surface of the capsid shell under the icosahedral five-fold vertex through interactions with the N-terminal regions and apical domains of the five shell protein dimers. The five-fold pore, which is formed by the tip loops of the shell protein apical domains, is covered by the RdRp bracelet domain in nontranscribing mature reoviruses. The helix bundle subdomain in the RdRp shifts at the elongation stage, resulting in the RNA-releasing tunnel connecting the transcript exit channel and the five-fold pore on the capsid shells of both turreted and nonturreted reoviruses.
In addition to the transcript-exiting pore, the shell protein regulates transcription through the conformational change of its apical domain. In transcribing cypovirus, the five copies of apical domains around the five-fold vertex tilt outward, resulting in an enlarged capsid chamber (Yang et al. 2012; Yu et al. 2015; Cui et al. 2019). In addition, the tilt of the apical domains is observed in the rotavirus DLP compared with those in the TLP (Jenni et al. 2019). The expanded capsid chamber provides genomic RNA with more flexibility to slide trough the RdRp within a densely packed capsid during transcription. The conformational change in the shell protein at the five-fold vertex is triggered by the binding of SAM and ATP with the turret protein in the cypovirus capsid (Yu et al. 2015). The same conformational change in the rotavirus shell protein is triggered by the removal of outer-layer proteins because the capsid structure of rotavirus TLP remains unchanged after incubation with nucleotides and SAM (Jenni et al. 2019). Most RdRps in the rotavirus TLP do not transcribe, suggesting that capsid expansion is required for reovirus RdRp transcription (Jenni et al. 2019). The conformational change in the apical domain may trigger the initiation of the reovirus RdRp. These structural results are consistent with the biochemical result that the shell protein is crucial for the RdRp catalytic activity (Starnes and Joklik 1993; Patton et al. 1997; Boyce et al. 2004).
The N-terminal regions of shell proteins present in different rope-like conformations adapt to the occupation of the RdRp–cofactor complex under the five-fold vertex of the turreted reovirus capsid (Zhang et al. 2015; Wang et al. 2018; Ding et al. 2018). These N-terminal regions play critical roles in recruiting the RdRp and cofactor protein during the virus assembly. Although no associated cofactor protein exists, the RdRp in nonturreted reoviruses is anchored to the inner capsid shell by the asymmetric N-terminal regions of shell proteins (He et al. 2019; Jenni et al. 2019; Ding et al. 2019), suggesting a conserved mechanism of the RdRp–capsid assembly among reoviruses. Deletion of the N-terminal region of the rotavirus shell protein inhibits the incorporation of the RdRp into recombinant virus-like particles but does not prevent the capsid-shell assembly (Zeng et al. 1998). The N-terminal regions of the shell protein may contribute to the regulation of the RdRp activity in rotavirus. An amphipathic helix (residues 78–84) in the N-terminal region of the rotavirus shell protein attaches to a hydrophobic pocket next to the helix–loop–helix subdomain in transcribing RdRp but detaches from this pocket in nontranscribing RdRp. This amphipathic helix anchors the helix–loop–helix subdomain and prevents unfavorable interactions with genomic RNA during transcription, thus promoting RdRp activity (Ding et al. 2019).
Reovirus RNA Capping
Viruses have evolved mechanisms to protect their 5′ ends of RNA with a covalently linked cap moiety that is indistinguishable from cellular mRNA cap structures. The cap is essential for the efficient translation of viral mRNA by using the host machinery as well as the evasion of the host’s antiviral innate immune response (Decroly et al. 2012).
Reoviruses encode their own enzymes for nascent RNA capping. The cap structure consists of a 7-methylguanosine moiety, which is linked to the first nucleotide (with a 2′-O-methyl group) of nascent RNA through a 5′–5′ triphosphate bridge. The RNA capping process consists of four sequential reactions: (1) an RNA triphosphatase (RTPase) hydrolyzes the 5′ triphosphate of a nascent RNA to a diphosphate; (2) a guanylyltransferase (GTase) transfers a GMP molecule to the RNA 5′ diphosphate to form GpppRNA; (3) a 7-N-methyltransferase (7-N-MTase) transfers a methyl group from SAM to GpppRNA to form m7GpppRNA and generates S-adenosyl-Lhomocysteine (SAH); and (4) a 2′-O-methyltransferase (2′-O-MTase) transfers a methyl group from SAM to the 2′-hydroxyl group of the first nucleotide ribose of the RNA and generates SAH.
Turreted Reovirus Capping
For all turreted reoviruses, the five copies of the turret protein form a five-fold hollow turret chamber at each five-fold vertex of the capsid. The structure of the cypovirus turret protein, which is the best characterized among turreted reoviruses, is similar to that of other turreted reoviruses except for the lack of a flap-like structure present in orthoreovirus and aquareovirus (Figs. 17.2 and 17.3). The cypovirus turret protein contains three conserved enzymatic domains of GTase (residues 1–366), MTase-1 (residues 419–461 and 526–725), and MTase-2 (residues 833–1058) and a bridge domain (residues 367–418 and 726–832) (Cheng et al. 2011; Reinisch et al. 2000; Fausnaugh and Shatkin 1990; Luongo et al. 1998, 2000; Yang et al. 2012; Zhu et al. 2014). In addition, the cypovirus turret protein has a small brace domain (residues 462–525) inserted in the MTase-1 domain (Fig. 17.3). The brace domain, which is not observed in the orthoreovirus and aquareovirus turret protein, holds the flexible spike-like complex (homology of the orthoreovirus fiber) on the turret (Cheng et al. 2011).
The GTase domain is a pocket-like structure located at the turret bottom and interacts with the innermost shell at the five-fold vertex. An ATP-binding site is located in a subdomain (residues 267–321) of GTase, which is probably an SAM-dependent ATPase (Yu et al. 2015). The two MTase domains are located at the top region of the turret. A conserved catalytic tetrad KDKE (Lys531, Asp616, Lys649, and Glu689), which is believed to be a characteristic of RNA 2′-O-MTases (Bujnicki and Rychlewski 2001; Decroly et al. 2012), was observed in MTase-1, suggesting that MTase-1 is 2′-O-MTase. This motif is not present in MTase-2, suggesting that MTase-2 is 7-N-MTase (Zhu et al. 2014). The bridge domain bridges the GTase and the two MTases.
From the nontranscribing to transcribing stages, the cypovirus turret protein undergoes a conformational change at the putative ATPase subdomain, resulting in a pivotal movement (around residue 355) that drives the rigid-body movement of the two MTases and the bridge. This conformational change in the ATPase domain may mediate the activation of RNA transcription and capping (Yu et al. 2015).
GTase and GTP form a covalent GTase–GMP complex (GTase guanylylation). Subsequently, GMP is transferred to the 5′-diphosphorylated RNA transcript. A density is observed in the pocket of the GTase domain in the transcribing cypovirus (Yang et al. 2012; Yu et al. 2015). Yang et al. interpreted the density as a GMP moiety that forms the phosphoamide bond with the adjacent Lys234 in the cypovirus GTase (Yang et al. 2012) based on a previous mutagenesis study of lysine residues, suggesting that Lys190 of the orthoreovirus GTase is responsible for the guanylation of GTases (Luongo et al. 2000). However, Yu et al. suggested that the density is a GTP and the His217 is the catalytic amino acid for the guanylation of the cypovirus GTase (Yu et al. 2015). The SAM structure, which is the methyl donor for mRNA methylation, defines the active site in the structures of 2′-O-MTase and 7-N-MTase in the transcribing cypovirus (Zhu et al. 2014; Yu et al. 2015).
Although the location at which the first capping reaction (hydrolysis of 5′ triphosphate) occurs in reoviruses remains unknown, the newly catalyzed transcript, which enters into the turret chamber through the five-fold pore, can reach GTase first according to the steric arrangement of capping enzymes. A channel, which is formed by the bridge and brace domains of one subunit and the brace domain of the adjacent subunit, can guide the transcript from GTase to the active site of 7-N-MTase (Zhu et al. 2014). This channel becomes wider in the transcribing cypovirus. After its N7-guanine methylation, the transcript reaches the adjacent active site of 2′-O-MTase for 2′-O methylation.
Nonturreted Reovirus Capping
Nonturreted reoviruses have neither the turret nor other homologous assembly on its outer capsid. The asymmetric reconstructions of the RdRps within nonturreted reoviruses have indicated that the RdRp is attached to the innermost capsid shell (Jenni et al. 2019; Ding et al. 2019; He et al. 2019); however, capping enzymes that mediate the four capping reactions of nascent transcripts were not visualized in these reconstructions. These results suggest that the complex of capping enzymes does not have a fixed position with respect to the RdRp in nonturreted reovirus particles.
The crystal structures of the rotavirus VP3 tetramer and the BTV VP4 dimer have been resolved (Kumar et al. 2020; Sutton et al. 2007). Both the rotavirus VP3 and the BTV VP4 contain four enzymes required for nascent transcript capping. The domain organizations of both VP3 and VP4 monomers are similar, and catalytic residues in the GTase and MTase domains are conserved. The major difference is that the BTV VP4 does not have the phosphodiesterase domain in the rotavirus VP3 (Kumar et al. 2020). The organizations of the capping enzyme domains of both VP3 and VP4 exhibit no similarity to their counterpart (turret protein) in turreted reoviruses. The fundamentally different structural assembly and strategies that turreted and nonturreted reoviruses adopt for RNA capping suggest that the split between the two Reoviridae subfamilies may be ancient.
Nonturreted reoviruses harbor their capping enzymes in the capsid shell, probably in a location close to the RdRp. However, on the basis of the structure of the transcribing rotavirus DLP (Ding et al. 2019), the nascent transcript directly exits from the five-fold pore on the capsid shell, and therefore is unlikely to reach the VP3 inside the capsid shell. The mechanism through which the transcript undergoes the capping process remains unknown. A hypothesis that the priming of transcription is by a VP3-produced, capped nucleotide, or a capped dinucleotide would provide a potential alternative mechanism for rotavirus (Jenni et al. 2019). Similarly, priming of transcription by a diphosphorylated nucleotide or dinucleotide may explain the absence of RTPase in the turret or another location between the RdRp template exit channel and GTase. The diphosphorylated nucleotide or the 5′ diphosphorylated dinucleotide could be catalyzed by NTPase in the cofactor protein attached to the RdRp in the capsid.
Conclusions and Perspectives
Reoviruses are nonenveloped viruses that encapsidate 9–12 genomic RNA segments in single-, double-, or triple-layered capsids. After reoviruses are delivered into a host cell’s cytoplasm, the outer capsids shed off, and ICPs remain intact and contain enzymes that can repeatedly transcribe and cap RNA in the cytoplasm. Recent substantial advances in cryo-EM technology have allowed the reconstruction of the in situ structures of the viral RdRps at different transcriptional stages within the capsid at near-atomic resolutions.
The in situ structural differences in RdRp complexes and outer capsid layers suggest the diversification of reoviruses. For example, all RdRp channels are open in the nontranscribing virion of aquareovirus, rotavirus, and BTV; by contrast, the channels are partially blocked in the cypovirus RdRp in the nontranscribing stage. Thus, the RdRp transition from the nontranscribing to transcriptional initiation stage does not involve channel opening, suggesting the presence of different transcriptional activation strategies in reoviruses. Transcription is activated by the binding of SAM and ATP in cypovirus (Furuichi 1974, 1978; Yu et al. 2015) and by the removal of outer shells in rotavirus (Jenni et al. 2019).
Notwithstanding these diversifications, the conserved RdRp structure in the conserved innermost shell in all reoviruses suggests that they share common transcriptional regulatory mechanisms. The recently reported in situ RdRp structures of reoviruses at different stages enable us to visualize sequential transcriptional steps that occur in reovirus capsids. The shell protein subunits around the five-fold vertex region expand in all reoviruses with known structures at the initiation or preinitiation stages, making space for RNA sliding in the crowed capsid. The RdRp in the preinitiation stage is observed in the rotavirus DLP. The dsRNA unwinds; the template RNA binds to the entrance of the template entry channel, and the capped nontemplate RNA interacts with the cap-binding site. In the initiation stage, the template RNA inserts into the catalytic center and makes base pairs with a dinucleotide product. The template and product are stabilized in the catalytic center by the priming and switch loops. The nontemplate RNA is immobilized in a closed clamp formed by the N-terminal and the thumb. In the following early-elongation stage, the template forms an eight-base-pair template–transcript complex. The switch loop moves away, and the priming loop retracts. The clamp remains closed and immobilizes nontemplate RNA. At the later elongation stage, the negatively charged residues in the bracelet domain separate the template and product and direct them to exit through their respective channels. The conformational changes in the RdRp bracelet domain and shell protein apical domain result in the formation of the tunnel connecting the transcript exit channel and the five-fold pore on the capsid shell, from which the transcript is released. The clamp is opened, and the released nontemplate RNA anneals with the exiting template RNA.
Although the structures of capsid proteins, enzymes, and RNA in reoviruses have provided abundant information regarding the virus assembly, RNA transcription, and capping, many questions still remain. How do reoviruses recognize and encapsidate the RdRps and the full set of plus-strand RNA segments during capsid assembly? How do the newly encapsided RdRp and the plus-strand RNA segments synthesize and organize dsRNA segments? How do reoviruses regulate the transcription efficiency of RNA segments in different length? What are the functions of the cofactor protein in turreted reoviruses? Future studies addressing these questions can further elucidate the details of the viral life cycle and aid in the rational design of drugs against reovirus infections.
References
Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D 3rd, Wetmore DR, McGrath ME, Ray AS, Sofia MJ, Swaminathan S, Edwards TE (2015) Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347(6223):771–775. https://doi.org/10.1126/science.1259210
Auguste AJ, Kaelber JT, Fokam EB, Guzman H, Carrington CV, Erasmus JH, Kamgang B, Popov VL, Jakana J, Liu X, Wood TG, Widen SG, Vasilakis N, Tesh RB, Chiu W, Weaver SC (2015) A newly isolated reovirus has the simplest genomic and structural organization of any reovirus. J Virol 89(1):676–687. https://doi.org/10.1128/JVI.02264-14
Bartlett NM, Gillies SC, Bullivan S, Bellamy AR (1974) Electron-microscopy study of reovirus reaction cores. J Virol 14(2):315–326
Borodavka A, Dykeman EC, Schrimpf W, Lamb DC (2017) Protein-mediated RNA folding governs sequence-specific interactions between rotavirus genome segments. eLife 6:e27453. https://doi.org/10.7554/eLife.27453
Borsa J, Graham AF (1968) Reovirus: RNA polymerase activity in purified virions. Biochem Biophys Res Commun 33(6):895–901. https://doi.org/10.1016/0006-291x(68)90396-3
Boyce M, Wehrfritz J, Noad R, Roy P (2004) Purified recombinant bluetongue virus VP1 exhibits RNA replicase activity. J Virol 78(8):3994–4002. https://doi.org/10.1128/jvi.78.8.3994-4002.2004
Bujnicki JM, Rychlewski L (2001) Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda 2 protein. Genome Biol 2(9):RESEARCH0038
Carstens EB (2010) Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2009). Arch Virol 155(1):133–146. https://doi.org/10.1007/s00705-009-0547-x
Chandran K, Walker SB, Chen Y, Contreras CM, Schiff LA, Baker TS, Nibert ML (1999) In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins mu1 and sigma3. J Virol 73(5):3941–3950. https://doi.org/10.1128/JVI.73.5.3941-3950.1999
Chang CT, Zweerink HJ (1971) Fate of parental reovirus in infected cell. Virology 46(3):544–555. https://doi.org/10.1016/0042-6822(71)90058-4
Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N (2009) Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A 106(26):10644–10648. https://doi.org/10.1073/pnas.0904024106
Cheng L (2015) Cryo-EM shows the polymerase structures and a non-spooled genome within a dsRNA virus. In: International Mini-Symposium of Structural Virology, Beijing, China, April 9th 2015
Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH (2008) Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol 382(1):213–222
Cheng L, Zhu J, Hui WH, Zhang XK, Honig B, Fang Q, Zhou ZH (2010) Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol 397(3):852–863. https://doi.org/10.1016/j.jmb.2009.12.027
Cheng L, Sun J, Zhang K, Mou Z, Huang X, Ji G, Sun F, Zhang J, Zhu P (2011) Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc Natl Acad Sci U S A 108(4):1373–1378. https://doi.org/10.1073/pnas.1014995108
Coulibaly F, Chiu E, Ikeda K, Gutmann S, Haebel PW, Schulze-Briese C, Mori H, Metcalf P (2007) The molecular organization of cypovirus polyhedra. Nature 446(7131):97–101. https://doi.org/10.1038/Nature05628
Crawford SE, Mukherjee SK, Estes MK, Lawton JA, Shaw AL, Ramig RF, Prasad BV (2001) Trypsin cleavage stabilizes the rotavirus VP4 spike. J Virol 75(13):6052–6061. https://doi.org/10.1128/JVI.75.13.6052-6061.2001
Cui Y, Zhang Y, Zhou K, Sun J, Zhou ZH (2019) Conservative transcription in three steps visualized in a double-stranded RNA virus. Nat Struct Mol Biol 26(11):1023–1034. https://doi.org/10.1038/s41594-019-0320-0
Dai X, Li Z, Lai M, Shu S, Du Y, Zhou ZH, Sun R (2017) In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 541(7635):112–116. https://doi.org/10.1038/nature20589
de Gennes PG, Prost J (1993) The physics of liquid crystals. Oxford University Press, Oxford
Decroly E, Ferron F, Lescar J, Canard B (2012) Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10(1):51–65. https://doi.org/10.1038/nrmicro2675.nrmicro2675[pii]
Ding K, Nguyen L, Zhou ZH (2018) In situ structures of the polymerase complex and RNA genome show how Aquareovirus transcription machineries respond to Uncoating. J Virol 92(21):e00774-18. https://doi.org/10.1128/JVI.00774-18
Ding K, Celma CC, Zhang X, Chang T, Shen W, Atanasov I, Roy P, Zhou ZH (2019) In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release. Nat Commun 10(1):2216. https://doi.org/10.1038/s41467-019-10236-7
Drayna D, Fields BN (1982) Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol 41(1):110–118. https://doi.org/10.1128/JVI.41.1.110-118.1982
Dryden KA, Wang GJ, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS (1993) Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation - analysis of virions and dubviral particles by cryoelectron microscopy and image-reconstruction. J Cell Biol 122(5):1023–1041
Farsetta DL, Chandran K, Nibert ML (2000) Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J Biol Chem 275(50):39693–39701. https://doi.org/10.1074/jbc.M004562200
Fausnaugh J, Shatkin AJ (1990) Active site localization in a viral mRNA capping enzyme. J Biol Chem 265(13):7669–7672
Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N (2006) A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol 16(1):27–34. https://doi.org/10.1016/j.sbi.2005.12.002
Furuichi Y (1974) “Methylation-coupled” transcription by virus-associated transcriptase of cytoplasmic polyhedrosis virus containing double-stranded RNA. Nucleic Acids Res 1(6):809–822. https://doi.org/10.1093/nar/1.6.809
Furuichi Y (1978) “Pretranscriptional capping” in the biosynthesis of cytoplasmic polyhedrosis virus mRNA. Proc Natl Acad Sci U S A 75(3):1086–1090. https://doi.org/10.1073/pnas.75.3.1086
Furuichi Y, Miura K (1975) A blocked structure at the 5′ terminus of mRNA from cytoplasmic polyhedrosis virus. Nature 253(5490):374–375
Gillies S, Bullivant S, Bellamy AR (1971) Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science 174(4010):694–696. https://doi.org/10.1126/science.174.4010.694
Gouet P, Diprose JM, Grimes JM, Malby R, Burroughs JN, Zientara S, Stuart DI, Mertens PPC (1999) The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97(4):481–490
Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PPC, Stuart DI (1998) The atomic structure of the bluetongue virus core. Nature 395(6701):470–478
Harrus D, Ahmed-El-Sayed N, Simister PC, Miller S, Triconnet M, Hagedorn CH, Mahias K, Rey FA, Astier-Gin T, Bressanelli S (2010) Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J Biol Chem 285(43):32906–32918. https://doi.org/10.1074/jbc.M110.151316
He Y, Shivakoti S, Ding K, Cui Y, Roy P, Zhou ZH (2019) In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating. Proc Natl Acad Sci U S A 116(33):16535–16540. https://doi.org/10.1073/pnas.1905849116
Ilca SL, Sun X, El Omari K, Kotecha A, de Haas F, DiMaio F, Grimes JM, Stuart DI, Poranen MM, Huiskonen JT (2019) Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Nature 570(7760):252–256. https://doi.org/10.1038/s41586-019-1229-9
Jenni S, Salgado EN, Herrmann T, Li Z, Grant T, Grigorieff N, Trapani S, Estrozi LF, Harrison SC (2019) In situ structure of rotavirus VP1 RNA-dependent RNA polymerase. J Mol Biol 431(17):3124–3138. https://doi.org/10.1016/j.jmb.2019.06.016
Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W (2020) Arrangement of the polymerase complexes inside a nine-segmented dsRNA virus. Structure 28 (6):604-612, e603. https://doi.org/10.1016/j.str.2020.01.011
Kim J, Parker JS, Murray KE, Nibert ML (2004) Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor mu2. J Biol Chem 279(6):4394–4403. https://doi.org/10.1074/jbc.M308637200
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2011) Virus taxonomy. Ninth report of the international committee on taxonomy of viruses. Elsevier Inc., London
Kumar D, Yu X, Crawford SE, Moreno R, Jakana J, Sankaran B, Anish R, Kaundal S, Hu L, Estes MK, Wang Z, Prasad BVV (2020) 2.7 a cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity. Sci Adv 6(16):eaay6410. https://doi.org/10.1126/sciadv.aay6410
Lawton JA, Estes MK, Prasad BVV (1997) Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol 4(2):118–121
Lee PW, Hayes EC, Joklik WK (1981) Protein sigma 1 is the reovirus cell attachment protein. Virology 108(1):156–163. https://doi.org/10.1016/0042-6822(81)90535-3
Levin DH, Mendelsohn N, Schonberg M, Klett H, Silverstein S, Kapuler AM, Acs G (1970) Properties of RNA transcriptase in reovirus subviral particles. Proc Natl Acad Sci U S A 66(3):890–897. https://doi.org/10.1073/pnas.66.3.890
Li X, Zhou N, Chen W, Zhu B, Wang X, Xu B, Wang J, Liu H, Cheng L (2017) Near-atomic resolution structure determination of a cypovirus capsid and polymerase complex using cryo-EM at 200kV. J Mol Biol 429(1):79–87
Li X, Wang L, Wang X, Chen W, Yang T, Song J, Liu H, Cheng L (2020) Structure of RdRps within a transcribing dsRNA virus provides insights into the mechanisms of RNA synthesis. J Mol Biol 432(2):358–366. https://doi.org/10.1016/j.jmb.2019.09.015
Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC (2002) Structure of the reovirus membrane-penetration protein, mu 1, in a complex with its protector protein, sigma 3. Cell 108(2):283–295
Liu H (2015) Symmetry-mismatch reconstruction for icosahedral viruses. In: International Mini-Symposium of Structural Virology, Beijing, China, April 9th 2015
Liu H, Cheng L (2015) Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science 349(6254):1347–1350. https://doi.org/10.1126/science.aaa4938
Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC (2008) Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16(11):1678–1688. https://doi.org/10.1016/j.str.2008.09.006
Luongo CL, Contreras CM, Farsetta DL, Nibert ML (1998) Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus lambda 2 protein - evidence for activities in mRNA CAP methylation. J Biol Chem 273(37):23773–23780
Luongo CL, Reinisch KM, Harrison SC, Nibert ML (2000) Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein lambda 2. J Biol Chem 275(4):2804–2810
McClain B, Settembre E, Temple BRS, Bellamy AR, Harrison SC (2010) X-ray crystal structure of the rotavirus inner capsid particle at 3.8 angstrom resolution. J Mol Biol 397(2):587–599. https://doi.org/10.1016/j.jmb.2010.01.055
McDonald SM, Tao YJ, Patton JT (2009) The ins and outs of four-tunneled Reoviridae RNA-dependent RNA polymerases. Curr Opin Struct Biol 19(6):775–782. https://doi.org/10.1016/j.sbi.2009.10.007
Mendez II, Weiner SG, She YM, Yeager M, Coombs KM (2008) Conformational changes accompany activation of reovirus RNA-dependent RNA transcription. J Struct Biol 162(2):277–289. https://doi.org/10.1016/j.jsb.2008.01.006
Mertens P (2004) The dsRNA viruses. Virus Res 101(1):3–13
Miyazaki N, Uehara-Ichiki T, Xing L, Bergman L, Higashiura A, Nakagawa A, Omura T, Cheng RH (2008) Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell. J Virol 82(22):11344–11353
Nibert ML, Baker TS (2003) CPV, a stable and symmetrical machine for mRNA synthesis. Structure 11(6):605–607. https://doi.org/10.1016/S0969-2126(03)00099-6
Nibert ML, Fields BN (1992) A carboxy-terminal fragment of protein mu 1/mu 1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol 66(11):6408–6418. https://doi.org/10.1128/JVI.66.11.6408-6418.1992
Nibert ML, Kim J (2004) Conserved sequence motifs for nucleoside triphosphate binding unique to turreted reoviridae members and coltiviruses. J Virol 78(10):5528–5530
Nibert ML, Furlong DB, Fields BN (1991a) Mechanisms of viral pathogenesis. Distinct forms of reoviruses and their roles during replication in cells and host. J Clin Invest 88(3):727–734. https://doi.org/10.1172/JCI115369
Nibert ML, Schiff LA, Fields BN (1991b) Mammalian reoviruses contain a myristoylated structural protein. J Virol 65(4):1960–1967. https://doi.org/10.1128/JVI.65.4.1960-1967.1991
Noble S, Nibert ML (1997) Core protein mu2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J Virol 71(10):7728–7735
Odegard AL, Chandran K, Zhang X, Parker JS, Baker TS, Nibert ML (2004) Putative autocleavage of outer capsid protein micro1, allowing release of myristoylated peptide micro1N during particle uncoating, is critical for cell entry by reovirus. J Virol 78(16):8732–8745. https://doi.org/10.1128/JVI.78.16.8732-8745.2004
Patton JT, Jones MT, Kalbach AN, He YW, Xiaobo J (1997) Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J Virol 71(12):9618–9626
Periz J, Celma C, Jing B, Pinkney JN, Roy P, Kapanidis AN (2013) Rotavirus mRNAS are released by transcript-specific channels in the double-layered viral capsid. Proc Natl Acad Sci U S A 110(29):12042–12047. https://doi.org/10.1073/pnas.1220345110
Pesavento JB, Lawton JA, Estes MK, Prasad BVV (2001) The reversible condensation and expansion of the rotavirus genome. P Natl Acad Sci USA 98(4):1381–1386
Reinisch KM, Nibert M, Harrison SC (2000) Structure of the reovirus core at 3.6 angstrom resolution. Nature 404(6781):960–967
Roy P (2017) Bluetongue virus structure and assembly. Curr Opin Virol 24:115–123. https://doi.org/10.1016/j.coviro.2017.05.003
Scrima N, Caillet-Saguy C, Ventura M, Harrus D, Astier-Gin T, Bressanelli S (2012) Two crucial early steps in RNA synthesis by the hepatitis C virus polymerase involve a dual role of residue 405. J Virol 86(13):7107–7117. https://doi.org/10.1128/JVI.00459-12
Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC (2011) Atomic model of an infectious rotavirus particle. EMBO J 30(2):408–416. https://doi.org/10.1038/emboj.2010.322
Shatkin AJ, Sipe JD (1968) RNA polymerase activity in purified reoviruses. Proc Natl Acad Sci U S A 61(4):1462–1469. https://doi.org/10.1073/pnas.61.4.1462
Silverstein SC, Schonberg M, Levin DH, Acs G (1970) The reovirus replicative cycle: conservation of parental RNA and protein. Proc Natl Acad Sci U S A 67(1):275–281. https://doi.org/10.1073/pnas.67.1.275
Skehel JJ, Joklik WK (1969) Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology 39(4):822–831. https://doi.org/10.1016/0042-6822(69)90019-1
Smith RE, Furuichi Y (1980) Gene mapping of cytoplasmic polyhedrosis virus of silkworm by the full-length mRNA prepared under optimized conditions of transcription in vitro. Virology 103(2):279–290
Starnes MC, Joklik WK (1993) Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology 193(1):356–366. https://doi.org/10.1006/viro.1993.1132
Sutton G, Grimes JM, Stuart DI, Roy P (2007) Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol 14(5):449–451. https://doi.org/10.1038/nsmb1225
Tao YZ, Farsetta DL, Nibert ML, Harrison SC (2002) RNA synthesis in a cage - structural studies of reovirus polymerase lambda 3. Cell 111(5):733–745
Trask SD, McDonald SM, Patton JT (2012) Structural insights into the coupling of virion assembly and rotavirus replication. Nat Rev Microbiol 10(3):165–177. https://doi.org/10.1038/nrmicro2673
Van Dijk AA, Huismans H (1980) The in vitro activation and further characterization of the bluetongue virus-associated transcriptase. Virology 104(2):347–356. https://doi.org/10.1016/0042-6822(80)90339-6
Wang X, Zhang F, Su R, Li X, Chen W, Chen Q, Yang T, Wang J, Liu H, Fang Q, Cheng L (2018) Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly. Proc Natl Acad Sci U S A 115(28):7344–7349. https://doi.org/10.1073/pnas.1803885115
Wehrfritz JM, Boyce M, Mirza S, Roy P (2007) Reconstitution of bluetongue virus polymerase activity from isolated domains based on a three-dimensional structural model. Biopolymers 86(1):83–94. https://doi.org/10.1002/bip.20706
Yan XD, Parent KN, Goodman RP, Tang JH, Shou JY, Nibert ML, Duncan R, Baker TS (2011) Virion structure of baboon reovirus, a fusogenic orthoreovirus that lacks an adhesion fiber. J Virol 85(15):7483–7495. https://doi.org/10.1128/Jvi.00729-11
Yang C, Ji G, Liu H, Zhang K, Liu G, Sun F, Zhu P, Cheng L (2012) Cryo-EM structure of a transcribing cypovirus. Proc Natl Acad Sci U S A 109(16):6118–6123. https://doi.org/10.1073/pnas.1200206109
Yu X, Jiang J, Sun J, Zhou ZH (2015) A putative ATPase mediates RNA transcription and capping in a dsRNA virus. eLife 4:e07901. https://doi.org/10.7554/eLife.07901
Zeng CQ, Estes MK, Charpilienne A, Cohen J (1998) The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. J Virol 72(1):201–208. https://doi.org/10.1128/JVI.72.1.201-208.1998
Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS (2003) Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 angstrom. Nat Struct Biol 10(12):1011–1018. https://doi.org/10.1038/Nsb1009
Zhang X, Ji Y, Zhang L, Harrison SC, Marinescu DC, Nibert ML, Baker TS (2005a) Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 angstrom resolution. Structure 13(10):1545–1557. https://doi.org/10.1016/j.str.2005.07.012
Zhang X, Tang JH, Walker SB, O’Hara D, Nibert ML, Duncan R, Baker TS (2005b) Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 343(1):25–35. https://doi.org/10.1016/j.virol.2005.08.002
Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH (2010) 3.3 angstrom Cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141(3):472–482
Zhang X, Ding K, Yu X, Chang W, Sun J, Zhou ZH (2015) In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527(7579):531–534. https://doi.org/10.1038/nature15767
Zhang X, Patel A, Celma CC, Yu X, Roy P, Zhou ZH (2016) Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry. Nat Struct Mol Biol 23(1):74–80. https://doi.org/10.1038/nsmb.3134
Zhou ZH, Zhang H, Jakana J, Lu XY, Zhang JQ (2003) Cytoplasmic polyhedrosis virus structure at 8 angstrom by electron cryomicroscopy: structural basis of capsid stability and mRNA processing regulation. Structure 11(6):651–663. https://doi.org/10.1016/S0969-2126(03)00091-1
Zhu B, Yang C, Liu H, Cheng L, Song F, Zeng S, Huang X, Ji G, Zhu P (2014) Identification of the active sites in the methyltransferases of a transcribing dsRNA virus. J Mol Biol 426(11):2167–2174. https://doi.org/10.1016/j.jmb.2014.03.013
Acknowledgements
This research was supported by the National Natural Science Foundation of China (12034006, 31971122, and 32071209).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Liu, H., Cheng, L. (2022). Viral Capsid and Polymerase in Reoviridae. In: Harris, J.R., Marles-Wright, J. (eds) Macromolecular Protein Complexes IV. Subcellular Biochemistry, vol 99. Springer, Cham. https://doi.org/10.1007/978-3-031-00793-4_17
Download citation
DOI: https://doi.org/10.1007/978-3-031-00793-4_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-00792-7
Online ISBN: 978-3-031-00793-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)