Abstract
Automatic polyp detection during colonoscopy is beneficial for reducing the risk of colorectal cancer. However, due to the various shapes and sizes of polyps and the complex structures in the intestinal cavity, some normal tissues may display features similar to actual polyps. As a result, traditional object detection models are easily confused by such suspected target regions and lead to false-positive detection. In this work, we propose a multi-branch spatial attention mechanism based on the one-stage object detection framework, YOLOv4. Our model is further jointly optimized with a top likelihood and similarity to reduce false positives caused by suspected target regions. A similarity loss is further added to identify the suspected targets from real ones. We then introduce a Cross Stage Partial Connection mechanism to reduce the parameters. Our model is evaluated on the private colonic polyp dataset and the public MICCAI 2015 grand challenge dataset including the CVC-Clinic 2015 and Etis-Larib, both of the results show our model improves performance by a large margin and with less computational cost.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Colorectal cancer is one of the most common malignancies of the digestive system in the world. Most colorectal cancers originate from adenomatous polyp, and colonoscopy is an important way to screen for colorectal cancer [1]. Colonoscopy-based polyp detection is a key task in medical image computing. In recent years, Deep learning detection models are widely used in polyp detection [2,3,4, 8, 16]. However, influenced by the complex environment of the intestinal tract, bubbles, lens reflection, residues, and shadows may display polyp-like features. Those features can form the suspected target and confuse the model. See Fig. 1 below.
Currently two-stage [2,3,4, 6] and one-stage [5, 8, 16, 23] models are the most widely used models in object detection. Faster R-CNN [6] as the most widely used two-stage object detection model, has been adopted in various polyp detection tasks. Mo et al. [2] provide the first evaluation for polyp detection using Faster R-CNN framework, which provides a good trade-off between efficiency and accuracy. Shin et al. [4] propose FP learning. They first trained a network with polyp images and generated FP samples with additional normal videos. Then retrained the network by adding back the generated FP samples. Sornapudi et al. [3] propose a modified region-based convolutional neural network (R-CNN) by generating masks around polyp detected from still frames. One stage model such as You only look once (YOLO) [5] is also widely used for lesion detection with the advantage of its efficiency. Wang et al. [8] propose a new anchor free polyp detector, which can achieve real-time performance. Liu et al. [23] investigated the potential of the single shot detector (SSD) [18] framework for detecting polyps in colonoscopy videos. Three different feature extractors, including ResNet50, VGG16, and InceptionV3 are assessed. Tian et al. [16] propose a one-stage detection and classification approach for a new 5-class polyp classification problem.
To deal with the suspected target regions, some mechanisms such as attention mechanism (CBAM) [7] propose to make the model more focused on true target regions. Recently, Xiao et al. [10] propose a new sampling method based on the Faster R-CNN model to automatically learn features from the suspected target regions directly and effectively reduce false positives. Guo et al. [24] propose a method based on active learning to tackle false positives detected by the CADe system. But both [24] and [4] methods add the FP samples to the training set after finding the false-positive region to retrain the network, this process is more complicated. We design a semi-supervised method to automatically learn suspicious targets to solve this problem.
In addition, there are other methods to detect polyps. Tajbakhsh et al. [22] is based on a hybrid context-shape approach, which utilizes context information to remove non-polyp structures and shape information to reliably localize polyps. Tian et al. [25] integrate few-shot anomaly detection methods designed to perform the detection of frames containing polyps from colonoscopy videos with a method that rejects frames containing blurry images, feces and water jet sprays. Liu et al. [26] propose a consolidated domain adaptive detection and localization framework to bridge the domain gap between different colonosopic datasets effectively.
In this paper, we propose a novel one-stage polyp detection model based on YOLOv4. Moreover, Our model is validated on both the private dataset and the public dataset of the MICCAI 2015 challenge [11] including CVC-Clinic 2015 and Etis-Larib, brings significant performance improvements and outperform most cutting-edge models. To summarize, our main contributions include: (i) A multi-branched spatial attention mechanism (MSAM) is proposed to make the model more focus on the polyp lesion regions. (ii) Design the Top likelihood loss (Tloss) with a multi-scale sampling strategy to reduce false positives by learning from suspected regions from the background. (iii) Further propose Cosine similarity loss (Csimloss) to improve the discrimination ability between positive and negative images. (iv) A cross stage partial connection mechanism is further introduced to make the model more efficient. (v) Finally, from the large amount of experiments using the private and public datasets, we demonstrate that our detection model shows improved detection performance compared with other recent studies in the colonoscopy image datasets.
2 Methods
Our detailed model is shown in Fig. 2. The proposed framework consists of three parts: (1) A multi-branch spatial attention mechanism (MSAM) is proposed to make the model pay more attention to the polyp lesion regions (Sect. 2.1); (2) Top likelihood loss and cosine similarity loss are designed to the one-stage model for false-positive reduction (Sect. 2.2); (3) Cross Stage Partial Connection is introduced to reduce model parameters through feature fusion (Sect. 2.3). During training, the proposed model jointly optimizes positive and negative images. The positive images are trained by the original loss function, the negative images are trained with the top likelihood loss added. The pairs of positive and negative images are further optimized by the cosine similarity loss.
The architecture of the model. C-Block is the structure after adding cross stage partial connection, and C-M-Block is the structure after adding cross stage partial connection and multi-branch spatial attention mechanism (MSAM), the number represent the convolution kernel size, setting \(\mathrm {k}^{\prime } \in \{5,7,9\}\) in our model, They correspond to the three scales in the model.
2.1 Multi-branch Spatial Attention Mechanism
In order to make the model pay more attention to the polyp lesion regions and eliminate the effect of background contents, inspired by the idea of spatial attention mechanism (SAM) [7] which locates the most important information on the feature map, we propose a multi-branch spatial attention mechanism (MSAM). We put them in the three output positions of feature fusion, as shown in C-M-Block in Fig. 2, MSAM is a concrete structure. There are three different scales of feature maps for feature fusion, the receptive fields of the three scales are targeted to different sizes of objects.
Given an input F, we compute the MSAM map \(A_{\mathrm {s}}=\sigma \left( \sum _{\mathrm {k}^{\prime }} f^{\mathrm {k}^{\prime } \times \mathrm {k}^{\prime }}(F)\right) \). Where, \(f^{\mathrm {k}^{\prime } \times \mathrm {k}^{\prime }}\) represents the convolution operation with the kernel size of \({\mathrm {k}^{\prime } \times \mathrm {k}^{\prime }} \), and \(\sigma \) represents the sigmoid activation function. Setting \(\mathrm {k}^{\prime } \in \{5,7,9\}\) in our model, They correspond to the three scales in the model. The \(\mathrm {9} \times \mathrm {9}\) convolution kernel corresponds to the smaller receptive field, the \(\mathrm {7} \times \mathrm {7}\) convolution kernel corresponds to the middle scale receptive field, and the \(\mathrm {5} \times \mathrm {5}\) convolution kernel corresponds to the larger receptive field.
2.2 Top Likelihood and Similarity Loss
We design the top likelihood loss and cosine similarity loss to reduce false positives. The implementation details of the loss can be summarized in Fig. 3.
The illustration of the multi-scale top likelihood loss and cosine similarity loss where the solid point represents the selected sample: (a) show top likelihood loss with multi-scale sampling strategy, the K of each scale is set to 50. (b) In the same batch, positive and negative samples of the same scale calculate cosine similarity loss.
Top Likelihood Loss. When optimizing negative samples, since those images do not have any annotation information, this means that all areas will be randomly sampled with equal chance. As a result, the suspected target regions will have a small chance to get trained since it usually only occupies a small portion of the image. The prediction result may bias towards normal features, leading to some false positive detection. To solve this problem, we design top likelihood loss with multi-scale sampling strategy in a one-stage model. When dealing with negative images, we use top likelihood loss and select the proposals with top confidence scores.
Different from two-stage models, YOLOv4 directly generates object confidence score, category probability, and border regression. When training negative images, we compute the confidence scores and select the top 50 anchor boxes score negative anchor boxes on each scale (150 in total) to calculate the loss. The boxes with high scores will be more likely to represent the suspected target region, and as long as the boxes with high scores are minimized, all the boxes would be optimized to be negative regions. This top likelihood loss is defined as:
Here, i represents the index of anchor in a batch, and \(p_{i}\) represents the predicted score of the i-th anchors. \(L_{obj}\) is the cross-entropy loss.
Cosine Similarity Loss. We further propose the cosine similarity loss to improve the discrimination ability between positive and negative images. To make our model trained sufficiently, we make use all of the pairs of positive and negative images for computing the cosine similarity loss. Specifically, in each batch, positive images and negative images are random. In order to fully learn the characteristics between positive and negative images, we design a program to let the positive and negative images in the same batch size calculate the similarity loss between each other, and finally take the average. When the network processes the positive images, we take the positive samples with top K scores. Then, when the network processes negative images, we select the highest predicted K classification scores and pair them with positive ones. Assume A positive images and B negative images within one batch, there are \(A\times {B}\) positive-negative pairs. The similarity loss is obtained by computing the cosine similarity of K paired eigen-vectors and summing over the \(A\times {B}\) pairs.
Where \(X_{1}^{i}, X_{2}^{i}\) are the feature vectors from positive and negative images, csim is cosine similarity loss, \({\text {csim}}\left( X_{1}^{i}, X_{2}^{i}\right) =\frac{X_{1}^{i} \cdot X_{2}^{i}}{\left\| X_{1}^{i}\right\| \left\| _{2}^{i}\right\| }=\frac{\sum _{i=1}^{n} X_{1}^{i}\times X_{2}^{i}}{\sqrt{\sum _{i=1}^{n}\left( X_{1}^{i}\right) ^{2}} \times \sqrt{\sum _{i=1}^{n}\left( X_{2}^{i}\right) ^{2}}}\).
2.3 Cross Stage Partial Connection
We further introduce the Cross Stage Partial Network (CSPNet) [13] in our model. By dividing the gradient flow, CSPNet can make the gradient flow propagate through different network paths, which can improve the reasoning speed. As shown in Fig. 2, the feature fusion part includes five modules: three up-sampling and two down-sampling. As shown in C-Block and C-M-Block in the bottom right of Fig. 2, the Block represents the original connection, C-Block and C-M-Block represents the connection after adding CSP. through the split and merge strategy, the number of gradient paths can be doubled. Because of the cross-stage strategy, which can alleviate the disadvantages caused by using explicit feature map copy for concatenation. As shown in Table 1, the number of parameters significantly decrease by adding such an operation.
3 Experiment
3.1 Datasets
In order to verify the effectiveness of the proposed method, we conduct experiments on two datasets, the private colonic polyp dataset and the public dataset including CVC-Clinic 2015 and Etis-Larib.
Private Polyp Dataset. A dataset of private colonic polyp dataset is collected and labeled from the Colorectal and Anorectal Surgery Department of a local hospital, which contains 175 patients with 1720 colon polyp images. The 1720 images are randomly divided into training and testing set with a ratio of 4:1. We simulate the actual application scenes of colonoscopy and expand the dataset accordingly, including the expansion of blur, brightness, deformation and so on, finally expanding to 3582 images. The colon polyp images are combined with 1000 normal images without annotation information to build the training set. The original image size is varied from \(612\times {524}\) to \(1280\times {720}\). And we resize all the images to \(512\times {512}\).
MICCAI 2015 Colonoscopy Polyp Automatic Detection Classification Challenge. The challenge contains two datasets, the model is trained on CVC-Clinic 2015 and evaluated on Etis-Larib. The CVC-Clinic 2015 dataset contains 612 standard well-defined images extracted from 29 different sequences. Each sequence consists of 6 to 26 frames and contains at least one polyp in a variety of viewing angles, distances and views. Each polyp is manually annotated by a mask that accurately states its boundaries. The resolution is 384 \(\times \) 288. The Etis-Larib dataset contains 196 high-resolution images with a resolution of 1225 \(\times \) 966, including 44 distinct polyps obtained from 34 sequences.
3.2 Evaluation and Results
Evaluation Criteria. We use the same evaluation metrics presented in the MICCAI 2015 challenge to perform the fair evaluation of our polyp detector performance.
Since the number of false negative in this particular medical application is more harmful, we also calculate the F1 and F2 scores as follows. The evaluation criteria are as follows:
where TP and FN denote the true positive and false negative patient cases. FP represents the false positive patient cases.
Implementation Details. Our model uses the Pytorch framework and runs on NVIDIA GeForce RTX 2080Ti GPU servers. We set the batch size to 8. During training, we use the SGD optimization method, we also perform random angle rotation and image scaling data for data augmentation. The training contains 2000 epochs with 574 iterations for each epoch, Normally the training process starts with a high learning rate and then decreases every certain as the training goes on. However, a large learning rate applies on a randomly initialized network may cause instability for training. To solve this problem, we apply a smooth cosine learning rate learner [12]. The learning rate \(\alpha _{t}\) is computed as \(\alpha _{t}=\frac{1}{2}\left( 1+\cos \left( \frac{t \pi }{T}\right) \right) \alpha \), where t represents the current epoch, T represents the epoch and \(\alpha \) represents initial learning rate.
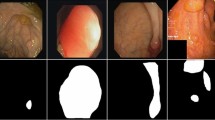
(a) Origin image with ground truth label (solid line box); (b) Heatmap generated by the original YOLOv4; (c) Heatmap generated by YOLOv4+MSAM; (d) Origin image with ground truth label (solid line box) and suspected target regions (dashed line box); (e) Heatmap generated by YOLOv4+MSAM; (f) Heatmap generated by YOLOv4+MSAM+Tloss (top likelihood loss);
Ablation Experiments on Private Dataset. In order to study the effect of MSAM and the new loss function, we conduct ablation experiments on our private dataset. As shown in Table 1, Compared to the YOLOv4 baseline, our proposed MSAM increases the Recall by 4.5%, resulting in a score increase of F1 and F2 by 2.2% and 4.0%, respectively. Adding the top likelihood loss only increases the Precision by 4.4%, and combining top likelihood loss together increases both Precision and Recall, leading to an increase of Precision by 2.9% and Recall by 3.1%. Finally, the model achieves the performance boosting over all the metrics when combining MSAM, Top likelihood and similarity loss, CSP module together, leading to increases of Precision by 4.4%, Recall by 3.7%, F1 by 4.0%, and F2 by 3.8%. It is also worth noting that CSP makes the model more efficient and leads decreases of FLOPs by 10.74% (8.66 to 7.73), and Parameters by 15.7% (63.94 to 53.9).
We also show some visualization results of the heatmap (last feature map of YOLOv4) for ablation comparison (shown in Fig. 4). The results demonstrate that MSAM makes the model more focus on the ground truth areas, and the top likelihood loss let the model better identify the suspected target regions and pay less attention to such areas.
Results and Comparisons on the Public Dataset. The results on the public dataset are shown in Table 2, we also test several previous models for the MICCAI 2015 challenges. The results show that our method improves performance on almost all metrics. Compare to the baseline, our proposed approach achieves a great performance boosting, yielding an increase of Precision by 11.8% (0.736 to 0.854), Recall by 7.5% (0.702 to 0.777), F1 by 9.5% (0.719 to 0.814), F2 by 8.2% (0.709 to 0.791). It is worth noting that the depth of CSPDarknet53 backbone for YOLOv4 is almost the same as Resnet50. However, our proposed approach even significantly outperforms the state-of-the-art model Sornapudi et al. [3] with a backbone of Resnet101 and Liu et al. [23] with a backbone of Inceptionv3. Comparison with Liu et al. [23], although it slightly decreases the Recall by 2.6% (0.803 to 0.777), it increases Precision by 11.5% (0.739 to 0.854), F1 by 4.6% (0.768 to 0.814), and F2 by 0.2% (0.789 to 0.791). We presented the Frame Per Second (FPS) for each model. It shows that our one-stage model is much faster than other models. It is 5.3 times faster than the Faster R-CNN (37.2 vs 7), 11.6 times faster than Sornapudi et al. [3] (37.2 vs 3.2) and 1.2 times faster than Liu et al. [23] (37.2 vs 32). Furthermore, The PR curve is plotted in Fig. 5. Comparison with baseline, our proposed approach increases the AP by 5.1% (0.728 to 0.779).
4 Conclusions
In this paper, we propose an efficient and accurate object detection method to detect colonoscopic polyps. We design a MSAM mechanism to make the model pay more attention to the polyp lesion regions and eliminate the effect of background content. To make our network more efficient, we develop our method based on a one-stage object detection model. Our model is further jointly optimized with a top likelihood and similarity loss to reduce false positives caused by suspected target regions. A Cross Stage Partial Connection mechanism is further introduced to reduce the parameters. Our approach brings performance boosting compare to the state-of-the-art methods, on both a private polyp detection dataset and public MICCAI 2015 challenge dataset. In the future, we plan to extend our model on more complex scenes, such as gastric polyp detection, lung nodule detection, achieving accurate and real-time lesion detection.
References
Zhang, P., Sun, X., Wang, D., Wang, X., Cao, Y., Liu, B.: An efficient spatial-temporal polyp detection framework for colonoscopy video. In: 2019 IEEE 31st International Conference on Tools with Artificial Intelligence, pp. 1252–1259. IEEE, Portland (2019). https://doi.org/10.1109/ICTAI.2019.00-93
Mo, X., Tao, K., Wang, Q., Wang, G.: An efficient approach for polyps detection in endoscopic videos based on faster R-CNN. In: 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 2018, pp. 3929–3934 (2018). https://doi.org/10.1109/ICPR.2018.8545174
Sornapudi, S., Meng, F., Yi, S.: Region-based automated localization of colonoscopy and wireless capsule endoscopy polyps. In: Applied Sciences (2019) . https://doi.org/10.3390/app9122404
Shin, Y., Qadir, H.A., Aabakken, L., Bergsland, J., Balasingham, I.: Automatic colon polyp detection using region based deep CNN and post learning approaches. In: IEEE Access, vol. 6, pp. 40950–40962 (2018). https://doi.org/10.1109/ACCESS.2018.2856402
Redmon, J., Divvala, S., Girshick, R., Farhadi, A.: You only look once: unified, real-time object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 779–788 (2016). https://doi.org/10.1109/CVPR.2016.91
Ren, S., He, K., Girshick, R., Sun, J.: Faster R-CNN: towards real-time object detection with region proposal networks. In: Advances in Neural Information Processing Systems, pp. 91–99 (2015)
Woo, S., Park, J., Lee, J.-Y., Kweon, I.S.: CBAM: convolutional block attention module. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11211, pp. 3–19. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01234-2_1
Wang, D., et al.: AFP-Net: realtime anchor-free polyp detection in colonoscopy. In: 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), Portland, OR, USA, pp. 636–643 (2019). https://doi.org/10.1109/ICTAI.2019.00094
Bochkovskiy, A., Wang, C.Y., Liao, H.Y.M.: YOLOv4: optimal speed and accuracy of object detection. In: arXiv:2004.10934 (2020)
Xiao, L., Zhu, C., Liu, J., Luo, C., Liu, P., Zhao, Y.: Learning from suspected target: bootstrapping performance for breast cancer detection in mammography. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 468–476. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_52
Bernal, J., Sanchez, F.J., Fernandez-Esparrach, G., Gil, D., Rodrguez, C., Vilarino, F.: WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. In: Computerized Medical Imaging and Graphics, vol. 43, pp. 99–111 (2015)
Loshchilov, I., Hutter, F.: SGDR: stochastic gradient descent with warm restarts. In: arXiv preprint arXiv:1608.03983 (2016)
Wang, C.Y., Liao, H.Y.M., Wu, Y.H., et al.: CSPNet: a new backbone that can enhance learning capability of CNN. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (2020)
Yuan, Z., IzadyYazdanabadi, M., Mokkapati, D., et al.: Automatic polyp detection in colonoscopy videos. In: Medical Imaging 2017: Image Processing, Orlando, Florida, USA, vol. 2017 (2017)
Tajbakhsh, S., Gurudu, R., Liang, J.: Automatic polyp detection in colonoscopy videos using an ensemble of convolutional neural networks. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, pp. 79–83 (2015). https://doi.org/10.1109/ISBI.2015.7163821
Tian, Y., Pu, L.Z.C.T., Singh, R., Burt, A.D., Carneiro, G.: One-stage five-class polyp detection and classification. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, pp. 70–73 (2019). https://doi.org/10.1109/ISBI.2019.8759521
Redmon, J., Farhadi, A.: YOLOv3: an incremental improvement. arXiv preprint arXiv:1804.02767 (2018)
Liu, W., et al.: SSD: single shot multibox detector. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9905, pp. 21–37. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46448-0_2
Zheng, Y., et al.: Localisation of colorectal polyps by convolutional neural network features learnt from white light and narrow band endoscopic images of multiple databases. In: EMBS, pp. 4142–4145 (2018)
Jia, X., et al.: Automatic polyp recognition in colonoscopy images using deep learning and two-stage pyramidal feature prediction. IEEE Trans. Autom. Sci. Eng. 17, 1570–1584 (2020). https://doi.org/10.1109/TASE.2020.2964827
Qadir, H.A., Shin, Y., Solhusvik, J., Bergsland, J., Aabakken, L., Balasingham, I.: Polyp detection and segmentation using mask R-CNN: does a deeper feature extractor CNN always perform better? In: 2019 13th International Symposium on Medical Information and Communication Technology (ISMICT), pp. 1–6. IEEE (2019)
Tajbakhsh, S., Gurudu, R., Liang, J.: Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imag. 35(2), 630–644 (2016). https://doi.org/10.1109/TMI.2015.2487997
Liu, M., Jiang, J., Wang, Z.: Colonic polyp detection in endoscopic videos with single shot detection based deep convolutional neural network. IEEE Access 7, 75058–75066 (2019). https://doi.org/10.1109/ACCESS.2019.2921027
Guo, Z., et al.: Reduce false-positive rate by active learning for automatic polyp detection in colonoscopy videos. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp. 1655–1658 (2020). https://doi.org/10.1109/ISBI45749.2020.9098500
Tian, Y., Pu, L., Liu, Y., et al.: Detecting, localising and classifying polyps from colonoscopy videos using deep learning. arXiv preprint arXiv:2101.03285v1 (2021)
Liu, X., Guo, X., Liu, Y., et al.: Consolidated domain adaptive detection and localization framework for cross-device colonoscopic images. Med. Image Anal. (2021). https://doi.org/10.1016/j.media.2021.102052
Xu, J., Zhao, R., Yu, Y., et al.: Real-time automatic polyp detection in colonoscopy using feature enhancement module and spatiotemporal similarity correlation unit. In: Biomedical Signal Processing and Control, vol. 66 (2021). https://doi.org/10.1016/j.bspc.2021.102503. ISSN 1746–8094
Acknowledgments
The research work supported by the National Key Research and Development Program of China under Grant No. 2018YFB1004300, the National Natural Science Foundation of China under Grant No. U1836206, U1811461, 61773361 and Zhengzhou collaborative innovation major special project (20XTZX11020).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, Z. et al. (2021). An Efficient Polyp Detection Framework with Suspicious Targets Assisted Training. In: Ma, H., et al. Pattern Recognition and Computer Vision. PRCV 2021. Lecture Notes in Computer Science(), vol 13022. Springer, Cham. https://doi.org/10.1007/978-3-030-88013-2_44
Download citation
DOI: https://doi.org/10.1007/978-3-030-88013-2_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-88012-5
Online ISBN: 978-3-030-88013-2
eBook Packages: Computer ScienceComputer Science (R0)