Abstract
The novel coronavirus disease 2019 (COVID-19) has been spreading rapidly around the world and caused a significant impact on public health and economy. However, there is still lack of studies on effectively quantifying the different lung infection areas caused by COVID-19. As a basic but challenging task of the diagnostic framework, distinguish infection areas in computed tomography (CT) images and help radiologists to determine the severity of the infection rapidly. To this end, we proposed a novel deep learning algorithm for automated infection diagnosis of multiple COVID-19 Pneumonia. Specifically, we use the aggregated residual network to learn a robust and expressive feature representation and apply the soft attention mechanism to improve the capability of the model to distinguish a variety of symptoms of the COVID-19. With a public CT image dataset, the proposed method achieves 0.91 DSC which is 14.6% higher than selected baselines. Experimental results demonstrate the outstanding performance of our proposed model for the automated segmentation of COVID-19 Chest CT images. Our study provides a promising deep learning-based segmentation tool to lay a foundation to facilitate the quantitative diagnosis of COVID-19 lung infection in CT images.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The novel coronavirus disease 2019, also known as COVID-19 outbreak first noted in Wuhan at the end of 2019, has been spreading rapidly worldwide [32]. As an infectious disease, COVID-19 is caused by severe acute respiratory syndrome coronavirus and presents with symptoms including fever, dry cough, shortness of breath, tiredness and so on. As the Jan 7th, over 87 million people around the world have been confirmed as COVID-19 infection with a case fatality rate of about 5.7% according to the statistic of World Health OrganizationFootnote 1.
So far, no specific treatment has proven effective for COVID-19. Therefore, accurate and rapid testing is extremely crucial for timely prevention of COVID-19 spread. Real-time reverse transcriptase polymerase chain reaction (RT-PCR) has been referred as the standard approach for testing COVID-19. However, RT-PCR testing is time-consuming and limited by the lack of supply test kits [17, 22]. Moreover, RT-PCR has been reported to suffer from low sensitivity and repeated checking is typically required for accurate confirmation of a COVID-19 case. This indicates that many patients will not be confirmed timely [1, 15], thereby resulting in a high risk of infecting a larger population.
In recent years, imaging technology has emerged as a promising tool for automatic quantification and diagnosis of various diseases. As a routine diagnostic tool for pneumonia, chest computed tomography (CT) imaging has been strongly recommended in suspected COVID-19 cases for both initial evaluation and follow-up. Chest CT scans play an indispensable role in detecting typical COVID-19 infections [11, 13, 19]. A systematic review [21] concluded that CT imaging of the chest was found to be sensitive when checking for COVID-19 cases even before some clinical symptoms were observed. Specifically, the typical radiographic features indicating ground class opacification, consolidation and pleural effusion have been frequently observed in the chest CT images scanned from COVID-19 patients [9, 24, 28].
Accurate segmentation of these important radiographic features is crucial for reliable quantification of COVID-19 infection in chest CT images. Segmentation of medical imaging needs to be manually annotated by well-trained expert radiologists. The rapidly increasing number of infected patients has caused a tremendous burden for radiologists and slowed down the labelling of ground-truth mask. Thus, there is an urgent need for automated segmentation of infection regions, which is a basic but arduous task in the pipeline of computer-aided disease diagnosis [6]. However, automatically delineating the infection regions from the chest CT scans is considerably challenging because of the large variation in both position and shape across different patients and low contrast of the infection regions in CT images [22].
Machine learning-based artificial intelligence provides a powerful technique for the design of data-driven methods in medical imaging analysis [24]. Developing advanced deep learning models would bring unique benefits to the rapid and automated segmentation of medical images [23]. So far, fully convolutional networks have proven superiority over other widely used registration-based approaches for segmentation [6]. In particular, U-Net models work decently well for most segmentation tasks in medical images [2, 3, 20, 22]. However, several potential limitations of U-Net have not been effectively addressed yet. For example, it is difficult for the U-net model to capture complex features such as multi-class image segmentation and recover the complex features into the segmentation image [18]. There are also a few successful applications that adopt U-Net or its variants to implement the CT image segmentation, including heart segmentation [30], liver segmentation [14], or multi-organ segmentation [5]. However, segmentation of COVID-19 infection regions with deep learning remains underexplored. The COVID-19 is a new disease but very similar to common pneumonia in the medical imaging side, which makes its accurate quantification considerably challenging. Recent advancement of the deep learning method provides heaps of insightful ideas about improving the U-Net architecture. The most popular one is the deep residual network (ResNet) [8]. ResNet provided an elegant way to stack CNN layers and demonstrate the strength when combined with U-Net [10]. On the other hand, attention was also applied to improve the U-Net and other deep learning models to boost the performance [18].
Accordingly, we propose a novel deep learning model for rapid and accurate segmentation of COVID-19 infection regions in chest CT scans. Our developed model is based on the U-Net architecture, inspired with recent advancement in the deep learning field. We exploit both the residual network and attention mechanism to improve the efficacy of the U-Net. Experimental analysis is conducted with a public CT image dataset collected from patients infected with COVID-19 to assess the efficacy of the developed model. The outstanding performance demonstrates that our study provides a promising segmentation tool for the timely and reliable quantification of lung infection, toward developing an effective pipeline for precious COVID-19 diagnosis.
Our aim is to develop a plausible segmentation model for automatically identifying the typical COVID-19 infection areas of lungs from chest CT images in order to facilitate COVID-19 diagnosis as the following: (i) our model provides a proper tool for determining the salient regions of CT images, and thus speed up the COVID-19 screening and diagnosis; (ii) comparing with existing approaches, our model is capable of producing fine-grained region of interest relating to COVID-19 infections. It would be useful to identify the different progression stages and therefore offer the groundings for further treatment plan.
The rest of the paper is summarized as follows. Our proposed new deep learning model is detailedly described in Sect. 2 Methodology, including the U-Net structure, the methods used to improve the encoder and decoder. The experimental study and performance assessment are described in Sect. 3, followed by a discussion and summary of our study.
2 Methodology
This section will introduce our proposed Residual Attention U-Net for the lung CT image segmentation in detail. We start by describing the overall structure of the developed deep learning model followed by explaining the two improved components including aggregated residual block and locality sensitive hashing attention as well as the training strategy. The overall flowchart is illustrated in Fig. 1.
Illustration of our developed residual attention U-Net model. The aggregated ResNeXt blocks are used to capture the complex feature from the original images. The left side of the U-Shape serves as encoder and the right side as decoder. Each block of the decoder receives the feature representation learned from the encoder, and concatenates them with the output of deconvolutional layer followed by LSH attention mechanism. The filtered feature representation after the attention mechanism is propagated through the skip connection.
2.1 Overview
U-Net was first proposed by Ronneberger et al. [20], which was basically a variant of fully convolutional networks (FCN) [16]. The traditional U-Net is a type of artificial neural network (ANN) containing a set of convolutional layers and deconvolutional layers to perform the task of biomedical image segmentation. The structure of U-Net is symmetric with two parts: encoder and decoder. The encoder is designed to extract the spatial features from the original medical image. The decoder is to construct the segmentation map from the extracted spatial features. The encoder follows a similar style like FCN with the combination of several convolutional layers. To be specific, the encoder consists of a sequence of blocks for down-sampling operations, with each block including two \(3\times 3\) convolution layers followed by a \(2 \times 2\) max-pooling layers with stride of 2. The number of filters in the convolutional layers is doubled after each down-sampling operation. In the end, the encoder adopts two \(3\times 3\) convolutional layers as the bridge to connect with the decoder.
Differently, the decoder is designed for up-sampling and constructing the segmentation image. The decoder first utilizes a \(2\times 2\) deconvolutional layer to up-sample the feature map generated by the encoder. The deconvolutional layer contains the transposed convolution operation and will half the number of filters in the output. It is followed by a sequence of up-sampling blocks which consists of two \(3\times 3\) convolution layers and a deconvolutional layer. Then, a \(1\times 1\) convolutional layer is used as the final layer to generate the segmentation result. The final layer adopted Sigmoid function as the activation function while all other layers used ReLU function. In addition, the U-Net concatenates part of the encoder features with the decoder. For each block in encoder, the result of the convolution before the max-pooling is transferred to decoder symmetrically. In decoder, each block receives the feature representation learned from encoder, and concatenates them with the output of deconvolutional layer. The concatenated result is then forwardly propagated to the consecutive block. This concatenation operation is useful for the decoder to capture the possible lost features by the max-pooling.
2.2 Aggregated Residual Block
As mentioned in the previous section, the U-Net only have four blocks of convolution layers to conduct the feature extraction. The conventional structure may not be sufficient for the complex medical image analysis such as multi-class image segmentation in the lung, which is the goal of this study. Although U-Net can easily separate the lung in a CT image, it may have limited ability to distinguish the difference infection regions of the lung which infected by COVID-19. Based on this case, the deeper network is needed with more layers, especially for the encoding process. However, when deeper network converges, a problem will be exposed: with increasing of the network depth, accuracy gets very high and then decreases rapidly. This problem is defined as the degradation problem [7]. He et al. proposed the ResNet [8] to mitigate the effect of network degradation on model learning. ResNet utilizes a skip connection with residual learning to overcome the degradation and avoid estimating a large number of parameters generated by the convolutional layer. The typical ResNet block can be defined as \(F(i) = \sum _{j=1}^D w_ji_j\) where \(i = [i_1, i_2, \cdots , i_D]\) and \(W= [w_1, w_2, \cdots , w_D]\) is the trainable weight for the weight layer. Different from the U-Net that concatenates the features map into the decoding process, ResNet adopts the shortcut to add the identity into the output of each block. The stacked residual block can better learn the latent representation of the input CT image. However, the ResNet normally have millions of parameters and may lead to under-fitting or over-fitting due to the model’s complexity. Regarding this, Xie et al. proposed the Aggregated Residual Network(ResNeXt) and showed that increasing the cardinality was more useful than increasing the depth or width [29]. The cardinality is defined as the set of the Aggregated Residual transformations with the formulation as \(F(i) = \sum _{j=1}^C \mathcal {T}_j(i)\) where C is the number of residual transformation to be aggregated and \(\mathcal {T}_j(i)\) can be any function. Considering a simple neuron, \(\mathcal {T}_j\) should be a transformation which projects i into a low-dimensional embedding ideally and then transforming it. Accordingly, we can extend it into the residual function \(y = \sum _{j=1}^C \mathcal {T}_j(i) + i\) where the y is the output. The ResNeXt block is visualized in Fig. 2. The weight layer’s size is smaller than ResNet as ResNeXt uses the cardinality to reduce the number of layers but keep the performance. One thing is worth to mention that the three small blocks inside the ResNeXt block need to have the same topology, in other words, they should be topologically equivalent.
An example ResNeXt Block. The variable i is the 256-dimension representation of the input image or features map. C represents the cardinality, which indicates that number of blocks inside. \(256,1\times 1,4\) represents the size of input image, filter and output’s channel. The dimension is determined by the input.
Similar to the ResNet, after a sequence of blocks, the learned features are fed into a global averaging pooling layer to generate the final feature map. Different from the convolutional layers and normal pooling layers, the global averaging pooling layers take the average of feature maps derived by all blocks. It can sum up all the spatial information captured by each step and is generally more robust than directly making the spatial transformation to the input. Mathematically, we can treat the global averaging pooling layer as a structural regularizer that is helpful for driving the desired feature maps [31].
Importantly, instead of using the encoder in the U-Net, our proposed deep learning model adopts the ResNeXt block (see Fig. 2) to conduct the features extraction. The ResNeXt provides a solution which can prevent the network from going very deeper but remain the performance. In addition, the training cost of ResNeXt is better than ResNet.
2.3 Locality Sensitive Hashing Attention
The decoder in U-Net is used to up-sampling the extracted feature map to generate the segmentation image. However, due to the capability of the convolutional neural network, it may not able to capture the complex features if the network structure is not deep enough. In recent years, transformers [26] have gained increasingly interest. The key to success is the attention mechanism [27]. Attention includes two different mechanisms: soft attention and hard attention. We adopt soft attention to improve model learning. Different from hard attention, soft attention can let model focus on each pixel’s relative position, but hard attention only can focus on the absolute position. There are two different types of soft attention: Scaled Dot-Product Attention and Multi-Head Attention as shown in Fig. 3. The scaled dot-product attention takes the inputs including a query Q, a key \(K_n\) of the n-dimension and value \(V_m\) of the m-dimension. The dot-product attention is defined as follows:
where \(K_n^T\) represents to the transpose of the matrix \(K_n\) and \(\sqrt{n}\) is a scaling factor. The softmax function \(\sigma (\mathbf{z} )\) with \(\mathbf{z} = [z_1, \cdots , z_n]\in \mathbb {R}^n\) is given by:
Vaswani et al. [27] mentioned that, performing the different linear project of the queries Q, keys K and values V in parallel h layers will benefit the attention score calculation. We can assume that Q, K and V have been linearly projected to \(d_k, d_k, d_v\) dimensions, respectively. It is worth noting that these linear projections are different and learnable. On each projection p, we have a pair of the query, key and value \(Q_p, K_p, V_p\) to conduct the attention calculation in parallel, which results in a \(d_v\)-dimensional output. The calculation can be formulated as:
where the the projections \(W_i^Q \in \mathbb {R}^{d_{model} \times d_k}\), \(W_i^K \in \mathbb {R}^{d_{model} \times d_k}\), \(W_i^V\in \mathbb {R}^{d_{model} \times d_v}\)are parameter matrices and \(W^O \in \mathbb {R}^{d_{model} \times hd_v}\) is the weight matrix used to balance the results of h layers.
However, the multi-head attention is memory inefficient due to the size of Q, K and V. Assume that the Q, K, V have the shape \([|batch|, length, d_{model}]\) where \(|\cdot |\) represents the size of the variable. The term \(QK^T\) will produce a tensor in shape \([length, length, d_{model}]\). Given the standard image size, the length \(\times \) length will take most of the memory. Kitaev et al. [12] proposed a Locality Sensitive Hashing(LSH) based Attention to address this issue. Firstly, we rewire the basic attention formula into each query position i, j in the partition form:
where the function z is the partition function, \(P_i\) is the set which query position i attends to and k, q, v are elements of the matrix K, Q, V. During model training, we normally conduct the batching and assume that there is a larger set \(P_i^L = \{0,1,\cdots ,l\}\supseteq P_i\) without considering elements not in \(P_i\):
Then, with a hash function \(h(\cdot )\): \(h(q_i) = h(k_j)\), we can get \(P_i\) as:
In order to guarantee that the number of keys can uniquely match with the number of quires, we need to ensure that \(h(q_i) = h(k_i)\) where \(k_i = \frac{q_i}{\Vert q_i\Vert }\). During the hashing process, some similar items may fall in different buckets because of the hashing. The multi-round hashing provides an effective way to overcome this issue. Suppose there is \(n_r\) round, and each round has different hash functions \(\{h_1, \cdots , h_{n_r}\}\), so we have:
Considering the batching case, we need to get the \(P_i^L\) for each round g:
where \(m=\frac{2l}{n_r}\). The last step is to calculate the LSH attention score in parallel. With the above formula , we can derive:
2.4 Training Strategy
The task of the lung CT image segmentation is to predict if each pixel of the given image belongs to a predefined class or the background. Therefore, the traditional medical image segmentation problem comes to a binary pixel-wise classification problem. However, in this study, we are focusing on the multi-class image segmentation, which can be concluded as a multi-classes pixel-wise classification. Hence, we choose the multi-class cross-entropy as the loss function:
where \(y_{o,c}\) is a binary value which is used to compare the correct class c and observation class o, \(p_{o,c}\) is a probability of the observation o to correct class c and M is the number of classes.
3 Experiment and Evolution Results
3.1 Data Description
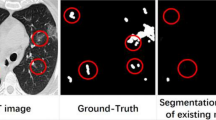
Visualization of segmentation results. The images (a) and (e) show the preprocessed chest CT images of two scans. The images (b) and (f) are the ground-truth masks for these two scans, where the yellow represents the consolidation, blue represents pleural effusion and green corresponds to ground-glass opacities. The images (c) and (g) are the segmentation results generated by our model where the blue represents the consolidation and brown represents the pleural effusion and sky-blue for the ground-glass opacities. The images (d) and (h) are the outputs of the U-Net. In order to make the visualization clear, we choose the light grey as the colour for the background segment. (Color figure online)
We used COVID-19 CT images collected by the Italian Society of Medical and Interventional Radiology (SIRM)Footnote 2 for our experimental study. The dataset included 110 axial CT images collected from 43 patients. These images were reversely intensity-normalized by taking RGB-values from the JPG-images from areas of air (either externally from the patient or in the trachea) and fat (subcutaneous fat from the chest wall or pericardial fat) and used to establish the unified Houndsfield Unit-scale (the air was normalized to –1000, fat to –100). The ground-truth segmentation was done by a trained radiologist using MedSegFootnote 3 with three labels: 1 = ground class opacification, 2 = consolidations, and 3 = pleural effusions. We split the dataset in both patient level and CT image levels to demonstrate the superior of our method. These data are publicly availableFootnote 4.
3.2 Data Preprocessing and Augmentation
The original CT images have a size of 512 \(\times \) 512 in matrix form. We use the opencv2Footnote 5 to transfer the matrix into gray-scale image to remove some random noises.
As our model is based on deep learning, the number of samples will affect the performance significantly. Consider the size of the dataset, data augmentation is necessary for training the neural network to achieve high generalizability. Our study implements parameterized transformations to realize data augmentation in the training set in this study. We rotate the existing images 90\(^\circ \), 180\(^\circ \) and 270\(^\circ \) to generate another sets of examples. We can easily generate the corresponding mask by rotating with the same degrees. Scaling has some property with the rotation, so we just scale the image to 0.5 and 1.5 separately to generate another sets of images and its corresponding masks.
3.3 Experiments Setting and Measure Metrics
For the model training, we use the Adma as the optimizer. For a fair comparison, we trained our model and the U-Net with the default parameter in 100 epochs, with learning rate 0.0001 and 3 as the kernel size. Both models are trained under data augmentation and non-augmentation cases. We conducted the experimental analyses on our server consisting of two 12-core/ 24-thread Intel(R) Xeon(R) CPU E5-2697 v2 CPUs, 6 NVIDIA TITAN X Pascal GPUs, 2 NVIDIA TITAN RTX, a total 768 GiB memory. In a segmentation task, especially for the multi-class image segmentation, the target area of interest may take a trivial part of the whole image. Thus, we adopt the Dice Score, accuracy, precision, recall, F1 score and hausdorff distance(HD) as the measure metrics. The dice score is defined as:
where X, Y are two sets, and \(|\cdot |\) calculates the number of elements in a set. Assume Y is the correct result of the test and X is the predicted result. We conduct the experimental comparison based on 10-fold cross-validation for performance assessment in patient level and image level. And the measure metric for multi-class classification can be calculated by averaging several binary classifications in the same task.
3.4 Results
The Fig. 4 provides two examples about the result images which have data augmentation. The Table 1 shows the measure metric for our proposed model and the U-Net in with data augmentation case and no data augmentation case. Based on this table, we can easily find that our proposed method is out-performed than U-Net that the improvement is at least 10% in all three measure metrics. As shown in Fig. 4(h), we find that the original U-Net almost failed to do the segmentation. The most possible reason is that the range of interest is very small, and the U-Net does not have enough capability to distinguish those trivial difference.
3.5 Ablation Study
In addition to the above-mentioned results, we are also interested in the effectiveness of each component in the proposed model. Accordingly, we conduct the ablation study about the ResNeXt and Attention separately to investigate how these components would affect the segmentation performance. To ensure a fair experimental comparison, we conduct the ablation study in the same experiment environment with our main experiments presented in Sect. 3.3. We implement the ablation study on two variants of our model: Model without Attention and Model without ResNeXt. Our model without ResNeXt is similar with literature [18]. We just use the M-R to represent it. The results are summarized in Table 3, where M-A represents the model without attention and M-R represents the model without ResNeXt block. We can observe that both the attention and ResNeXt blocks play important roles in our model and contribute to derive improved segmentation performance in comparison with U-Net (Tables 3 and 4).
4 Discussion and Conclusions
Up to now, the most common screening tool for COVID-19 is CT imaging. It can help the community to accelerate the speed of diagnosing and accurately evaluate the severity of COVID-19 [22]. In this paper, we presented a novel deep learning-based algorithm for automated segmentation of COVID-19 CT images, and its proved that such algorithm is plausible and superior comparing to a series of baselines. We proposed a modified U-Net model by exploiting the residual network to enhance the feature extraction. An efficient attention mechanism was further embedded in the decoding process to generate high-quality multi-class segmentation results. Our method gained more than 10% improvement in multi-class segmentation when comparing against U-Net and a set of baselines.
A recent study shows that the early detection of COVID-19 is very important [4]. If the infection in chest CT image can be detected at an early stage, the patients would have a higher chance to survive [25]. Our study provides an effective tool for the radiologist to precisely determine the lung’s infection percentage and diagnose the progression of COVID-19. It also shed some light on how deep learning can revolutionize the diagnosis and treatment in the midst of COVID-19.
Our future work would be generalizing the proposed model into a wider range of practical scenarios, such as facilitating with diagnosing more types of diseases from CT images. In particular, in the case of a new disease, such as the coronavirus, the amount of ground truth data is usually limited given the difficulty of data acquisition and annotation. The model is capable of generalizing and adapting itself using only a few available ground-truth samples. Another line of future work lies in the interpretability, which is especially critical for the medical domain applications. Although deep learning is widely accepted to its limitation in interpretability, the attention mechanism we proposed in this work can produce the interpretation of internal decision process at some levels. To gain deeper scientific insights, we will keep working along with this direction and explore the hybrid attention model for generating meaningfully semantic explanations.
References
Ai, T., et al.: Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology 200642 (2020)
Alom, M.Z., Hasan, M., Yakopcic, C., Taha, T.M., Asari, V.K.: Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation (2018). arXiv:1802.06955
Baumgartner, C.F., Koch, L.M., Pollefeys, M., Konukoglu, E., et al.: An exploration of 2D and 3D deep learning techniques for cardiac MR image segmentation. In: Pop, M. (ed.) STACOM 2017. LNCS, vol. 10663, pp. 111–119. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75541-0_12
Bernheim, A., et al.: Chest ct findings in coronavirus disease-19 (covid-19): relationship to duration of infection. Radiology, 200463 (2020)
Dong, X., et al.: Automatic multiorgan segmentation in thorax ct images using u-net-gan. Med. Phys. 46(5), 2157–2168 (2019)
Gaál, G., Maga, B., Lukács, A.: Attention u-net based adversarial architectures for chest x-ray lung segmentation (2020). arXiv:2003.10304
He, K., Sun, J.: Convolutional neural networks at constrained time cost. In: IEEE CVPR, pp. 5353–5360 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE CVPR, pp. 770–778 (2016)
Huang, C., et al.: Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. The Lancet 395(10223), 497–506 (2020)
Ibtehaz, N., Rahman, M.S.: Multiresunet: rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw. 121, 74–87 (2020)
Jiang, X., et al.: Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Comput. Mater. Continua 62(3), 537–551 (2020)
Kitaev, N., Kaiser, L., Levskaya, A.: Reformer: the efficient transformer. In: International Conference on Learning Representations (2020). https://openreview.net/forum?id=rkgNKkHtvB
Li, Y., Xia, L.: Coronavirus disease 2019 (covid-19): role of chest ct in diagnosis and management. Am. J. Roentgenol 214, 1–7 (2020)
Liu, Z., et al.: Liver ct sequence segmentation based with improved u-net and graph cut. Expert Syst. Appl. 126, 54–63 (2019)
Long, C., et al.: Diagnosis of the coronavirus disease (covid-19): rrt-pcr or ct? Eur. J. Radiol. 126, 108961 (2020)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE CVPR, pp. 3431–3440 (2015)
Narin, A., Kaya, C., Pamuk, Z.: Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks (2020). arXiv:2003.10849
Oktay, O., et al.: Attention u-net: Learning where to look for the pancreas (2018). arXiv:1804.03999
Raptis, C.A., et al.: Chest ct and coronavirus disease (covid-19): a critical review of the literature to date. Am. J. Roentgenol. 215(1), 1–4 (2020)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Salehi, S., Abedi, A., Balakrishnan, S., Gholamrezanezhad, A.: Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am. J. Roentgenol 215, 1–7 (2020)
Shan, F., et al.: Lung infection quantification of covid-19 in ct images with deep learning (2020). arXiv:2003.04655
Shen, D., Wu, G., Suk, H.I.: Deep learning in medical image analysis. Ann. Rev. Biomed. Eng. 19, 221–248 (2017)
Shi, F., et al.: Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19 (2020). arXiv:2004.02731
Song, F., et al.: Emerging 2019 novel coronavirus (2019-ncov) pneumonia. Radiology, 200274 (2020)
Sutskever, I., Vinyals, O., Le, Q.V.: Sequence to sequence learning with neural networks. In: NIPS, pp. 3104–3112 (2014)
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, pp. 5998–6008 (2017)
Wang, L.S., Wang, Y.R., Ye, D.W., Liu, Q.Q.: A review of the 2019 novel coronavirus (covid-19) based on current evidence. Int. J. Antimicrob. Agents 55, 105948 (2020)
Xie, S., Girshick, R., Dollár, P., Tu, Z., He, K.: Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE CVPR, pp. 1492–1500 (2017)
Ye, C., Wang, W., Zhang, S., Wang, K.: Multi-depth fusion network for whole-heart ct image segmentation. IEEE Access 7, 23421–23429 (2019)
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., Torralba, A.: Learning deep features for discriminative localization. In: Proceedings of the IEEE CVPR, pp. 2921–2929 (2016)
Zhu, N., et al.: A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 382, 727–733 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, X., Yao, L., Zhang, Y. (2021). RAU: An Interpretable Automatic Infection Diagnosis of COVID-19 Pneumonia with Residual Attention U-Net. In: Zhang, W., Zou, L., Maamar, Z., Chen, L. (eds) Web Information Systems Engineering – WISE 2021. WISE 2021. Lecture Notes in Computer Science(), vol 13081. Springer, Cham. https://doi.org/10.1007/978-3-030-91560-5_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-91560-5_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-91559-9
Online ISBN: 978-3-030-91560-5
eBook Packages: Computer ScienceComputer Science (R0)