Abstract
Recently, developments have been made towards modelling patient-specific deformable mitral valves from transesophageal echocardiography (TEE). Thus far, a major limitation in the workflow has been the manual process of segmentation and model profile definition. Completing a manual segmentation from 3D TEE can take upwards of two hours, and existing automated segmentation approaches have limitations in both computation time and accuracy. Streamlining the process of segmenting the valve and generating a surface mold is important for the scalability and accuracy of patient-specific mitral valve modelling. We present DeepMitral, a fully automatic, deep learning based mitral valve segmentation approach that can quickly and accurately extract the geometry of the mitral valve directly from TEE volumes. We developed and tested our model on a data set comprising 48 diagnostic TEE volumes with corresponding segmentations from mitral valve intervention patients. Our proposed pipeline is based on the Residual UNet architecture with five layers. Evaluation of our proposed pipeline was assessed using manual segmentations performed by two clinicians as a gold-standard. The comparisons are made using the mean absolute surface distance (MASD) between the boundaries of the complete segmentations, as well as the 95% Hausdorff distances. DeepMitral achieves a MASD of \({0.59 \pm 0.23}\mathrm{mm}\) and average 95% Hausdorff distance of \({1.99 \pm 1.14}\mathrm{mm}\). Additionally, we report a Dice score of 0.81. The resulting segmentations from our approach successfully replicate gold-standard segmentations with improved performance over existing state-of-the-art methods. DeepMitral improves the workflow of the mitral valve modelling process by reducing the time required for completing an accurate mitral valve segmentation, and providing more consistent results by removing user variability from the segmentation process.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Mitral valve (MV) disease is a common pathologic problem occurring in approximately 2% of the general population but climbing to 10% in those over the age of 75 [3]. Of this group, approximately 20% have a sufficiently severe form of the disease that may require surgical intervention to restore normal valve function and prevent early mortality [21]. The preferred intervention for mitral regurgitation is valve repair, due to superior patient outcomes compared to those following valve replacement [1, 17]. However, the repair must be tailored to the patient-specific anatomy and pathology, which requires expert training and experience. Consequently, there is a need for patient-specific models that can permit the training and procedure-planning of patient-specific repairs to minimize its learning curve and preventable errors [7, 9]. Heart simulator technology has been adopted widely by both industry for evaluation of technologies for imaging heart valves [14], and academia for the assessment of modelled heart valves [16]. Recently, developments have been made on a workflow to create 3D, patient-specific valve models directly from trans-esophageal echocardiography (TEE) images. When viewed dynamically using TEE within a pulse duplicator simulator, it has been demonstrated that these models result in pathology-specific TEE images similar to those acquired from the patient’s valves in-vivo [8].
A key step in patient-specific modelling workflows is delineating the mitral valve leaflets in patient ultrasound image data, a necessary operation to extract the patient-specific leaflet geometry, which will be used to form the basis of the model. Performing manual segmentations is very time consuming, taking upwards of 2 hours, which is a serious bottleneck in modelling workflows. Several mitral leaflet segmentation methods have been proposed, targeting a number of different applications. These methods focus on varying goals between deriving quantitative valve measurements and extracting annular and leaflet geometry from 3D TEE images. These methods can be divided into two categories: semi-automatic and fully automatic approaches. The semi-automatic approaches all require some level of user intervention during the segmentation process, while fully automatic methods do not. Scheinder et.al. proposed a semi-automatic method for segmenting the mitral leaflets in 3D TEE over all phases of the cardiac cycle [23]. This method utilizes geometric priors and assumptions about the mechanical properties of the valve to model the leaflets through coaptation with a reported surface error of 0.84mm. However, their method only represents the mitral leaflets as a single medial surface, rather than structures with thickness. Burlina et.al. [4] proposed a semi-automatic segmentation method based on active contours and thin tissue detection for the purpose of computational modelling, reporting errors in the range of 4.00 mm to 5.00 mm . An additional semi-automatic approach designed for patient specific valve modelling reported a surface error of 1.4mm overall, and an surface error of 1.01mm for the atrial surface critical in mitral valve model creation [5]. Several fully automatic methods have been proposed that are based on population average atlases. Ionasec et.al. [10] describe a technique which uses a large database of manually labelled images and machine learning algorithms to locate and track valve landmarks, reporting a surface error of 1.54mm. While this method is fully automatic, the use of sparse landmarks potentially limits the amount of patient-specific detail that can be extracted. Pouch et.al. [20] also describe a fully automatic method that employs a set of atlases to generate a deformable template, which is then guided to the leaflet geometry using joint label fusion. The surface error of this method is reported as 0.7mm, however this is only achieved on healthy valves and performance is reduced when segmenting diseased valves. Atlas based methods relying on sparse deformation could be biased towards the atlas geometry, limiting the potential for capturing patient specific detail, which is not ideal for valve modelling applications.
Automatic 3D segmentation methods offer significant implications for the feasibility of patient-specific modelling in clinical use. While existing methods have demonstrated the ability to accurately segment the mitral valve structure,they remain highly time-intensive. Furthermore, some of these published methods show decreased performance when applied to highly diseased valves, demonstrating limitations in patient-specificity. Convolutional neural networks (CNNs) have been widely demonstrated to be effective for segmentation tasks. However, to our knowledge no CNN segmentation approaches have been reported for mitral valve segmentation in 3D TEE imaging, although 3D Unet based approaches have been used in other cardiac ultrasound applications such as automatic annulus detection [2]. Working in 2D, UNet has been used for mitral leaflet segmentation [6]. In this paper, we present DeepMitral, a 3D segmentation pipeline for mitral valve segmentation based on the 3D Residual UNet architecture [12]. We demonstrate the feasibility of CNN based segmentation for 3D TEE images, and establish a baseline of performance for future methods. DeepMitral will have applications in patient specific valve modelling, enabling improvements in the workflow. DeepMitral has been made open source including our trained model and is freely available on GitHubFootnote 1.
2 Methods
2.1 Data Acquisition
Patients with mitral valve regurgitation undergoing clinical interventions were imaged preoperatively as per clinical protocol with appropriate ethics approval using a Philips Epiq system with an X8-2T transducer,. The 3D TEE images were exported into Cartesian format, and the SlicerHeart module was used to import the Cartesian DICOM files into 3D SlicerFootnote 2 [22]. Images at end-diastole were selected for image analysis. The exported Cartesian format images have an axial resolution of approximately 0.5mm. We collected a total of 48 volumes, which were divided into training, validation and testing partitions with 36, 4, and 8 volumes respectively. Annotations for the training and validation sets were performed in 3D Slicer by multiple different trained users. These segmentations were performed via manual refinement of the output of a semi-automatic segmentation tool [5], and were then reviewed and modified as necessary by a single experienced user to ensure consistency. The test set was annotated entirely manually by two cardiac imaging clinicians using 3D Slicer.
2.2 Model Selection
Our training and validation sets were used to perform model selection and hyper-parameter tuning on a variety of network architectures including Residual UNet, VNet, AHNet and SegResNetVAE [12, 15, 18, 19]. We trained each network with a selection of hyper-parameters and computed mean Dice coefficient scores, mean surface error scores, and mean 95% Hausdorff distance on the validation set. The best performing version of each network architecture is shown in Table 1. Of the models, the Residual UNet architecture achieved the best performance with respect to all validation scores, so it was chosen as our final network for use in the DeepMitral pipeline.
2.3 DeepMitral Pipeline
Our 3D TEE volume segmentation platform was built using the MONAIFootnote 3 framework, that provides domain-optimized foundational capabilities for developing healthcare imaging training workflows. This platform includes the implementation of many common network architectures for both 2D and 3D data, as well as a number of medical imaging focused pre-processing methods.
Our workflow begins with a sequence of pre-processing operations from the MONAI framework. First, we load the images, and add a channel along the first dimension to transform them into channel-first representation. Next, we isotropically re-sample the volumes to 0.3mm spacing, using bilinear re-sampling for the image data and nearest neighbour re-sampling for the label. Following re-sampling, we re-scale the image intensities to the range of 0.00 to 1.00, then crop the images to the foreground using the smallest possible bounding box that includes all non-zero voxels. Finally, random sampling is performed on the volumes, taking 4 samples of size 96 \(\times \) 96 \(\times \) 96, centered on voxels labelled as leaflet. The final random sampling step is recomputed at every epoch during network training. No data-augmentation was performed as adding rigid or deformable spatial transformations resulted in no improvements in validation metrics, and a reduction of training speed.
Our network uses a Residual UNet architecture [12], implemented by the MONAI framework, with 5 layers of 16, 32, 64, 128 and 256 channels respectively. Each of these layers is created using a residual unit with 2 convolutions and a residual connection. Convolutions are performed with stride 2 at every residual unit for up-sampling and down-sampling.
We trained our model using batch sizes of 32, composed of 8 different volumes, with 4 random samples being taken from each volume. Training was performed for 2000 epochs using an Nvidia GTX 1080 graphics card with 8GB of ram, and took roughly 5 hours. We employed the Adam optimizer, with an initial learning rate of \(1.00 \times 10^{-3}\), which is reduced to \(1.00 \times 10^{-3}\) after 1000 epochs [13]. Batch normalization is used to help prevent over-fitting of the model.
2.4 Evaluation
Final evaluation of our pipeline is performed using a separate test set consisting of eight volumes with ground truth annotations that were performed manually by two cardiac clinicians. Prior to evaluating our model, we retrained the network using combined training and validation sets. The primary comparison metrics are the mean absolute surface distance (MASD) between the boundaries of the complete segmentations, as well as the 95% Hausdorff distances. We also report the Dice coefficient scores.
3 Results
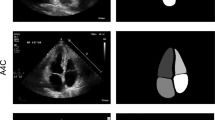
DeepMitral achieves a MASD of \({0.59 \pm 0.23}\mathrm{mm}\), average 95% Hausdorff distance of \({1.99 \pm 1.14}\mathrm{mm}\), and a Dice score of 0.81. In all 8 volumes used for testing the mitral leaflets were successfully segmented, with no cases of complete failure to identify the leaflets. Overall, the scores are consistent among 6 of the 8 examples, with 2 examples exhibiting lower performance. In one instance, case P3, the leaflets are under-segmented near the leaflet tips, and in another instance, P8, the chordae tendineae are mis-labelled as leaflet, as seen in Fig. 1. We see the corresponding metrics for these cases in Fig. 2, which show higher surface distance errors and worse dice scores than the other cases.
3.1 Inference Runtime Performance
Deep learning segmentation methods enable predictions to be performed for low computational cost. We evaluated DeepMitral’s inference speed on our test set using both CPU only (Intel i7-6700K) and GPU acceleration (Nvidia GeForce GTX 1080). The size of the volumes range from approximately voxels. Using only the CPU for inference, DeepMitral takes on average \({9.5 \pm 2.26}\mathrm{s}\) to perform the inference itself, and \({11.40 \pm 2.28}\mathrm{s}\) for overall runtime including startup overhead. When using GPU acceleration, these times are reduced on average to \({3.5 \pm 0.53}\mathrm{s}\) for inference and \({5.69 \pm 0.66}\mathrm{s}\) for overall runtime. DeepMitral achieves fast inference times on both CPU and GPU, with GPU acceleration reducing runtime by a factor of two on average. Additionally, startup overhead is consistently around 2s for both CPU and GPU. This overhead would only occur once in the case of performing inference on multiple volumes in a single run.
4 Discussion
These results demonstrate the feasibility of CNN based techniques for mitral valves segmentations in 3D TEE volumes. Trained with a relatively small dataset, DeepMitral achieves an improvement in accuracy over the existing state of the art approaches. Average surface error is reduced to 0.6mm on average, where the best performing existing methods report an error of 0.7mm. Additionally, our reported error is almost equal to typical inter-user variability, which was previously reported as \({0.6\pm 0.17}\mathrm{mm}\) [11]. Our reported MASD is approaching the axial resolution of the ultrasound volumes, which is approximately 0.5mm on average. These results indicate that while DeepMitral is accurately labelling the valve leaflets overall, we note the 95% Hausdorff distances are typically larger, in the range of 1.00mm to 4.00mm. We can see in Fig. 2 that there are small regions of the leaflets where accuracy is worse, contributing to these larger 95% Hausdorff errors, while the majority of the leaflet surface maintains sub-millimetre error. For P8, we see that the leaflets themselves are well identified, however the large protrusion where the chordae were mis-identified contributes to the poor error metrics in the case.
Our results suggest deep learning based approaches perform better than atlas based approaches for capturing unique valve features. Although both methods use a collection of prior data to inform the segmentation process, CNNs are much more flexible at applying this prior knowledge to new problems. Atlas based methods, however, can transfer particular geometric patterns to a new segmentation, and thus tend to perform best only on healthy valves [20]. We tested DeepMitral on exclusively diseased valves, but as demonstrated by our test data, there is a range of distinct valve geometries, all of which are accurately identified by our model. The areas of poor performance are not due to systematic geometrical bias, but are instead caused by poor image data and mis-identified structures.
CNN based approaches are particularly beneficial for use in valve modelling applications since they eliminate the computational time that prior methods have reported, ranging from 15 minutes to 3 hours for a single segmentation. Deep learning methods instead can perform a segmentation in seconds, which removes a large bottleneck in valve modelling workflows. Additionally, since these methods are fully automatic, the resulting segmentations will be more consistent than semi-automatic or manual approaches, where individual users can vary greatly on how much of the atrial wall they label as leaflet. DeepMitral produces accurate segmentations in most cases, however in instances where the segmentation is sub-optimal manual editing of the result is still possible. DeepMitral can be easily integrated into 3D Slicer, which would allow for an initial segmentation to be created very quickly, and then be verified and edited if necessary before being used in any downstream applications.
4.1 Limitations and Future Work
DeepMitral fails to differentiate between chordae and leaflet in some images where the chordae are very clear, as seen in case P8 and to a lesser extent case P6 in Fig. 1. Currently we have a lack of training data where chordae are strongly delineated in the image, as this rarely occurs in TEE imaging. As a result, our model tends to classify the chordae as leaflet, as it presents as a similar image feature when visible. This will be addressed in future work by including an additional label for the chordae in our training set. This will allow our model to learn how to differentiate between leaflet and chordae. Additionally, sub-optimal image quality can cause the segmentation to perform poorly. This is a fundamental limitation when working with cardiac ultrasound, as it is possible for acquisitions to be very noisy, or lack detail of the mitral leaflets due to signal dropout. Expanding our data-set to include wider variations in image quality will allow us to better evaluate the conditions in which this approach can be successful. We plan on expanding our results beyond single frames to a 4D segmentation problem, that will allow us to incorporate the cyclical nature of cardiac motion into a segmentation workflow. Since different structures are better imaged at different phases of the cardiac cycle, this technique has the potential to further improve our results and overcome limitations due to image quality. Our methods could also be extended to adult tricuspid valves through transfer learning, enabling improved results as data availability for the tricuspid valve is more limited and image quality is generally poor.
5 Conclusions
The results from DeepMitral successfully replicate the gold standard segmentations with improved performance over existing state-of-the-art methods. Sub-millimetre average surface error in the segmentation stage are sufficient for use in patient specific valve modelling without manual intervention. We demonstrate the effectiveness of CNN based segmentation approaches for mitral valves from 3D TEE volumes. Improved mitral valve segmentation methods have wide applications including basic valve research and improved patient diagnostics. DeepMitral improves the workflow of the mitral valve modelling process by reducing the time required for completing an accurate mitral valve segmentation and providing more consistent and accurate results. Improvements in the mitral valve modelling workflow will lead to easier clinical translation, and will have implications in both surgical planning and training.
Conflict of interest
The authors declare that they have no conflict of interest.
References
Ailawadi, G., et al.: Is mitral valve repair superior to replacement in elderly patients? Ann. Thorac. Surg. 86(1), 77–86 (2008)
Andreassen, B.S., Veronesi, F., Gerard, O., Solberg, A.H.S., Samset, E.: Mitral annulus segmentation using deep learning in 3-D transesophageal echocardiography. IEEE J. Biomed. Health Inf. 24(4), 994–1003 (2020)
Benjamin, E.J., et al.: Heart disease and stroke statistics—2018 update: a report from the American heart association. Circulation 137(12), E67–E492 (2018)
Burlina, P., et al.: Patient-specific modeling and analysis of the mitral valve using 3D-TEE. In: Navab, N., Jannin, P. (eds.) IPCAI 2010. LNCS, vol. 6135, pp. 135–146. Springer, Heidelberg (2010). https://doi.org/10.1007/978-3-642-13711-2_13
Carnahan, P., et al.: Interactive-automatic segmentation and modelling of the mitral valve. In: Coudière, Y., Ozenne, V., Vigmond, E., Zemzemi, N. (eds.) FIMH 2019. LNCS, vol. 11504, pp. 397–404. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-21949-9_43
Costa, E., et al.: Mitral valve leaflets segmentation in echocardiography using convolutional neural networks. In: 2019 IEEE 6th Portuguese Meeting on Bioengineering (ENBENG), IEEE (February 2019)
Eleid, M.F., et al.: The learning curve for transcatheter mitral valve repair with MitraClip. J. Interv. Cardiol. 29(5), 539–545 (2016)
Ginty, O.K., et al.: Dynamic, patient-specific mitral valve modelling for planning transcatheter repairs. Int. J. Comput. Assist. Radiol. Surg. 14(7), 1227–1235 (2019)
Holzhey, D.M., Seeburger, J., Misfeld, M., Borger, M.A., Mohr, F.W.: Learning minimally invasive mitral valve surgery. Circulation 128(5), 483–491 (2013)
Ionasec, R.I., et al.: Patient-specific modeling and quantification of the aortic and mitral valves from 4-D cardiac CT and TEE. IEEE Trans. Med. Imaging 29(9), 1636–1651 (2010)
Jassar, A.S., et al.: Quantitative mitral valve modeling using real-time three-dimensional echocardiography: technique and repeatability. Ann. Thorac. Surg. 91(1), 165–171 (2011)
Kerfoot, E., Clough, J., Oksuz, I., Lee, J., King, A.P., Schnabel, J.A.: Left-Ventricle Quantification Using Residual U-Net. In: Pop, M., et al. (eds.) STACOM 2018. LNCS, vol. 11395, pp. 371–380. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-12029-0_40
Kingma, D.P., Ba, J.L.: Adam: A Method for Stochastic Optimization. In: CoRR, vol. 1412.6980 (2014)
Kozlowski, P., Bandaru, R.S., D’hooge, J., Samset, E.: Real-time catheter localization and visualization using three-dimensional echocardiography. In: Webster, R.J., Fei, B. (eds.) Medical Imaging 2017: Image-Guided Procedures, Robotic Interventions, and Modeling. SPIE (Mar 2017)
Liu, S., et al.: 3D anisotropic hybrid network: transferring convolutional features from 2D images to 3D anisotropic volumes. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 851–858. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_94
Mashari, A., et al.: Hemodynamic testing of patient-specific mitral valves using a pulse duplicator: a clinical application of three-dimensional printing. J. Cardiothorac. Vasc. Anesth. 30(5), 1278–1285 (2016)
McNeely, C.A., Vassileva, C.M.: Long-term outcomes of mitral valve repair versus replacement for degenerative disease: a systematic review. Curr. Cardiol. Rev. 11(2), 157–62 (2015). http://www.ncbi.nlm.nih.gov/pubmed/25158683
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional Neural Networks for Volumetric Medical Image Segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571. IEEE (October 2016)
Myronenko, A.: 3D MRI brain tumor segmentation using autoencoder regularization. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 311–320. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_28
Pouch, A.M., et al.: Fully automatic segmentation of the mitral leaflets in 3D transesophageal echocardiographic images using multi-atlas joint label fusion and deformable medial modeling. Med. Image Anal. 18(1), 118–129 (2014)
Ray, S.: Changing epidemiology and natural history of valvular heart disease. Clin. Med. 10(2), 168–171 (2010)
Scanlan, A.B., et al.: Comparison of 3D echocardiogram-derived 3D printed valve models to molded models for simulated repair of pediatric atrioventricular valves. Pediatr. Cardiol. 39(3), 538–547 (2017)
Schneider, R.J., Tenenholtz, N.A., Perrin, D.P., Marx, G.R., del Nido, P.J., Howe, R.D.: Patient-specific mitral leaflet segmentation from 4D ultrasound. In: Fichtinger, G., Martel, A., Peters, T. (eds.) MICCAI 2011. LNCS, vol. 6893, pp. 520–527. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-23626-6_64
Acknowledgements
We would like to acknowledge the following sources of funding: Canadian Institutes for Health Research, Natural Sciences and Engineering Research Council of Canada; Canadian Foundation for Innovation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Carnahan, P., Moore, J., Bainbridge, D., Eskandari, M., Chen, E.C.S., Peters, T.M. (2021). DeepMitral: Fully Automatic 3D Echocardiography Segmentation for Patient Specific Mitral Valve Modelling. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12905. Springer, Cham. https://doi.org/10.1007/978-3-030-87240-3_44
Download citation
DOI: https://doi.org/10.1007/978-3-030-87240-3_44
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87239-7
Online ISBN: 978-3-030-87240-3
eBook Packages: Computer ScienceComputer Science (R0)