Abstract
Breast Cancer is one of the most prevalent malignancies amongst men and women. Currently, it has become the common health issue all over the world with its drastic increase in death rate every year. Early detection of breast cancer provides high treatment efficiency and better healing chances. The main contribution of this paper is to find the model which is most accurate for predicting the type of tumour cell (Benign or Malignant). ANOVA f-test Feature Selection is applied to the Wisconsin Breast Cancer dataset to select the subsets of input features that are most relevant to the target variable. We compared various machine learning algorithms like Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Decision Tree, Logistic Regression, Gaussian Naive Bayes, Random Forest and XG boost Classifier algorithms. We obtained the highest accuracy of 98.25% in XGboost classifier as it uses ensemble techniques and is a very powerful classifier.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
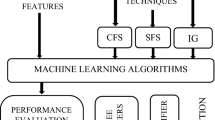
Globally, Breast Cancer has remained the second most common disease which causes death among women [1]. Breast cancer classification of tumors accurately helps in curing the disease at the early stage itself. Breast cancer tumors are mainly classified into malignant (Cancerous) and Benign (Non-Cancerous). To discriminate amid these tumors, doctors require a reliable and safe diagnostic system. However sometimes, even the specialists find it challenging to identify the tumors correctly. So the early prediction of the disease is the need of the hour to reduce the risk of death in this case. Breast cancer malignancy is the most prevalent disease among women; it has consistently high mortality and frequency rates. We collected data from Wisconsin breast cancer dataset and applied ANOVA f-test Feature Selection method to decrease the high data dimensionality of the feature space before the classification process. It also helped in selecting the subsets of input features that are more relevant to the target variable so that we can get better results. After computing all the models, we compared them based on eight parameters such as Accuracy, Precision, Recall, F1-Score, Sensitivity, Specificity, False Negative Rate and False Positive Rate (Fig. 1).
2 Related Works
This research paper has gathered the information from various papers who researched the prediction of breast cancer on different datasets, including Wisconsin breast cancer dataset. Paper by Anusha [7], compared Support Vector Machine (SVM), Decision Tree (CART), Naive Bayes (NB) and k Nearest Neighbours (kNN) based on accuracy, Another paper by Naveen [6], compared ensemble machine learning models which gave 100% accuracy in KNN and decision tree on Coimbra breast cancer split train-test dataset in a ratio of 90:10. Fabiano Teixeira [8] evaluated different classification methods: Multilayer Perceptron, Decision Tree, Random Forest, Support Vector Machine and Deep Neural Network and got a good performance in accuracy level of 92%. Gilbert Gutabaga Hungilo [12] in his paper compared AdaBoost, Random Forest, and XGBoostwhose result indicates that the random forest is the best predictive model and has the following performance measure, accuracy 97%, sensitivity 96%, and specificity 96%. Another paper by Quang H. Nguyen [14] analysed prediction models using Feature Selection and Ensemble Voting which returned with the accuracy of at least 98%.
Till now, people compared the three-four algorithms [2, 5, 6] of their choice mainly based on the accuracy. Although accuracy is the main factor, they could get the highest accuracy, not more than 97% even after applying feature selection. So in this paper, We compared seven commonly used algorithms such as Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Decision Tree, Logistic Regression, Gaussian Naive Bayes, Random Forest and XG-boost.
3 Proposed Methodology
We have collected the Breast Cancer malignant growth instances from the benchmark database Wisconsin Breast Cancer diagnosis data set. We compared various ML algorithms like Decision Tree, K Nearest Neighbor, Gaussian Naive Bayes, Random Forests, Logistic Regression, Support Vector Machine. In this paper, we use named XG Boost classifier, which is an ensemble learning algorithm (aggregate of predictive powers of multiple algorithms) for acquiring the best results. Below is the flowchart representing the proposed model Fig. 2.
3.1 Data Collection
Data is collected from the Wisconsin Breast Cancer data set publicly available in UCI Machine Learning Repository [17]. Data set contains 569 occurrences with 30 attributes. It consists of 32 segments, with ‘ID number’, ‘diagnosis’ result (“Benign” or “Malignant”), and the ‘mean’, ‘standard deviation’ and the ‘mean of the worst estimations’ of 10 features. The class distribution is shown in the Fig. 3.
3.2 Data Processing
Every row in the dataset which is incomplete or has some missing attribute values is removed, and attributes such as ‘id’ are also deleted as it is of no use.
3.3 Data Manipulation
As the target attribute ‘diagnosis’ is a categorical data which machine can’t read, so is converted into numerical data.
3.4 Data Visualization
After data collection and manipulation, we performed data visualization of all the remaining 31 attributes to identify areas that needed attention or improvement. We can easily interpret data using Fig. 4.
3.5 Feature Selection
Feature selection, it is the most important as the final result values are dependent on the pattern of feature selection. So we choose the feature in such a manner so that we get the best accuracy and other parameters. In this paper, we used ANOVA f-test Feature Selection method.
ANOVA F-Test Feature Selection. ANOVA stands for Analysis of Variance. It is a popular numerical feature selection method. It compares mean between more than two groups. An F-test is a class of statistical tests that compute the relationship between the values of the variances, e.g., the variance of two different samples or the variance that is explained and unexplained by a statistical test here referring to as the ANOVA f-test. We select the features having the best variance using an object and applying the fit. Transform over the features and target variable. The static F is given as Eq. 1
Z-Score Normalization. The purpose of normalisation is to equalise the scale of all data points such that each attribute is equally important. The Min-Max normalization fail in handling outliers. This outlier issue can be solved by using Z-score normalization. The formula for this technique is given below Eq. 2:
Where \(\mu \) and \(\sigma \) are mean value and standard deviation value of the feature respectively. The value will be normalised to 0 if it is exactly equal to the mean of all the values of the feature. It will be a negative number if it is below the mean, and a positive number if it is above the mean.
3.6 Data Splitting and Feature Scaling
In this paper, we use 75% training data and 25% of data for testing. Since attributes vary in magnitudes, units and range, we have scaled features using z-score normalization to bring all characteristics to a similar degree level. The feature distribution after feature scaling is shown in the Fig. 4.
4 Background
This paper aims to select the machine learning algorithm that best suits for developing our model to the fullest. Machine learning algorithms classified into two types: Supervised and Unsupervised learning. We need Supervised learning for our breast cancer prediction model [8] (Fig. 5).
4.1 Supervised Machine Learning
[7] In this learning, we train the machine using data which is “labelled”. This learning algorithm predicts outcomes after learning from the labelled training data. This learning uses regression and other classification techniques to develop predictive models.
-
Logistic Regression: This algorithm exhibits a direct connection between a dependant (y) and at least one independent (y)’s factor. Since linear regression uncovers a linear relationship, it decides how the dependent variable’s value changes with the independent factor’s value.
-
KNN: KNN algorithm which is the short form for K-nearest neighbours, utilizes the given information to predict and allots the new information point dependent on how intently it coordinates with the focuses in the preparation set, i.e., depending on the similarity.
-
SVM: An SVM model is a data characterization algorithm for predictive analysis, that allocates new data components to one of the known gatherings; it works by defining a straight boundary between two classes. The data points that fall on the right side are considered one class, and the opposite side is regarded as the other.
-
Gaussian Naive Bayes: Gaussian Naive Bayes is widely used as a classifier and also with few alterations it can be used for regression too. In this algorithm, values are distributed based on Gaussian distribution. And this distribution is also called as a normal distribution. The classification is done based on Bayes Theorem.
-
Decision Tree: This algorithm identifies different ways to split data. It is used for both classification and regression. Using tree representation, it tries to resolve the error.
-
Random Forest: Random Forest classifier assembles different decision trees which represent different factual probabilities. Then it combines these decision trees to acquire a steady and precise prediction as shown pictorially in Fig. 10. Trees mapped to a solitary tree known as Classification and Regression Trees (CART) model. This calculation utilized for both regression and classification issues.
-
XGBoost: XGboost or eXtreme gradient Boosting algorithm is the application of gradient boosted decision trees developed for high speed and better performance. It is an ensemble learning method. Implies that each new model is prepared to rectify the error of the previous model, and the arrangement gets halted when there is no further improvement. In boosting the base learners are weak learners and do not have high predictive power, whereas the final one is a strong learner with high predictive power. The strong learner is a combination of the weak learners that provide some information for prediction (Fig. 6).
5 Experimental Results
In our results, we are considering the confusion matrix which gives a conclusion of the results of the classification problem prediction. It shows how much your model or algorithm classifier is in dilemma when we make predictions. We have also found out Accuracy, Precision, Recall, F1-Score, Sensitivity, Specificity, False Negative Rate and False positive Rate of all the algorithms [6].
-
Accuracy: XGBoost gives the Highest accuracy of 98.25% which is best for our model, whereas Decision tree gives the lowest accuracy of 88.81% as shown in Fig. 7 (Table 1).
$$\begin{aligned} Accuracy = \frac{TP + TN}{TP + FP + TN + FN} \end{aligned}$$(3)Here, TP = True Positive
TN = True Negative
FP = False Positive
FN = False Negative
-
Precision: SVM and Random forest shows the highest precision value of 96.23% and XGBoost has a precision of 95.83%, whereas the decision tree has the lowest precision of 86.23% as shown in Fig. 8 (Table 2).
$$\begin{aligned} Precision = \frac{TP}{TP + FP} \end{aligned}$$(4) -
Recall: XGBoost has highest recall value of 100%, other algorithms also gave good results but decision tree shows the lowest value of 78.46% as shown in Fig. 9 (Table 3).
$$\begin{aligned} Recall = \frac{TP}{TP + FN} \end{aligned}$$(5) -
F1-Score: XGBoost gives the Highest F1 score of 97.87% whereas Decision tree gives the lowest accuracy of 86.44% as shown in Fig. 10 (Table 4).
$$\begin{aligned} F1 = \frac{2 \times ( Precision \times Recall)}{Precision + Recall} \end{aligned}$$(6) -
Sensitivity: XGBoost shows the excellent and highest sensitivity value of 100%, whereas decision tree shows the lowest value of 78.46% as shown in Fig. 11.
$$\begin{aligned} Sensitivity = \frac{TP}{TP + FN} \end{aligned}$$(7) -
Specificity: Support Vector Machine gave the highest specificity of 97.75%, others also gave good results but Gaussian Naive Bayes shows the lowest value of 93.41%. XGBoost has shown 97.06% specificity as shown in Fig. 12 (Tables 5 and 6).
$$\begin{aligned} Specificity = \frac{TN}{FP + TN} \end{aligned}$$(8) -
False Negative Rate: XGBoost shown the lowest False Negative of 0.00% which is great and Decision tree showed the highest value of 0.21% as shown in Fig. 13.
$$\begin{aligned} False\ Negative\ Rate = 100 *\frac{FN}{FN + TP} \end{aligned}$$(9) -
False Positive Rate: XGBoost, Random Forest, Decision Tree and Support Vector Machine (SVM) have shown the least false positive rate of 0.02% whereas Gaussian Naive Bayes gave a maximum of 0.06% as shown in Fig. 14 (Tables 7 and 8).
$$\begin{aligned} False\ Positive\ Rate = 100 *\frac{FP}{FP + TN} \end{aligned}$$(10)
6 Results and Conclusion
This research analysis offered a new plan of action applying Feature Selection based on ANOVA F-test, z-score normalization, and XGBoost classifier algorithm for Prediction of breast cancer. This proposed action offers the following advantages: Improved classification accuracy, better recall, boosted sensitivity, increased precision, reducing the false-positive rate and false-negative rate. The classification accuracy of the new strategy is obtained as 98.25%, the recall is 100%, the f1-score is 97.87%, the sensitivity is 100%, the precision is 95.83%, false positive is 0.02%, and a false-negative rate as 0.00%.
The result of this new strategy using a hybrid approach of ‘feature selection and XGBoost’ was compared to predict breast malignancy with distinct algorithms. It yielded a more reliable performance in terms of various parameters. As XGBoost is an ensemble machine learning algorithm (Ensemble model is a combination of multiple models). It is able to give the best results. So in this paper we successfully created a prediction model for breast cancer.
For future research, we intend to execute the Feature selection based upon differential evolution algorithm to provide reasonably practical and more precise results. Furthermore, we also plan to deploy the same using different datasets and compare the performance of the hybrid approach using optimal features.
References
Breast cancer: prevention and control (2020). https://www.who.int/cancer/detection/breastcancer/en/index1.html
Sinha, N.K., Khulal, M., Gurung, M., Lal, A.: Developing a web based system for breast cancer prediction using XGboost classifier. Int. J. Eng. Res. Technol. (IJERT) 9, June 2020. http://dx.doi.org/10.17577/IJERTV9IS060612
Jadhav, M., Thakkar, Z., Chawan, P.: Breast cancer prediction using supervised machine learning algorithms. Int. Res. J. Eng. Technol. (IRJET) 07(08), October 2019. e-ISSN: 2395–0056
Karthikeyan, B.: Breast cancer detection using machine learning. Int. J. Adv. Trends Comput. Sci. Eng. 9, 981–984 (2020). https://doi.org/10.30534/ijatcse/2020/12922020
Shravya, C., Pravalika, K., Subhani, S.: Prediction of breast cancer using super-vised machine learning techniques. Int. J. Innov. Technol. Explor. Eng. (IJITEE) 8, 1106–1110 (2019)
Naveen, Sharma, R.K., Nair, A.R.: Efficient breast cancer prediction using ensemble machine learning models. In: 2019 4th International Conference on Recent Trends on Electronics, Information, Communication and Technology (RTEICT), Bangalore, India, pp. 100–104 (2019). https://doi.org/10.1109/RTEICT46194.2019.9016968
Bharat, A., Pooja, N., Reddy, R.A.: Using machine learning algorithms for breast cancer risk prediction and diagnosis. In: 2018 3rd International Conference on Circuits, Control, Communication and Computing (I4C), Bangalore, India, pp. 1–4 (2018). https://doi.org/10.1109/CIMCA.2018.8739696
Teixeira, F., Montenegro, J.L.Z., da Costa, C.A., da Rosa Righi, R.: An analysis of machine learning classifiers in breast cancer diagnosis. In: XLV Latin American Computing Conference (CLEI), Panama, pp. 1–10 (2019). https://doi.org/10.1109/CLEI47609.2019.235094
Das, S., Biswas, D.: Prediction of breast cancer using ensemble learning. In: 2019 5th International Conference on Advances in Electrical Engineering (ICAEE), Dhaka, Bangladesh, pp. 804–808 (2019). https://doi.org/10.1109/ICAEE48663.2019.8975544
Chandrasegar, T., Nikhilesh Vutukuri, S.B.: Optimized machine learning model using Decision Tree for cancer prediction. In: Innovations in Power and Advanced Computing Technologies (i-PACT), Vellore, India, pp. 1–4 (2019). https://doi.org/10.1109/i-PACT44901.2019.8960129
Dhanya, R., Paul, I.R., Sindhu Akula, S., Sivakumar, M., Nair, J.J.: A comparative study for breast cancer prediction using machine learning and feature selection. In: 2019 International Conference on Intelligent Computing and Control Systems (ICCS), Madurai, India, pp. 1049–1055 (2019). https://doi.org/10.1109/ICCS45141.2019.9065563
Hungilo, G.G., Emmanuel, G., Emanuel, A.W.R.: Performance evaluation of ensembles algorithms in prediction of breast cancer. In: International Biomedical Instrumentation and Technology Conference (IBITeC). Special Region of Yogyakarta, Indonesia, pp. 74–79 (2019). https://doi.org/10.1109/IBITeC46597.2019.9091718
Laghmati, S., Tmiri, A., Cherradi, B.: Machine Learning based system for prediction of breast cancer severity. In: 2019 International Conference on Wireless Networks and Mobile Communications (WINCOM), Fez, Morocco, pp. 1–5 (2019). https://doi.org/10.1109/WINCOM47513.2019.8942575
Nguyen, Q.H., et al.: Breast cancer prediction using feature selection and ensemble voting. In: 2019 International Conference on System Science and Engineering (ICSSE), Dong Hoi, Vietnam, pp. 250–254 (2019). https://doi.org/10.1109/ICSSE.2019.8823106
Suryachandra, P., Reddy, P.V.S.: Comparison of machine learning algorithms for breast cancer. In: 2016 International Conference on Inventive Computation Technologies (ICICT), Coimbatore, pp. 1–6 (2016). https://doi.org/10.1109/INVENTIVE.2016.7830090
Liu, B., et al.: Comparison of machine learning classifiers for breast cancer diagnosis based on feature selection. In: 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Miyazaki, Japan, pp. 4399–4404 (2018). https://doi.org/10.1109/SMC.2018.00743
https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Likitha, B., Nakka, J., Verma, J., Naik, N.S. (2021). Prediction of Breast Cancer Analysis Using Machine Learning Algorithms and XGBoost Technique. In: Venugopal, K.R., Shenoy, P.D., Buyya, R., Patnaik, L.M., Iyengar, S.S. (eds) Data Science and Computational Intelligence. ICInPro 2021. Communications in Computer and Information Science, vol 1483. Springer, Cham. https://doi.org/10.1007/978-3-030-91244-4_24
Download citation
DOI: https://doi.org/10.1007/978-3-030-91244-4_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-91243-7
Online ISBN: 978-3-030-91244-4
eBook Packages: Computer ScienceComputer Science (R0)