Abstract
Heart disease is one of the leading causes of fatality. A reliable and robust prediction system is needed for people to take preventive measures and medication beforehand and develop a proactive lifestyle accordingly. Various vital features determine human heart health, and it is important to recognize the critical ones that could be determining the chances of getting heart disease in the future. The various machine learning algorithms based on the critical features could predict heart disease more accurately. This article employs evolutionary algorithms like Genetic Algorithm (GA) and Particle Swarm Optimization (PSO) for the feature selection to improve the accuracy of machine learning algorithms further. GA and PSO combined with Naïve Bayes (NB), Support Vector Machine (SVM), and J48 have been applied for feature selection. After selecting the significant features, the effectiveness of the feature selection algorithm is evaluated by applying machine learning approaches on the complete dataset and reduced dataset. Five different machine learning approaches, viz., NB, SVM, Decision Tree (DT), Logistic Regression (LR), and Random Forest (RF) algorithm, have been used to predict heart disease and thus measure the effectiveness of the feature selection approaches. The results indicate that the GA has been the most effective algorithm for feature selection as it enhances the prediction accuracy most.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Feature selection
- Genetic Algorithm (GA)
- Heart disease prediction
- Classification
- Particle Swarm Optimization (PSO)
1 Introduction
According to the WHO, cardiovascular diseases (CVDs) are cause of death of 17.9 million people each year, which accounts for 32% of all fatalities worldwide [1]. Heart disease prediction and prevention is one of the major clinical research areas, and even a tiny improvement in this area is significant for medical science. Most of the patients suffering from CVDs are detected when the disease becomes severe. Thus, detecting CVDs at an earlier stage is necessary for saving the life of a patient. Manual observation of the patient‘s record in order to find disease is not only difficult but also takes time. In a study conducted by Richens et al. [2], it is found that machine learning-based approaches can outperform doctors. Predicting whether a person is prone to CVDs depends on many features such as Chest Pain type, Exercise-induced Angina, Thal evolved, etc. Also, using statistical threshold-based approaches for these large number of features may lead to lower accuracy. Therefore, applying machine learning approaches can significantly boost the performance of the prediction.
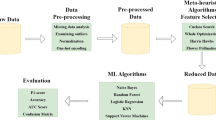
In this article, we have used the UCI Hungarian dataset [3] to find relevant features vital for detecting CVDs. The selection of features is made using feature selection approaches which select a features‘ subset by ranking them. After selecting features, the new dataset (reduced dataset) is formed by removing all the other features. Now this reduced dataset can be used for the prediction. The feature selection approach not only minimizes the dataset but also improves the performance. Feature selection is an NP-hard problem; therefore, we have applied metaheuristics for selecting the most appropriate features. This paper uses two metaheuristics approaches, i.e., PSO [4] and GA [5], along with Naïve Bayes [6], SVM [7], and J48 classifier [8] to select relevant features. Here, Naïve Bayes, SVM, and J48 are used as an objective function to compare the performance of two different subsets of features. After selecting relevant features, machine learning based approaches can be applied to evaluate the performance of feature selection approaches.
Genetic algorithm for improving feature set is a revolutionary approach and used when the traditional algorithms are not working out well. This type of approach is derived from the biological theory of evolution. GA is generally used for optimization purposes. Every new state of the instance is denoted as a chromosome made up of genes, and using these chromosomes further, a better state can be created. Genetic algorithm have four main functions as follows:
-
1.
Roulette Wheel Selection: Its a method for choosing the genomes as the next parent. Figure 1 shows an arbitrary Roulette wheel. All the genomes are given an area on the circle in proportion to their fitness value from the last state. The higher fitness value corresponds higher area for the genome that it will get on the circle/wheel. The wheel is rotated with a fixed pointer to select the genome. In programming, it is implemented with a random number generator and mod function as the pointer to select. Higher the fitness value better probability to be selected as a parent for the next state. Other methods used for selection are Rank selection, Random selection, etc.
-
2.
Crossover: It is a process to get the new genomes by crossing over the selected parents on a random basis, cutting feature subset string with a certain probability. Some of the methods are single point crossover, two-point crossover (shown in Fig. 2), uniform crossover, N-point crossover, etc.
-
3.
Mutation: It is randomly tweaking the genomes values to get some newer population. It is used to maintain some diversity. Mutation in the genetic algorithm is used with significantly lower probability; otherwise, this algorithm reduces to random searching.
-
4.
Fitness Model: Finally, using the various classification methods for the role of judging the fitness of the genomes, which is basically the average of accuracy, precision, and recall for that feature set. Using these values again, parent selection for the next generation is made using the selection method.
This article aims to improve the accuracy of prediction through a suitable feature selection approach. Many feature selection approaches have been applied in the past, but none of them have been done using evolutionary algorithms like GA and PSO. This article uses five machine learning approaches, viz., SVM, DT, NB, LR, and RF, for the CVD prediction. Nonetheless, these approaches have been applied on two datasets, i.e., the original and reduced datasets after feature selection. These approaches are compared via prediction accuracy and the four well-known metrics - precision, recall, ROC area, and accuracy have been computed for the proposed approach. The result shows that a better result is achieved by applying feature selection, and out of all the approaches, the GA-based feature selection approach performs better in terms of prediction accuracy.
This article has been written in five sections. The first section introduced the problem and discussed the probable solution. The rest of the article proceeds as follows. The second section presents the background and related work on heart disease prediction and feature selection. The third section presents the proposed work along with the explanation of the approaches applied. The fourth section presents the experimental work and results along with a brief explanation of the utilized metrics. The fifth section provides the results and analyses it. Finally, the sixth and last section concludes this article and provides direction for future work.
2 Related Work
Many research works have been done for CVD detection using machine learning approaches. Few of the recent and significant contributions have been discussed here to present the background. Kanika and Shah [9] in 2016 proposed an approach to predict CVD considering attributes such as Age, Sex, Weight, Chest pain type, etc., for the prediction. The authors applied preprocessing techniques like noise removal, discarding records with missing data, filling default values, and level-wise classification of attributes to make decisions. SVM and NB had been employed for the prediction, among which SVM is found better.
Mirmozaffari et al. [10] in 2016 used clustering algorithms for feature selection. The authors have applied clustering approaches like K-means, hierarchical clustering, and density-based clustering. The best algorithm has been chosen using multilayer filtering preprocessing and a quantitative evaluation method. The accuracies, error functions, and building times of clusters are compared. Density-Based Clustering and K-Means functioned perform quite well, based on the results.
Jabbar et al. [11] in 2017 applied the genetic algorithm for selecting the optimal feature subset for heart disease prediction. This method works well for pruning redundant and irrelevant features by actually applying every new generation to the test. In this case, KNN is used as a supervised algorithm to check the accuracy for every generation. It is repeated until the performance starts to stabilize. The authors have got 4–5% improved accuracy after applying GA based feature selection approach on various UCI repository datasets.
Gokulnath et al. [12] in 2019 used SVM as the fitness function for the GA, which performs better than the KNN and works on data that has a less linear dependency. The SVM model was 83.70% accurate when classifying CVD with the full features. However, using the framework for feature reduction, an improvement of 5% in the accuracy is seen.
Gárate-Escamila et al. [13] in 2020, the authors have used Principal Component Analysis (PCA), an unsupervised method of feature reduction (filter method), based on a non-parametric statistical technique. PCA had also been utilized by Santhanam and Ephzibah [14] in 2013. The authors used PCA on the UCI dataset (total of 297 samples, 13 input, and one output attribute), along with the regression technique used for feature reduction and ranking. The features selected using the PCA method were further utilized for classifying and predicting through regression and Feedforward neural network models.
Bashir et al. [15] in 2019 used a hybrid approach of various feature selection methods and ensemble learning with Minimum Redundancy Maximum Relevance Feature (MRMR) selection. Senthil et al. [16] in 2019 also used the hybrid approach of random forest and Linear Model for optimal performance. ANN with backpropagation is used for HRFLM.
All these research works are inspired by some natural phenomena of optimizing the performance in general and have their own sets of advantages as well as limitations. This article goes a step further and involves evolutionary algorithms like GA and PSO for feature reduction in combination with traditional machine learning algorithms. The improvement in prediction accuracy opens the door for the employment of evolutionary algorithms for feature selection before predicting a utility value.
3 Proposed Work
The objective is to improve the accuracy of classification models that predict heart disease when applied to heart datasets. For an accurate prediction model, a dataset is needed that has the best feature set, which has noise and redundancies removed. Wrapper-Method for feature selection is one of the wise choices. It is applied using the genetic algorithm. It could not perform well if proper parameters for the algorithm are not set. A fitness function plays a significant role in choosing the next generation for the algorithm. Choosing the right fitness function could improve GA further. Parameters like crossover and mutation probability values are optimized by trial and test only. The goal is to optimize feature selection using various classification functions as a fitness function in the genetic algorithm for finding a better next state to reach the optimized subset of features. Since the motive is to remove redundant features, Naïve Bayes is one of the stronger candidates. Naïve Bayes also resonates with the same principle because if there is some redundancy left in the feature, the fitness value will be meager as compared to others. Due to this, Naïve Bayes will give more accuracy than other classification methods. The proposed algorithm has been shown in Algorithm 1, and its flow has been explained in Fig. 3. The genomes for the next state will be that features-set only which has a higher fitness value. This hypothesis is further verified with experiments and other classification methods as fitness functions in the analysis section.

4 Experimental Details
WEKA is used as a tool to build a predictive model and further increase the accuracy of models. It is a software tool to analyze and work on different machine learning models. It has all the package to build a classification model based on the provided data set. In the attribute selection section, various wrapper and filter methods are available to choose an optimal subset. Ranking of attributes can be done with other methods like ReliefF algorithm, Pearson’s correlation, etc. Initially, the data set is loaded into the WEKA tool, then various filter methods and selection methods can be applied. The tool allows changing various parameters for the input of algorithms like kernel function for SVM or number of generations for Genetic algorithm. The description of the utilized dataset is provided in Subsect. 4.1 and the flow of experimental activities is discussed in Subsect. 4.2
4.1 Dataset Description
The dataset used is a standard UCI Hungarian dataset which has 14 attributes that describe various factors like age, chest pain, exercise-induced angina, etc. The description of attributes is given in Table 1. It has 294 instances. The motive is to design a framework that gives a better performance in most of the predictive models and produces higher accuracy than others. The data set is split into the training set and testing set. (70% for training) and (30% for testing).
4.2 Flow of Experimental Activities
The experiments have been done on the UCI dataset using the WEKA tool. The value for crossover probability is taken as 0.6, max generation as 20, mutation probability as 0.333, and population size as 20. Classification models like NB, SVM, etc., are deployed one by one as a fitness function to find the feature subset that works best. The flow of experimental activities carried out is as follows:
-
1.
Data Pre-processing: Data is normalized to bring every data point to the same scale for fewer errors in the classification.
-
2.
Feature Selection: An optimal subset of features is found out using various attribute selector methods, which is less redundant and more relevant for contribution in the model.
-
3.
Model Building: Various classification models are build using the curated data subset found in step-2. Models like NB, LR, SVM, DT, and RF are used for classification purposes, as they performed better in comparison to other models.
-
4.
Performance Comparison: Comparison of accuracy is made for utilized classification models with and without attribute selection methods.
5 Results and Analysis
Accuracy and performance for the classification models have been compared before and after the feature selection using various methods like ReliefF algorithm, Pearson‘s coefficient, GA and PSO. For the GA and PSO, accuracy with three “Fitness-functions” have been considered corresponding to NB, SVM, and J48. Table 2 shows the prediction accuracy for various classification models corresponding to different feature selection techniques. The first column shows the feature selection (FS) method applied before executing the classification of the dataset. All the models were tested without feature selection also. The first row depicts the prediction accuracy of models in such a condition. Rest rows show the result for other FS techniques applied, viz., ReliefF algorithm, Pearson‘s co-efficient, GA+NB, GA+SVM, GA+J48, PSO+NB, PSO+SVM, and PSO+J48.
Without feature selection, the accuracy recorded is lower. It ranges from 77% to 84%. These results have been recorded using UCI Hungarian dataset, which already has lesser noise. However, when the engaged datasets are just raw data having hundreds of attributes, accuracy will be a lot lesser, and the time for computing the models will be much more. As the number of dimensions increases, the performance would be degraded very quickly due to the overfitting of the model usually caused by unwanted attributes. Naïve Bayes gave the best result in terms of processing time. The highest accuracy value is 83.67% without feature selection, and it improved to 87.36% by just applying the feature selection. The accuracy could be improved further by combining various other optimizations.
Figure 4 shows the bar graph for the accuracy of five models utilized corresponding to the various FS techniques considered. Naïve Bayes is the best performing model in 7 out of 8 cases with all the FS techniques. It can be seen that for all the classification models, the feature selection technique of GA along with Naïve Bayes outperforms others. The best result is achieved using GA+NB as the FS technique and NB as the classification model. The results also infer that the traditional feature selection techniques only marginally improve the classification accuracy. Machine learning makes the feature selection more useful and delivers better results. The improvement in the result is obviously due to the selection of best features that are based on the Fitness-function used in the GA and other machine learning-based techniques.
GA with Naïve Bayes as a fitness function selected the relevant attributes as (3,9,11,13), which is Chest Pain type, Exercise-induced Angina, Slope of Peak exercise ST segment, and Thal. These contribute more than any other attributes, which help build a simple and accurate model with lesser features and decide better for CVD. The results also show an increase in the accuracy from around 1.5% to 6% in different classification models. Various performance measures like precision, recall, F-measure, and ROC curve area for the best-established feature selection technique (GA+NB) corresponding to all the models have been shown in Table 3. The confusion matrices for the testing phase of the NB model using no FS technique, using GA+SVM, and using GA+NB as FS techniques are shown in Fig. 4.
6 Conclusion and Future Work
This article proposed a feature selection method based on the evolutionary technique of GA. Using the Naïve Bayes as a fitness function, the feature selection method helped enhance the accuracy of CVD prediction. The experiments for prediction have been done using five machine learning techniques - NB, SVM, LR, RF, and J48. Each of the classifications had been done with eight different feature selection mechanisms. The results established NB as the best classifier employed after the feature selection through GA+NB. Hence, Naïve Bayes comes as a decent option for heuristic methods as a next-gen selector and optimizes search even further. It is also established that feature selection in CVD prediction studies has an improving role whenever the right combinations are used. In the future, newer optimizations techniques based on advanced machine learning approaches like deep neural networks could be utilized for feature selection.
References
Organization, W.H.: Cardiovascular diseases (2021). www.who.int/health-topics/cardiovascular-diseases/tab/tab/1 Accessed 02 Sept 2021
Richens, J.G., Lee, C.M., Johri, S.: Improving the accuracy of medical diagnosis with causal machine learning. Nat. Commun. 11(1), 1–9 (2020)
UCI Machine Learning Repository: Heart disease data set. archive.ics.uci.edu/ml/datasets/heart+disease Accessed 02 Sept 2021
Poli, R., Kennedy, J., Blackwell, T.: Particle swarm optimization. Swarm Intell. 1(1), 33–57 (2007)
Mirjalili, S.: Genetic algorithm. In: Evolutionary Algorithms and Neural Networks. Studies in Computational Intelligence, vol. 780. Springer, Cham (2019). https://doi.org/10.1007/978-3-319-93025-1_4
Murphy, K.P., et al.: Naive bayes classifiers. Univ. British Columbia 18(60), 1–8 (2006)
Noble, W.S.: What is a support vector machine? Nat. Biotechnol. 24(12), 1565–1567 (2006)
Mathuria, M.: Decision tree analysis on j48 algorithm for data mining. Int. J. Adv. Res. Comput. Sci. Softw. Eng. 3(6) (2013)
Kanikar, P., Shah, D.R.: Prediction of cardiovascular diseases using support vector machine and bayesien classification. Int. J. Comput. Appl. 156(2) (2016)
Mirmozaffari, M., Alinezhad, A., Gilanpour, A.: Heart disease prediction with data mining clustering algorithms. Int. J. Comput. Commun. Instrument. Eng. 4(1), 16–19 (2017)
Deekshatulu, B., Chandra, P., et al.: Classification of heart disease using k-nearest neighbor and genetic algorithm. Proc. Technol. 10, 85–94 (2013)
Gokulnath, C.B., Shantharajah, S.: An optimized feature selection based on genetic approach and support vector machine for heart disease. Cluster Comput. 22(6), 14777–14787 (2019)
Gárate-Escamila, A.K., El Hassani, A.H., Andrès, E.: Classification models for heart disease prediction using feature selection and pca. Inf. Med. Unlocked 19, 100330 (2020)
Santhanam, T., Ephzibah, E.P.: Heart disease classification using PCA and feed forward neural networks. In: Prasath, R., Kathirvalavakumar, T. (eds.) Mining Intelligence and Knowledge Exploration. LNCS, vol. 8284. Springer, Cham (2013). https://doi.org/10.1007/978-3-319-03844-5_10
Bashir, S., Khan, Z.S., Khan, F.H., Anjum, A., Bashir, K.: Improving heart disease prediction using feature selection approaches. In: 2019 16th International Bhurban Conference on Applied Sciences and Technology (IBCAST), pp. 619–623. IEEE (2019)
Mohan, S., Thirumalai, C., Srivastava, G.: Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 7, 81542–81554 (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 Springer Nature Switzerland AG
About this paper
Cite this paper
Aleem, A., Prateek, G., Kumar, N. (2022). Improving Heart Disease Prediction Using Feature Selection Through Genetic Algorithm. In: Woungang, I., Dhurandher, S.K., Pattanaik, K.K., Verma, A., Verma, P. (eds) Advanced Network Technologies and Intelligent Computing. ANTIC 2021. Communications in Computer and Information Science, vol 1534. Springer, Cham. https://doi.org/10.1007/978-3-030-96040-7_57
Download citation
DOI: https://doi.org/10.1007/978-3-030-96040-7_57
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-96039-1
Online ISBN: 978-3-030-96040-7
eBook Packages: Computer ScienceComputer Science (R0)