Abstract
The most common type of multivariate data collected in ecology is also one of the most challenging types to analyse—when some abundance-related measure (e.g. counts, presence–absence, biomass) is simultaneously collected for all taxa or species encountered in a sample, as in Exercises 14.1–14.3. The rest of the book will focus on the analysis of these multivariate abundances.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
The most common type of multivariate data collected in ecology is also one of the most challenging types to analyse—when some abundance-related measure (e.g. counts, presence–absence, biomass) is simultaneously collected for all taxa or species encountered in a sample, as in Exercises 14.1–14.3. The rest of the book will focus on the analysis of these multivariate abundances .
In his revegetation study (Exercise 10.3), Anthony classified anything that fell into his pitfall traps into orders, and thus counted the abundance of each of 24 invertebrate orders across 10 sites. He wants to know:
Is there evidence of a change in invertebrate communities due to revegetation efforts?
What type of response variable(s) does he have? How should Anthony analyse his data?
FormalPara Exercise 14.2: Invertebrates Settling on SeaweedIn Exercise 1.13, David and Alistair looked at invertebrate epifauna settling on algal beds (seaweed) with different levels of isolation (0, 2, or 10 m buffer) from each other, at two sampling times (5 and 10 weeks). They observed presence–absence patterns of 16 different types of invertebrate (across 10 replicates).
They would like to know if there is any evidence of a difference in invertebrate presence–absence patterns with distance of isolation. How should they analyse the data?
FormalPara Exercise 14.3: Do Offshore Wind Farms Affect Fish Communities?As in Exercise 10.2, Lena studied the effects of an offshore wind farm on fish communities by collecting paired data before and after wind farm construction, at 36 stations in each of 3 zones (wind farm, north, and south). She counted how many fish were caught at each station, classified into 16 different taxa.
Lena wants to know if there is any evidence of a change in fish communities at wind farm stations, compared to others, following construction of the wind farm. How should she analyse the data?
This type of data goes by lots of other names—“species by site data” , “community composition data” , even sometimes “multivariate ecological data” , which sounds a bit too broad, given that there are other types of multivariate data used in ecology (such as allometric data, see Chap. 13). The term multivariate abundances is intended to put the focus on the following key statistical properties.
- Multivariate::
-
There are many correlated response variables, sometimes more variables than there are observations :
- Abundance::
-
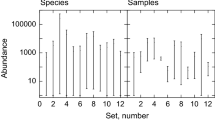
Abundance or presence–absence data usually exhibits a strong mean–variance relationship, as in Fig. 14.1.
You need to account for both properties in your analysis.
Multivariate abundance data are especially common in ecology, probably for two reasons. Firstly, it is often of interest to say something collectively about a community, e.g. in environmental impact assessment, we want to know if there is any impact of some event on the ecological community. Secondly, this sort of data arises naturally in sampling—even when you’re interested in some target species, others will often be collected incidentally along the way, e.g. pitfall traps set specifically for ants will inevitably capture a range of other types of invertebrate also. So even when they are interested in something else, many ecologists end up with multivariate abundances and feel like they should do something with them. In this second case we do not have a good reason to analyse multivariate abundances. Only bother with multivariate analysis if the primary research question of interest is multivariate, i.e. if a community or assemblage of species is of primary interest. Don’t go multivariate just because you have the data.
There are a few different types of questions one might wish to answer using multivariate abundances. The most common type of question, as in each of Exercises 14.1–14.3, is whether or not the community is associated with some predictor (or set of predictors) characterising aspects of the environment—whether looking at the effect on a community of an experimental treatment, testing for environmental impact (Exercise 14.3), or something else again.
In Chap. 11 some multivariate regression techniques were introduced, and model-based inference was used to study the effects of predictors on response. If there were only a few taxa in the community, those methods would be applicable. But (as flagged in Table 11.1) a key challenge with multivariate abundances is that typically there are many responses. It’s called biodiversity for a reason! There are lots of different types of organisms out there. The methods discussed in this chapter are types of high-dimensional regression , intended for when you have many responses, but if you only have a few responses, you might be better off back in Chap. 11. High-dimensional regression is technically difficult and is currently a fast-moving field.
In this chapter we will use design-based inference (as in Chap. 9). Design-based inference has been common in ecology for this sort of problem for a long time as a way to handle the multivariate property, and the focus in this chapter will be on applying design-based inference to models that appropriately account for the mean–variance relationship in data (to also handle the abundance property). There are some potential analysis options beyond design-based inference, which we will discuss later.
Multivariate abundance data (also “species by site data”, “community composition data”, and so forth) has two key properties: a multivariate property, that there are many correlated response variables, and an abundance property, a strong mean–variance relationship. It is important to account for both properties in your analysis.
1 Generalised Estimating Equations
Generalised estimating equations (GEEs, Liang & Zeger, 1986; Zeger & Liang, 1986) are a fast way to fit a model to correlated counts, compared to hierarchical models (Chaps. 11–12). Design-based inference techniques like the bootstrap (Chap. 9) tend to be computationally intensive, especially when applied to many correlated response variables, GEEs are a better choice when planning to use design-based inference. Parameters from GEEs are also slightly easier to interpret than those of a hierarchical model, because they specify marginal rather than conditional models, so parameters in the mean model have direct implications for mean abundance (see Maths Box 11.4 for problems with marginal interpretation of hierarchical parameters).
GEEs are ad hoc extensions of equations used to estimate parameters in a GLM, defined by taking the estimating equations from a GLM, forcing them to be multivariate (Maths Box 14.1), and hoping for the best. An assumption about the correlation structure of the data is required for GEEs. Independence of responses is commonly assumed, sometimes called independence estimating equations , which simplifies estimation to a GLM problem, and then correlation in the data is adjusted for later (using “sandwich estimators” for standard errors, Hardin & Hilbe, 2002).
Maths Box 14.1:
 Generalised Estimating Equations
Generalised Estimating Equations
As in Maths Box 10.2, maximum likelihood is typically used to estimate parameters in a GLM, which ends up meaning that we need to find the values of parameters that solve the following score equations:
(where \(\boldsymbol {d}_i=\frac {\partial \mu _i}{\partial \boldsymbol {\beta }}=\frac {x_i}{g^\prime (\mu _i)}\), as in Eq. 10.4). The GEE approach involves taking these estimating equations and making them multivariate, by replacing the response with a vector of correlated responses, replacing the variance with a variance–covariance matrix, and hoping for the best:
V (μ i) is now a variance–covariance matrix, requiring a “working correlation” structure to be specified.
A similar fitting algorithm is used as for GLMs, which means that GEEs are typically relatively quick to fit.
Notice that whereas the score equations for GLMs are derived as the gradient of the log-likelihood function, that is not how GEEs are derived. In fact, unless responses are assumed to be normally distributed, or they are assumed to be independent of each other, there is no GEE likelihood function. This complicates inference, because standard likelihood-based tools such as AIC, BIC, and likelihood ratio tests cannot be used because we cannot calculate a GEE likelihood.
Some difficulties arise when using GEEs, because of the fact that they are motivated from equations for estimating parameters, rather than from a parametric model for the data. GEEs define marginal models for data, but (usually) not a joint model, with the estimating equations no longer corresponding to the derivative of any known likelihood function. One difficulty that this creates is that we cannot simulate data under a GEE “model”. A second difficulty is that without a likelihood, a likelihood ratio statistic can’t be constructed. Instead, another member of the “Holy Trinity of Statistics” (Rao, 1973) could be used for inference, a Wald or score statistic. Maybe this should be called the Destiny’s Child of Statistics (Maths Box 14.2), because while the Wald and score statistics are good performers in their own right, the likelihood ratio statistic is the main star (the Beyoncé). Wald statistics have been met previously, with the output from summary for most R objects returning Wald statistics. These statistics are based on parameter estimates under the alternative hypothesis, by testing if parameter estimates are significantly different from what is expected under the null hypothesis. A score statistic (or Rao’s score statistic) is based on the estimating equations themselves, exploiting the fact that plausible estimates of parameters should give values of the estimating equations that are close to zero. Specifically, parameter estimates under the null hypothesis are plugged into the estimating equations under the alternative hypothesis, and a statistic constructed to test for evidence that the expression on the right-hand side of Eq. 14.1 is significantly different from zero.
Maths Box 14.2: The Destiny’s Child of Statistics
Consider testing for evidence against a null model (\(\mathcal {M}_0\)) with parameter θ 0, in favour of a more general alternative model (\(\mathcal {M}_1\)) with parameter \(\hat {\theta }_1\). There are three main types of likelihood-based test statistics, the Destiny’s Child of Statistics (or the Holy Trinity of Statistics, according to Rao, 1973). These can be visualised in a plot of the log-likelihood function against θ:

The

\( -2\log \varLambda (\mathcal {M}_0,\mathcal {M}_1) = 2\ell _{\mathcal {M}_1}(\hat {\theta }_1;\mathbf {y}) - 2\ell _{\mathcal {M}_0}(\theta _0;\mathbf {y})\) focuses on whether the likelihoods of the two models are significantly different (vertical axis).
The

focuses on the parameter (horizontal axis) of \(\mathcal {M}_1\) and whether \(\hat {\theta }_1\) is significantly far from what would be expected under \(\mathcal {M}_0\), using \(\frac {\hat {\theta }_1-\theta _0}{\hat {\sigma }_{\hat {\theta }_1}}\).
The

focuses on the score equation u(θ), the gradient of the log-likelihood at \(\mathcal {M}_0\). The likelihood should be nearly flat for a model that fits the data well. So if \(\mathcal {M}_0\) is the correct model, u(θ 0) should be near zero, and we can use as a test statistic \(\frac {u(\theta _0)}{\hat {\sigma }_{u(\theta _0)}}\).
In GEEs, u(θ) is defined, hence θ can be estimated, but the likelihood is not defined (unless assuming all variables are independent). So for correlated counts we can use GEEs to calculate a Wald or score statistic, but not a likelihood ratio statistic. Sorry, no Beyoncé!
2 Design-Based Inference Using GEEs
A simple GEE model for abundance at site i of taxon j is
An offset or a row effect term can be added to account for variation in sampling intensity, which is useful for diversity partitioning, as discussed later (Sect. 14.3).
A working correlation matrix (R) is needed, specifying how abundances are associated with each other across taxa. The simplest approach is to use independence estimating equations, ignoring correlation for the purposes of estimation (assuming R = I, a diagonal matrix of ones, with all correlations equal to zero), so that the model simplifies to fitting a GLM separately to each response variable. This is pretty much the simplest possible model that will account for the abundance property, and by choosing a simple model, we hope that resampling won’t be computationally prohibitive.
We need to handle the multivariate property of the data to make valid multivariate inferences about the effects of predictors (environmental associations), and this can be done by resampling rows of data. Resampling rows keeps site abundances for all taxa together in resamples, to preserve the correlation between taxa. This is a form of block resampling (Sect. 9.7.1). Correlation can also be accounted for in constructing the test statistic.
The manyglm function in the mvabund package was written to carry out the preceding operation, and it behaves a lot like glm, so it is relatively easy to use if you are familiar with the methods of Chap. 10 (Code Box 14.1). It does, however, take longer to run (for anova or summary), so sometimes you have to be patient. Unlike the glm function, manyglm defaults to family="negative.binomial". This is done because the package was designed to analyse multivariate abundances (hence the name), and these are most commonly available as overdispersed counts.
Code Box 14.1: Using mvabund to Test for an Effect of Revegetation in Exercise 12.2
> library(ecostats) > library(mvabund) > data(reveg) > reveg$abundMV=mvabund(reveg$abund) > ft_reveg=manyglm(abundMV~treatment+offset(log(pitfalls)), family="negative.binomial", data=reveg) # offset included as in Ex 10.9 > anova(ft_reveg) Time elapsed: 0 hr 0 min 9 sec Analysis of Deviance Table Model: manyglm(formula = abundRe ~ treatment + offset(log(pitfalls)), Model: family = "negative.binomial") Multivariate test: Res.Df Df.diff Dev Pr(>Dev) (Intercept) 9 treatment 8 1 78.25 0.024 ∗ --- Signif. codes: 0 ‘∗∗∗' 0.001 ‘∗∗' 0.01 ‘∗' 0.05 ‘.' 0.1 ‘ ' 1 Arguments: Test statistics calculated assuming uncorrelated response (for faster computation)P-value calculated using 999 iterations via PIT-trap resampling.
Exercise 14.4: Testing for an Effect of Isolation on Invertebrates in Seaweed
Consider David and Alistair’s study of invertebrate epifauna settling on algal beds with different levels of isolation (0, 2, or 10 m buffer) at different sampling times (5 and 10 weeks), with varying seaweed biomass in each patch.
What sort of model is appropriate for this dataset? Fit this model and call it ft_epiAlt and run anova(ft_epiAlt). (This might take a couple of minutes to run.)
Now fit a model under the null hypothesis that there is no effect of distance of isolation, and call it ft_epiNull. Run anova(ft_epiNull, ft_epiAlt). This second anova took much less time to fit—why?
Is there evidence of an effect of distance of isolation on presence–absence patterns in the invertebrate community?
2.1 Mind Your Ps and Qs
The manyglm function makes the same assumptions as for GLMs, plus a correlation assumption:
-
1.
The observed y ij-values are independent across observations (across i), after conditioning on x i.
-
2.
The y ij-values come from a known distribution (from the exponential family) with known mean–variance relationship V (μ ij) .
-
3.
There is a straight-line relationship between some known function of the mean of y j and x
$$\displaystyle \begin{aligned} g(\mu_{ij}) = \beta_{0j} + {\mathbf{x}}_i^T \boldsymbol{\beta}_j \end{aligned}$$ -
4.
Residuals have a constant correlation matrix across observations.
Check assumptions 2 and 3 as for GLMs—using Dunn-Smyth residual plots (Code Box 14.2). As usual, the plot function could be used to construct residual plots for mvabund objects, or the plotenvelope function could be used to add simulation envelopes capturing the range of variation to expect if assumptions were satisfied. For large datasets, plotenvelope could take a long time to run, unless using sim.method="stand.norm" (as in Code Box 14.3) to simulate standard normal random variables, instead of simulating new responses and refitting the model for each. As usual, we want no trend in the residuals vs fits plot and would be particularly worried by a U shape (non-linearity) or a fan shape (problems with the mean–variance assumption, as in Code Box 14.3), and in the normal quantile plot we expect residuals to stay close to the trend line. The meanvar.plot function can also be used to plot sample variances against sample means, by taxon and optionally by treatment (Code Box 14.3).
Code Box 14.2: Checking Assumptions for the Revegetation Model of Code Box 14.1
par(mfrow=c(1,3)) ft_reveg=manyglm(abundMV~treatment,offset=log(pitfalls), family="negative.binomial", data=reveg) plotenvelope(ft_reveg, which=1:3)

What do you reckon?
Code Box 14.3: Checking Mean–Variance Assumptions for a Poisson Revegetation Model
ft_revegP=manyglm(abundMV~treatment, offset=log(pitfalls), family="poisson", data=reveg) par(mfrow=c(1,3)) plotenvelope(ft_revegP, which=1:3, sim.method="stand.norm")

Plotting sample variances against sample means for each taxon and treatment:
meanvar.plot(reveg$abundMV~reveg$treatment) abline(a=0,b=1,col="darkgreen")

How’s the Poisson assumption looking?
Exercise 14.5: Checking Assumptions for the Habitat Configuration Data
Consider the multivariate analysis of the habitat configuration study (Exercise 14.4).
What assumptions were made?
Where possible, check these assumptions.
Do the assumptions seem reasonable?
Exercise 14.6: Checking Assumptions for Wind Farm Data
Consider Lena’s offshore wind farm study (Exercise 14.3). Fit an appropriate model to the data. Make sure you include a Station main effect (to account for the paired sampling design).
What assumptions were made?
Where possible, check these assumptions.
Do the assumptions seem reasonable? In particular, think about whether there is evidence that the counts are overdispersed compared to the Poisson.
2.2 Test Statistics Accounting for Correlation
When using anova on a manyglm object, a “sum-of-LR” statistic (Warton et al., 2012b) is the default—a likelihood ratio statistic computed separately for each taxon, then summed across taxa for a community-level measure. By summing across taxa, the sum-of-LR statistic is calculated assuming independent responses (and the job of accounting for the multivariate property is left to row resampling).
If you want to account for correlation between variables in the test statistic, you need to change both the type of test statistic (via the test argument) and the assumed correlation structure (via the cor.type argument). The test statistic to use is a score (test="score") or Wald (test="wald") statistic, as described previously. The type of correlation structure to assume is controlled by the cor.type argument. Options currently available include the following:
- cor.type="I" :
-
(Default) Assumes independence for test statistic calculation—sums across taxa for a faster fit.
- cor.type="R" :
-
Assumes unstructured correlation between all variables, i.e. estimates a separate correlation coefficient between each pair of responses. Not recommended if there are many variables compared to the number of observations, because it will become numerically unstable (Warton, 2008).
- cor.type="shrink" :
-
A middle option between the previous two. Use this to account for correlation unless you have only a few variables. This method shrinks an unstructured correlation matrix towards the matrix you would use if assuming independence, using the data to work out how far to move towards independence (Warton, 2011, as in Code Box 14.4).
Note that even if you ignore correlation when constructing a test statistic, it is accounted for in the P-value because rows of observations are resampled. This means the procedure is valid even if the independence assumption used in constructing the test statistic is wrong. But recall that valid≠ efficient —while this procedure is valid, the main risk when using this statistic is that if there are correlated variables you can miss structure in the data (the scenario depicted in Fig. 11.1). So one approach, as when making inferences from multivariate linear models (Sect. 11.2), is to try a couple of different test statistics, in case the structure in the data is captured by one of these but not another. This approach would be especially advisable if using Wald statistics, because they can be insensitive when many predicted values for a taxon are zero (as in Chap. 10).
Code Box 14.4: A manyglm Analysis of Revegetation Data Using a Statistic Accounting for Correlation
> anova(ft_reveg,test="wald",cor.type="shrink") Time elapsed: 0 hr 0 min 6 sec Analysis of Variance Table Model: abundMV ~ treatment + offset(log(pitfalls)) Multivariate test: Res.Df Df.diff wald Pr(>wald) (Intercept) 9 treatment 8 1 8.698 0.039 ∗ --- Signif. codes: 0 ‘∗∗∗' 0.001 ‘∗∗' 0.01 ‘∗' 0.05 ‘.' 0.1 ‘ ' 1 Arguments: Test statistics calculated assuming correlated response via ridge regularization P-value calculated using 999 iterations via PIT-trap resampling.
You can also use the summary function for manyglm objects, but the results aren’t quite as trustworthy as for anova. The reason is that resamples are taken under the alternative hypothesis for summary, where there is a greater chance of fitted values being zero, especially for rarer taxa (e.g. if there is a treatment combination in which a taxon is never present). Abundances don’t resample well if their predicted mean is zero.
2.3 Computation Time
One major difference between glm and manyglm is in computation time. Analysing your data using glm is near instantaneous, unless you have a very large dataset. But in Code Box 14.1, an anova call to a manyglm object took almost 10 s, on a small dataset with 24 responses variables. Bigger datasets will take minutes, hours, or sometimes days! The main problem is that resampling is computationally intensive —by default, this function will fit a glm to each response variable 1000 times, so there are 24,000 GLMs in total. If an individual GLM were to take 1s to fit, then fitting 24,000 of them would take almost 7 h (fortunately, that would only happen for a pretty large dataset).
For large datasets, try setting nBoot=49 or 99 to get a faster but less precise answer. Then scale it up to around 999 when you need a final answer for publication. You can also use the show.time="all" argument to get updates every 100 bootstrap samples, e.g. anova(ft_reveg,nBoot=499,show.time="all").
If you are dealing with long computation times, parallel computing is a solution to this problem—if you have 4 computing cores to run an analysis on, you could send 250 resamples to each core then combine, to cut computation down four-fold. If you have access to a computational cluster, you could even send 1000 separate jobs to 1000 nodes, each consisting of just one resample, and reduce a hard problem from days to minutes. By default, mvabund will split operations up across however many nodes are available to it at the time.
Another issue to consider with long computation times is whether some of the taxa can be removed from the analysis. Most datasets contain many taxa that are observed very few times (e.g. singletons, seen only once, and doubletons or tripletons), and these typically provide very little information to the analysis, while slowing computation times. The slowdown due to rarer taxa can be considerable because they are more difficult to fit models to. So an obvious approach to consider is removing rarer taxa from the analysis—this rarely results in loss of signal from the data but removes a lot of noise, so typically you will get faster (and better) results from removing rare species (as in Exercise 14.7). It is worth exploring this idea for yourself and seeing what effect removing rarer taxa has on results. Removing species seen three or fewer times is usually a pretty safe bet.
Exercise 14.7: Testing for an Effect of Offshore Wind Farms (Slowly)
Consider Lena’s offshore wind farm study (Exercise 14.3). The data contain a total of 179 rows of data and a Station main effect (to account for the paired sampling) that has lots of terms in it. Analysis will take a while.
Fit models under the null and alternative hypotheses of interest. Run an anova to compare these 2 models, with just 19 bootstrap resamples, to estimate computation time.
Remove zerotons and singletons from the dataset using
windMV = mvabund(windFarms$abund[,colSums(windFarms$abund>0)>1])
Now fit a model to this new response variable, again with just 19 bootstrap resamples. Did this run take less time? How do the results compare? How long do you think it would take to fit a model with 999 bootstrap resamples, for an accurate P-value?
2.4 The manyany Function
The manyglm function is currently limited to just a few choices of family and link function to do with count or presence–absence data, focusing on distributions like the negative binomial, Poisson, and binomial. An extension of it is the manyany function (Code Box 14.5), which allows you to fit (in principle) any univariate function to each column of data and use anova to resample rows to compare two competing models. The cost of this added flexibility is that this function is very slow—manyglm was coded in C (which is much faster than R) and optimised for speed, but manyany was not.
Code Box 14.5: Analysing Ordinal Data from Habitat Configuration Study Using manyany
Regression of ordinal data is not currently available in the manyglm function, but it can be achieved using manyany:
> habOrd = counts = as.matrix( round(seaweed[,6:21]∗seaweed$Wmass)) > habOrd[counts>0 & counts<10] = 1 > habOrd[counts>=10] = 2 > library(ordinal) > summary(habOrd) # Amphipods are all "2" which would return an error in clm > habOrd=habOrd[,-1] #remove Amphipods > manyOrd=manyany(habOrd~Dist∗Time∗Size,"clm",data=seaweed) > manyOrdNull=manyany(habOrd~Time∗Size,"clm",data=seaweed) > anova(manyOrdNull, manyOrd) LR Pr(>LR) sum-of-LR 101.1 0.12 --- Signif. codes: 0 ‘∗∗∗' 0.001 ‘∗∗' 0.01 ‘∗' 0.05 ‘.' 0.1 ‘ ' 1
What hypothesis has been tested here? Is there any evidence against it?
3 Compositional Change and Partitioning Effects on α- and β-Diversity
In the foregoing analyses, the focus was on modelling mean abundance, but sometimes we wish to focus on relative abundance or composition . The main reason for wanting to do this is if there are changes in sampling intensity for reasons that can’t be directly measured. For example, pitfall traps are often set in terrestrial systems to catch insects, but some will be more effective than others because of factors unrelated to the abundance of invertebrates, such as how well pitfall traps were placed and the extent to which ground vegetation impedes movement in the vicinity of the trap (Greenslade, 1964). A key point here is that some of the variation in abundance measurements is due to changes in the way the sample was taken rather than being due to changes in the study organisms—variation is explained by the sampling mechanism as well as by ecological mechanisms. In this situation, only relative abundance across taxa is of interest, after controlling for variation in sampling intensity. In principle, it is straightforward to study relative abundances using a model-based approach—we simply add a term to the model to account for variation in abundance across samples. So the model for abundance at site i of taxon j becomes

The new term in the model, α 0i, accounts for variation across samples in total abundance, so that remaining terms in the model can focus on change in relative abundance. The optional term \(\boldsymbol {x}_i^\prime \boldsymbol {\alpha }\) quantifies how much of this variation in total abundance can be explained by environmental variables. The terms in Eq. 14.3 have thus been partitioned into those studying total abundance (the α) and those studying relative abundance (the β). Put another way, the effects of environmental variables have been split into main effects (the α) and their interactions with taxa (the β, which take different values for different taxa). The model needs additional constraints for all the terms to be estimable, which R handles automatically (e.g. by setting α 01 = 0).
Key Point
Often the primary research interest is in studying the effects of environmental variables on community composition or species turnover. This is especially useful if some variation in abundance is explained by the sampling mechanism, as well as ecological mechanisms. This can be accounted for in a multivariate analysis by adding a “row effect” to the model, a term that takes a different value for each sample according to its total abundance. Thus, all remaining terms in the model estimate compositional effects (β-diversity), after controlling for effects on total abundance (α-diversity).
A classic paper by Whittaker (1972) described the idea of partitioning species diversity into α-diversity , “the community’s richness of species”, and β-diversity, the “extent of differentiation in communities along habitat gradients”.Footnote 1 The parameters of Eq. 14.3 have been written as either α or β to emphasise their connections to α-diversity and β-diversity. Specifically, larger α coefficients in Eq. 14.3 correspond to samples or environmental variables that have larger effects on abundance of all species (and hence on species richness), whereas larger β coefficients in Eq. 14.3 correspond to taxa that differ from the overall α-trend in terms of how their abundance relates to the environmental variable, implying greater species turnover along the gradient. The use of statistical models to tease apart effects of environmental variables on α- and β-diversity is a new idea that has a lot of potential.
A model along the lines of Eq. 14.3 can be readily fitted via manyglm using the composition argument, as in Code Box 14.6. The manyany function also has a composition argument, which behaves similarly.
Code Box 14.6: A Compositional Analysis of Anthony’s Revegetation Data
> ft_comp=manyglm(abundMV~treatment+offset(log(pitfalls)), data=reveg, composition=TRUE) > anova(ft_comp,nBoot=99) Time elapsed: 0 hr 0 min 21 sec Model: abundMV ~ cols + treatment + offset(log(pitfalls)) + rows + cols:(treatment + offset(log(pitfalls))) Res.Df Df.diff Dev Pr(>Dev) (Intercept) 239 cols 216 23 361.2 0.01 ∗∗ treatment 215 1 14.1 0.01 ∗∗ rows 206 9 25.5 0.02 ∗ cols:treatment 184 23 56.7 0.01 ∗∗
In this model, coefficients of cols, treatment, rows, and cols:treatment correspond in Eq. 14.3 to β 0j, α, α 0i, and β j, respectively.
Which term measures the effect of treatment on relative abundance? Is there evidence of an effect on relative abundance?
Fitting models using composition=TRUE is currently computationally slow. Data are re-expressed in long format (along the lines of Code Box 11.5) to fit a single GLM as in Eq. 14.3, with abundance treated as a univariate response and row and column factors used as predictors to distinguish different samples and responses (respectively). This is fitted using the manyglm computational machinery, but keeping all observations from the same site together in resamples, as previously. A limitation of this approach is that the model is much slower to fit than in short format and may not fit at all for large datasets because of substantial inefficiencies that a long format introduces. In particular, the design matrix storing x variables has p times as many rows in it and nearly p times as many columns! Computation times for anova can be reduced by using it to compare just the null and alternative models for the test of interest, manually fitted in long format, as in Code Box 14.7. Further limitations, related to treating the response as univariate, are that the model is unable to handle correlation across responses (so cor.type="I" is the only option for compositional analyses), multiple testing across responses (see following section) is unavailable, and any overdispersion parameters in the model are assumed constant across responses (i.e. for each j, we assumed ϕ j = ϕ in Code Box 14.6). Writing faster and more flexible algorithms for this sort of model is possible and a worthwhile avenue for future research, whether using sparse design matrices (Bates & Maechler, 2015) or short format.
Code Box 14.7: A Faster Compositional Analysis of Anthony’s Revegetation Data
In Code Box 14.6, every term in ft_comp was tested, even though only the last term was of interest. The data used to fit this model are stored in long format in ft_comp$data, so we can use this data frame to specify the precise null and alternative models we want to test so as to save computation time:
> ft_null = manyglm(abundMV~cols+rows+offset(log(pitfalls)), data=ft_comp$data) > ft_alt = manyglm(abundMV~cols+rows+treatment:cols +offset(log(pitfalls)), data=ft_comp$data) > anova(ft_null, ft_alt, nBoot=99, block=ft_comp$rows) Time elapsed: 0 hr 0 min 5 sec ft_null: abundMV ~ cols + rows + offset(log(pitfalls)) ft_alt: abundMV ~ cols + rows + treatment:cols + offset(log(pitfalls)) Res.Df Df.diff Dev Pr(>Dev) ft_null 207 ft_alt 184 23 56.74 0.01 ∗∗ --- Signif. codes: 0 ‘∗∗∗' 0.001 ‘∗∗' 0.01 ‘∗' 0.05 ‘.' 0.1 ‘ ' 1 Arguments: P-value calculated using 99 iterations via PIT-trap resampling.
The results are the same (same test statistic and similar P-value) but take about a quarter of the computation time. Notice also that a main effect for the treatment term was left out of the formulas. Why didn’t exclusion of the treatment term change the answer?
3.1 Quick-and-Dirty Approach Using Offsets
For large datasets it may not be practical to convert data to long format, in which case the composition=TRUE argument is not a practical option. In this situation a so-called quick-and-dirty alternative for count data is to calculate the quantity
and use this as an offset (Code Box 14.8). The term \(\hat {\mu }\) refers to the predicted value for y ij from the model that would be fitted if you were to exclude the compositional term. The s i estimate the row effect for observation i as the difference in log-row sums between the data and what would be expected for a model without row effects. The best way to use this approach would be to calculate a separate offset for each model being compared, as in Code Box 14.8. If there was already an offset in the model, it stays there, and we now add a second offset as well (Code Box 14.8).
Code Box 14.8: Quick-and-Dirty Compositional Analysis of Anthony’s Revegetation Data
We can approximate the row effects α 0i using the expression in Eq. 14.4:
> # calculate null model offset and fit quick-and-dirty null model > ft_reveg0 = manyglm(abundMV~1+offset(log(pitfalls)), data=reveg) > QDrows0 = log(rowSums(reveg$abundMV)) - log(rowSums(fitted(ft_reveg0))) > ft_row0 = manyglm(abundMV~1+offset(log(pitfalls))+ offset(QDrows0), data=reveg) > # calculate alt model offset and fit quick-and-dirty alt model > ft_reveg = manyglm(abundMV~treatment+offset(log(pitfalls)), data=reveg) > QDrows = log(rowSums(reveg$abundMV)) - log(rowSums(fitted(ft_reveg))) > ft_row = manyglm(abundMV~treatment+offset(log(pitfalls))+ offset(QDrows), data=reveg) > anova(ft_row0,ft_row) Time elapsed: 0 hr 0 min 7 sec Analysis of Deviance Table ft_row0: abundMV ~ 1 + offset(QDrows0) ft_row: abundMV ~ treatment + offset(QDrows) Multivariate test: Res.Df Df.diff Dev Pr(>Dev) ft_row0 9 ft_row 8 1 50.26 0.048 ∗ --- Signif. codes: 0 ‘∗∗∗' 0.001 ‘∗∗' 0.01 ‘∗' 0.05 ‘.' 0.1 ‘ ' 1 Arguments: Test statistics calculated assuming uncorrelated response (for faster computation)P-value calculated using 999 iterations via PIT-trap resampling.
This was over 10 times quicker than Code Box 14.7 (note that it used 10 times as many resamples), but the results are slightly different—the test statistic is slightly smaller and the P-value larger. Why do you think this might be the case?
The approach of Code Box 14.8 is quick (because it uses short format), and we will call it quick-and-dirty for two reasons. Firstly, unless counts are Poisson, it does not use the maximum likelihood estimator of α 0i, so in this sense it can be considered sub-optimal. Secondly, when resampling, the offset is not re-estimated for each resample when it should be, so P-values become more approximate. Simulations suggest this approach is conservative, so perhaps the main cost of using the quick-and-dirty approach is loss of power—test statistics are typically slightly smaller and less significant, as in Code Box 14.8. Thus the composition argument should be preferred, where practical.
Key Point
A key challenge in any multivariate analysis is understanding what the main story is and communicating it in a simple way. Some tools that can help with this include the following:
-
Identifying a short list of indicator taxa that capture most of the effect.
-
Visualisation tools—maybe an ordination, but especially looking for ways to see the key results using the raw data.
4 In Which Taxa Is There an Effect?
In Code Box 14.1, Anthony established that revegetation does affect invertebrate communities. This means that somewhere in the invertebrate community, there is evidence that some invertebrates responded to revegetation—maybe all taxa responded, or maybe just one did. The next step is to think about which of the response variables most strongly express the revegetation effect. This can be done by adding a p.uni argument as in Code Box 14.9.
Code Box 14.9: Posthoc Testing for Bush Regeneration Data
> an_reveg = anova(ft_reveg,p.uni="adjusted") > an_reveg Analysis of Deviance Table Model: manyglm(formula = dat ~ treatment + offset(log(pitfalls)), family = "negative.binomial") Multivariate test: Res.Df Df.diff Dev Pr(>Dev) (Intercept) 9 treatment 8 1 78.25 0.022 ∗ Univariate Tests: Acarina Amphipoda Araneae Blattodea Dev Pr(>Dev) Dev Pr(>Dev) Dev Pr(>Dev) Dev (Intercept) treatment 8.538 0.208 9.363 0.172 0.493 0.979 10.679 Coleoptera Collembola Dermaptera Pr(>Dev) Dev Pr(>Dev) Dev Pr(>Dev) Dev (Intercept) treatment 0.117 9.741 0.151 6.786 0.307 0.196
⋮
The p.uni argument allows univariate test statistics to be stored for each response variable, with P-values from separate tests reported for each response, to identify taxa in which there is statistical evidence of an association with predictors. The p.uni="adjusted" argument uses multiple testing, adjusting P-values to control family-wise Type I error, so that the chance of a false positive is controlled jointly across all responses (e.g. for each term in the model, if there were no effect of that term, there would be a 10% chance, at most, of at least one response having a P-value less than 0.1). The more response variables there are, the bigger the P-value adjustment and the harder it is to get significant P-values, after adjusting for multiple testing. It is not uncommon to get global significance but no univariate significance—e.g. Anthony has good evidence of an effect on invertebrate communities but can’t point to any individual taxon as being significant. This comes back to one of the original arguments for why we do multivariate analysis (Chap. 11, introduction)—it is more efficient statistically than separately analysing each response one at a time.
The size of univariate test statistics can be used as a guide to indicator taxa , those that contribute most to a significant multivariate result. A test statistic constructed assuming independence (cor.type="I") is a sum of univariate test statistics for each response, so it is straightforward to work out what fraction of it is due to any given subset of taxa. For example, the “top 5” taxa from Anthony’s revegetation study account for more than half of the treatment effect (Code Box 14.10). This type of approach offers a short list of taxa to focus on when studying the nature of a treatment effect, which can be done by studying their coefficients (Code Box 14.10), plotting the subset, and (especially for smaller datasets) looking at the raw data.
Code Box 14.10: Exploring Indicator Taxa Most Strongly Associated with Treatment Effect in Anthony’s Revegetation Data
Firstly, sorting univariate test statistics and viewing the top 5:
> sortedRevegStats = sort(an_reveg$uni.test[2,],decreasing=T, index.return=T) > sortedRevegStats$x[1:5] Blattodea Coleoptera Amphipoda Acarina Collembola 10.679374 9.741038 9.362519 8.537903 6.785946
How much of the overall treatment effect is due to these five orders of invertebrates? The multivariate test statistic across all invertebrates, stored in an$table[2,3], is 78.25. Thus, the proportion of the difference in deviance due to the top 5 taxa is
> sum(sortedRevegStats$x[1:5])/an_reveg$table[2,3] [1] 0.5764636
So about 58% of the change in deviance is due to these five orders.
The model coefficients and corresponding standard errors for these five orders are as follows:
> coef(ft_reveg)[,sortedRevegStats$ix[1:5]] Blattodea Coleoptera Amphipoda Acarina Collembola (Intercept) -0.3566749 -1.609438 -16.42495 1.064711 5.056246 treatmentReveg -3.3068867 5.009950 19.42990 2.518570 2.045361 > ft_reveg$stderr[,sortedRevegStats$ix[1:5]] Blattodea Coleoptera Amphipoda Acarina Collembola (Intercept) 0.3779645 1.004969 707.1068 0.5171539 0.4879159 treatmentReveg 1.0690450 1.066918 707.1069 0.5713194 0.5453801
Note that a log-linear model was fitted, so the exponent of coefficients tells us the proportional change when moving from the control to the treatment group. For example, cockroach abundance (Blattodea) decreased by a factor of about e 3.3 = 27 on revegetation, while the other four orders increased in abundance with revegetation. Can you construct an approximate 95% confidence interval for the change in abundance of beetles ( Coleoptera ) on revegetation?
Exercise 14.8: Indicator Species for Offshore Wind Farms?
Which fish species are most strongly associated with offshore wind farms in Lena’s study?
Reanalyse the data to obtain univariate test statistics and univariate P-values that have been adjusted for multiple testing. Recall that the key term of interest, in terms of measuring the effects of offshore wind farms on fish communities, is the interaction between Zone and Year. Is there evidence that any species clearly have a Zone:Year interaction, after adjusting for multiple testing? What proportion of the total Zone:Year effect is attributable to these potential indicator species?
Plot the abundance of each potential indicator species against Zone and Year. What is the nature of the wind farm effect for each species? Do you think these species are good indicators of an effect of wind farms?
5 Random Factors
One limitation of design-based inference approaches like mvabund is computation time; another is difficulties dealing with mixed models to account for random factors (as in Chap. 6). There are additional technical challenges associated with constructing resampling schemes for mixed models, but the main obstacle is that resampling mixed models is computationally intensive, to the point that resampling a “manyglmm” function would not be practical for most datasets. A model-based approach, making use of hierarchical models, holds some promise, and we hope that this can address the issue in the near future.
6 Other Frameworks for Making Inferences About Community–Environment Associations
This chapter has focused on design-based inference using GEEs. What other options are there for making inferences about community–environment associations? A few alternative frameworks could be used; their key features are summarised in Table 14.1. Copula models are mentioned in the table and will be discussed in more detail later (Chap. 17).
6.1 Problems with Dissimilarity-Based Algorithms
Multivariate analysis in ecology has a history dating back to the 1950s (Bray & Curtis, 1957, for example), whereas the other techniques mentioned in Table 14.1 are modern advances using technology not available in most of the twentieth century, and only actually introduced to ecology in the 2010s (Walker and Jackson, 2011; Wang et al., 2012; Popovic et al., 2019). In the intervening years, ecologists developed some algorithms to answer research questions using multivariate abundance data, which were quite clever considering the computational and technological constraints of the time. These methods are still available and widely used in software like PRIMER (Anderson et al., 2008), CANOCO (ter Braak & Smilauer, 1998), and free versions such as in the ade4 (Dray et al., 2007) or vegan (Oksanen et al., 2017) packages.
The methods in those packages (Clarke, 1993; Anderson, 2001, for example) tend to be stand-alone algorithms that are not motivated by an underlying statistical model for abundance,Footnote 2 in contrast to GEEs and all other methods in this book (so-called model-based approaches). The algorithmic methods are typically faster than those using a model-based framework because they were developed a couple of decades ago to deal with computational constraints that were much more inhibiting than they are now. However, these computational gains come at potentially high cost in terms of statistical performance, and algorithmic approaches are difficult to reconcile conceptually with conventional regression approaches used elsewhere in ecology (Chaps. 2–11). So while at the time of writing many algorithmic techniques are still widely used and taught to ecologists, a movement has been gathering pace in recent years towards model-based approaches to multivariate analysis in ecology, whether a GEE approach or another model-based framework for analysis. A few of the key issues that arise when using the algorithmic approach are briefly reviewed below.
Recall that a particular issue for multivariate abundances is the abundance property, with strong mean–variance patterns being the rule rather than the exception and a million-fold range of variances across taxa not being uncommon (e.g. Fig. 10.3). Algorithmic methods were not constructed in a way that can account for the abundance property; instead, data are typically transformed or standardised in pre-processing steps to try to address this issue, rather than addressing it in the analysis method itself. However, this approach is known to address the issue ineffectively (Warton, 2018) and can lead to undesirable and potentially misleading artefacts in ensuing analyses (for example Warton et al., 2012b, or Fig. 14.2).
Simulation results showing that while algorithmic approaches may be valid, they are not necessarily efficient when testing no-effect null hypotheses. In this simulation there were counts in two independent groups of observations (as in Anthony’s revegetation study, Exercise 12.2), with identical means for all response variables except for one, which had a large (10-fold) change in mean. Power (at the 0.05 significance level) is plotted against the variance of this one “effect variable” when analysing (a) untransformed counts; (b) \(\log (y+1)\)-transformed counts using dissimilarity-based approaches, compared to a model-based approach (“mvabund”). A good method will have the power to detect a range of types of effects, but the dissimilarity-based approaches only detect differences when expressed in responses with high variance
Another issue with algorithmic approaches is that because they lack an explicit mean model, they have difficulty capturing important processes affecting the mean, such as variation in sampling intensity. Adjusting for changes in sampling intensity is essential to making valid inferences about changes in community composition. It is relatively easy to do in a statistical model using an offset term (as in Sect. 10.5) or a row effect (Sect. 14.3), but algorithmic approaches instead try to adjust for this using data pre-processing steps like row standardisation. This can be problematic and can lead to counter-intuitive results (Warton & Hui, 2017), because the effects of changing sampling intensity can differ across datasets and depend on data properties (e.g. the effects of sampling intensity on variance are governed by the mean–variance relationship). Related difficulties are encountered when algorithmic approaches are used to try to capture interactions or random factors.
A final issue worthy of mention is that we always need to mind our Ps and Qs. But without a model for abundance, the assumptions of algorithmic approaches are not made explicit, making it harder to understand what data properties we should be checking. This also makes it more difficult to study how these methods behave for data with different types of data properties, but the results we do have are less than encouraging (Warton et al., 2012b; Warton & Hui, 2017).
6.2 Why Not Model-Based Inference?
Design-based inference was used in this chapter to make inferences from models about community–environment associations. As in Chap. 9, design-based inference is often used in place of model-based inference when the sampling distribution of a statistic cannot be derived without making assumptions that are considered unrealistic or when it is not possible to derive the sampling distribution at all. A bit of both is happening here, with high dimensionality making it difficult to specify good models for multivariate abundances and to work out the relevant distribution theory. However, progress is being made on both fronts.
A specific challenge for model-based inference procedures is that the number of parameters of interest needs to be small relative to the size of the dataset. For a model that has different parameters for each taxon (as in Eq. 14.2), this means that the number of taxa would need to be small. For example, Exercise 11.6 took Petrus’s data and analysed just the three most abundant genera of hunting spiders. This model used six parameters (in β) to capture community–environment associations. If such an approach were applied to all 12 hunting spider species, 24 parameters would be needed to make inferences about community–environment associations, and standard approaches for doing this would not be reliable (especially considering that there are only 28 observations). A parametric bootstrap could be used instead, but as previously, this is computationally intensive and not a good match for a hierarchical GLM, unless you are very patient.
There are a few ways forward to deal with this issue that involve simplifying the regression parameters, β, e.g. assuming they come from a common distribution (Ovaskainen & Soininen, 2011) or have reduced rank (Yee, 2006). Using hierarchical GLMs for inference about community–environment associations is an area of active research, but at the time of writing, issues with model-based inference had not been adequately resolved. Although they may well be soon!
Notes
- 1.
He also defined γ-diversity, the richness of species in a region, but this is of less interest to us here.
- 2.
Although the methods in CANOCO have some connections to Poisson regression.
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecology, 26, 32–46.
Anderson, M. J., Gorley, R. N., & Clarke, K. R. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth: PRIMER-E.
Bates, D., & Maechler, M. (2015). MatrixModels: Modelling with sparse and dense matrices. R package version 0.4-1.
Bray, J. R., & Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs, 27, 325–349.
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Australian Journal of Ecology, 18, 117–143.
Dray, S., Dufour, A.-B., et al. (2007). The ade4 package: Implementing the duality diagram for ecologists. Journal of Statistical Software, 22, 1–20.
Greenslade, P. (1964). Pitfall trapping as a method for studying populations of carabidae (coleoptera). The Journal of Animal Ecology, 33, 301–310.
Hardin, J. W., & Hilbe, J. M. (2002). Generalized estimating equations. Boca Raton: Chapman & Hall.
Liang, K. Y., & Zeger, S. L. (1986). Longitudinal data analysis using generalized linear models. Biometrika, 73, 13–22.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O’Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., & Wagner, H. (2017). vegan: Community ecology package. R package version 2.4-3.
Ovaskainen, O., & Soininen, J. (2011). Making more out of sparse data: Hierarchical modeling of species communities. Ecology, 92, 289–295.
Popovic, G. C., Warton, D. I., Thomson, F. J., Hui, F. K. C., & Moles, A. T. (2019). Untangling direct species associations from indirect mediator species effects with graphical models. Methods in Ecology and Evolution, 10, 1571–1583.
Rao, C. R. (1973). Linear statistical inference and its applications (2nd ed.). New York: Wiley.
ter Braak, C. J. F., & Smilauer, P. (1998). CANOCO reference manual and user’s guide to CANOCO for Windows: Software for canonical community ordination (version 4). New York: Microcomputer Power.
Walker, S. C., & Jackson, D. A. (2011). Random-effects ordination: Describing and predicting multivariate correlations and co-occurrences. Ecological Monographs, 81, 635–663.
Wang, Y., Naumann, U., Wright, S. T., & Warton, D. I. (2012). mvabund—an R package for model-based analysis of multivariate abundance data. Methods in Ecology and Evolution, 3, 471–474.
Warton, D. I. (2008). Penalized normal likelihood and ridge regularization of correlation and covariance matrices. Journal of the American Statistical Association, 103, 340–349.
Warton, D. I. (2011). Regularized sandwich estimators for analysis of high dimensional data using generalized estimating equations. Biometrics, 67, 116–123.
Warton, D. I. (2018). Why you cannot transform your way out of trouble for small counts. Biometrics, 74, 362–368
Warton, D. I., & Hui, F. K. C. (2017). The central role of mean-variance relationships in the analysis of multivariate abundance data: A response to Roberts (2017). Methods in Ecology and Evolution, 8, 1408–1414.
Warton, D. I., Wright, S. T., & Wang, Y. (2012b). Distance-based multivariate analyses confound location and dispersion effects. Methods in Ecology and Evolution, 3, 89–101.
Whittaker, R. H. (1972). Evolution and measurement of species diversity. Taxon, 21, 213–251.
Yee, T. (2006). Constrained additive ordination. Ecology, 87, 203–213.
Zeger, S. L., & Liang, K. Y. (1986). Longitudinal data analysis for discrete and continuous outcomes. Biometrics, 42, 121–130.
Author information
Authors and Affiliations
Rights and permissions
Copyright information
© 2022 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Warton, D.I. (2022). Multivariate Abundances—Inference About Environmental Associations. In: Eco-Stats: Data Analysis in Ecology. Methods in Statistical Ecology. Springer, Cham. https://doi.org/10.1007/978-3-030-88443-7_14
Download citation
DOI: https://doi.org/10.1007/978-3-030-88443-7_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-88442-0
Online ISBN: 978-3-030-88443-7
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)





 Generalised Estimating Equations
Generalised Estimating Equations
