Abstract
Being accountable for the signed reports, pathologists may be wary of high-quality deep learning outcomes if the decision-making is not understandable. Applying off-the-shelf methods with default configurations such as Local Interpretable Model-Agnostic Explanations (LIME) is not sufficient to generate stable and understandable explanations. This work improves the application of LIME to histopathology images by leveraging nuclei annotations, creating a reliable way for pathologists to audit black-box tumor classifiers. The obtained visualizations reveal the sharp, neat and high attention of the deep classifier to the neoplastic nuclei in the dataset, an observation in line with clinical decision making. Compared to standard LIME, our explanations show improved understandability for domain-experts, report higher stability and pass the sanity checks of consistency to data or initialization changes and sensitivity to network parameters. This represents a promising step in giving pathologists tools to obtain additional information on image classification models. The code and trained models are available on GitHub.
M. Graziani and I.P. de Sousa—Equal contribution (a complex randomization process was employed to determine the order of the first and second authors).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Convolutional Neural Networks (CNNs) can propose with very high accuracy regions of interest and their relative tumor grading in Whole Slide Images (WSIs), gigapixel scans of pathology glass slides [24]. This can support pathologists in clinical routine by reducing the size of the areas to analyze in detail and eventually highlighting missed or underestimated anomalies [3]. Without justifications for the decision-making, there is an opaque barrier between the model criteria and the clinical staff. Reducing such opaqueness is important to ensure the uptake of CNNs for sustained clinical use [22]. An already wide variety of off-the-shelve toolboxes has been proposed to facilitate the explanation of CNN decisions while keeping the performance untouched [2, 5, 14, 17, 19]. Among these, Local Interpretable Model-agnostic Explanations (LIME) are widely applied in radiology [16] and histopathology [15, 20].
As argued by Sokol and Flach [19], enhancements of existing explainability tools are needed to provide machine learning consumers with more accessible and interactive technologies. Existing visualization methods present pitfalls that urge for improvement, as pointed out by the unreliability shown in [1, 11]. LIME outputs for histopathology, for example, do not indicate any alignment of the explanations to clinical evidence and show high instability and scarce reproducibility [6]. Optimizing and reformulating this existing approach is thus a necessary step to promote its realistic deployment in clinical routines.
In this work, we propose to employ a better segmentation strategy that leads to sharper visualizations, directly highlighting relevant nuclei instances in the input images. The proposed approach brings improved understandability and reliability. Sharp-LIME heat maps appear more understandable to domain experts than the commonly used LIME and GradCAM techniques [18]. Improved reliability is shown in terms of result consistency over multiple seed initializations, robustness to input shifts, and sensitivity to weight randomizations. Finally, Sharp-LIME allows for direct interaction with pathologists, so that areas of interest can be chosen for explanations directly. This is desirable to establish trust [19]. In this sense, we propose a relevant step towards reliable, understandable and more interactive explanations in histopathology.
2 Methods
2.1 Datasets
Three publicly available datasets are used for the experiments, namely Camelyon 16, Camelyon 17 [13] and the breast subset of the PanNuke dataset [4]Footnote 1. Camelyon comprises 899 WSIs of the challenge collection run in 2017 and 270 WSIs of the one in 2016. Slide-level annotations of metastasis type (i.e. negative, macro-metastases, micro-metastases, isolated tumor cells) are available for all training slides, while a few manual segmentations of tumor regions are available for 320 WSIs. Breast tissue scans from the PanNuke dataset are included in the analysis. For these images, the semi-automatic instance segmentation of multiple nuclei types is available, allowing to identify neoplastic, inflammatory, connective, epithelial, and dead nuclei in the images. No dead nuclei are present, however, in the breast tissue scans [4]. Image patches of \(224 \times 224\) pixels are extracted at the highest magnification level from the WSIs to build training, validation and test splits as in Table 1. To balance the under-representation, PanNuke input images were oversampled by five croppings, namely in the center, upper left, upper right, bottom left and bottom right corners. The pre-existing PanNuke folds were used to separate the patches in the splits. Reinhard normalization is applied to all the patches to reduce the stain variability.
2.2 Network Architectures and Training
Inception V3 [21] with ImageNet pre-trained weights is used for the analysis. The network is fine-tuned on the training images to classify positive patches containing tumor cells. The fully connected classification block has four layers, with 2048, 512, 256 and 1 neurons. A dropout probability of 0.8 and L2 regularization were used to avoid overfitting. This architecture was trained with mini-batch Stochastic Gradient Descent (SGD) optimization with standard parameters (learning rate of \(1e^{-4}\), Nesterov momentum of 0.9). For the loss function, class-weighted binary cross-entropy was used. Network convergence is evaluated by early stopping on the validation loss with patience of 5 epochs. The model performance is measured by the average Area Under the ROC Curve (AUC) over ten runs with multiple initialization seeds, reaching \(0.82 \pm {0.0011}\) and \(0.87 \pm {0.005}\) for the internal and external test sets respectively.
Nuclei contours of the Camelyon input are extracted by a Mask R-CNN model [7] fine-tuned from ImageNet weights on the Kumar dataset for the nuclei segmentation task [12]. The R-CNN model identifies nuclei entities and then generates pixel-level masks by optimizing the Dice score. ResNet50 [7] is used for the convolutional backbone as in [10]. The network is optimized by SGD with standard parameters (learning rate of 0.001 and momentum of 0.9).
2.3 LIME and Sharp-LIME
LIME for Image Classifiers Defined by Ribeiro et al. [17] for multiple data classifiers, a general formulation of LIME is given by:
Eq. (1) represents the minimization of the explanatory infidelity \(\mathcal {L}(f,g,\pi _{x})\) of a potential explanation g, given by a surrogate model G, in a neighborhood defined by \(\pi _{x}(z)\) around a given sample of the dataset (x). The neighborhood is obtained by perturbations of x around the decision boundary.
For image classifiers, that are the main focus of this work, an image x is divided into representative image sub-regions called super-pixels using a standard segmentation algorithm, e.g. Quickshift [23]. Perturbations of the input image are obtained by filling random super-pixels with black pixels. The surrogate linear classifier G is a ridge regression model trained on the perturbed instances weighed by the cosine similarity (\(\pi _{x}(z)\)) to approximate the prediction probabilities. The coefficients of this linear model (referred to as explanation weights) explain the importance of each super-pixel to the model decision-making. Explanation weights are displayed in a symmetrical heatmap where super-pixels in favor of the classification (positive explanation weights) are in blue, and those against (negative weights) in red.
Previous improvements of LIME for histopathology proposed a systematic manual search for parameter heuristics to obtain super-pixels that visually correspond to expert annotations [20]. Consistency and super-pixel quality were further improved by genetic algorithms in [15]. Both solutions are impractical for clinical use, being either too subjective or too expensive to compute.
Sharp-LIME The proposed implementation of Sharp-LIME, as illustrated in Fig. 1, uses nuclei contours as input super-pixels for LIME rather than other segmentation techniques. Pre-existing nuclei contour annotations may be used. If no annotations are available, the framework suggests automatic segmentation of nuclei contours by the Mask R-CNN. Manual annotations of regions of interest may also be drawn directly by end-users to probe the network behavior for specific input areas. For the super-pixel generation, the input image is split into nuclei contours and background. The background is further split into 9 squares of fixed size. This splitting reduces the difference between nuclei and background areas, since overly large super-pixels may achieve large explanation weights by sheer virtue of their size. The code to replicate the experiments (developed with Tensorflow \(>2.0\) and Keras 2.4.0) is available at github.com/maragraziani/sharp-LIME, alongside the trained CNN weights. Experiments were run using a GPU NVIDIA V100. A single Sharp-LIME explanation takes roughly 10 s to generate in this setting. 200 perturbations were used, as it already showed low variability in high explanation weight super-pixels, as further discussed in Sect. 3.
2.4 Evaluation
Sharp-LIME is evaluated against the state-of-the-art LIME by performing multiple quantitative evaluations. Not having nuclei type labels for Camelyon, we focused on the PanNuke data. We believe, however, that the results would also apply to other inputs. Sanity checks are performed, testing for robustness to constant input shifts and sensitivity to network parameter changes as in [1, 11]. Spearman’s Rank Correlation Coefficient (SRCC) is used to evaluate the similarity of the ranking of the most important super-pixels. The cascading randomization test in [1] is performed by assigning random values to the model weights starting from the top layer and progressively descending to the bottom layer. We already expect this test to show near-zero SRCC for both techniques, since by randomizing the network weights, the network output is randomized as well as LIME and Sharp-LIME explanations. The repeatability and consistency for multiple seed initializations are evaluated by the SRCC, the Intraclass Correlation Coefficient (ICC) (two-way model), and the coefficient of variation (CV) of the explanation weights.
Additionally, we quantify domain appropriateness as the alignment of the explanations with relevant clinical factors [22]. The importance of a neoplastic nucleus, an indicator of a tumor [4], is measured by the sign and magnitude of the explanation weight. Descriptive statistics of the explanation weights are compared across the multiple types of nuclei in PanNuke. Pairwise non-parametric Kruskal tests for independent samples are used for the comparisons. A paired t-test is used to compare LIME weights obtained from a randomly initialized and a trained network, as suggested in [6].
3 Results
3.1 Improved Understandability
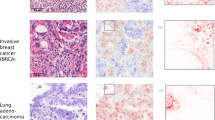
Qualitative Evaluation By Domain Experts. Figure 2 shows a qualitative comparison of LIME and Sharp-LIME for PanNuke and Camelyon inputs. For conciseness, only two examples are provided. An extended set of results can be inspected in the GitHub repositoryFootnote 2.
a) Comparison between Sharp-LIME explanation weights for a trained and a randomly initialized CNN; b) Zoom on the random CNN in a). These results can be compared to those obtained for standard LIME in [6].
Five experts in the digital pathology domain with experience in CNN-based applications for clinical research purposes compared LIME, Sharp-LIME and Gradient Weighted Class Activation Mapping (Grad-CAM) [18] for a few images in this work. The experts generally use these visualizations to improve their model understanding, particularly if the suggested diagnosis is different from theirs. Sharp-LIME was assessed as easier to understand than Grad-CAM and LIME by 60% of them. Two of the five experts further confirmed that these explanations help increasing their confidence in the model’s decision-making. While it is difficult to obtain quantitative comparisons, we believe this expert feedback, although subjective, is an essential evaluation.
3.2 Improved Reliability
Quantification of Network Attention. We quantify the Sharp-LIME explanation weights for each of the functionally diverse nuclei types of the PanNuke dataset in Fig. 3. As Fig. 3a shows, the explanation weights of the neoplastic nuclei, with average value \(0.022 \pm 0.03\), are significantly larger than those of the background squared super-pixels, with average value \(-0.018 \pm 0.05\). Explanation weights of the neoplastic nuclei are also significantly larger than those of inflammatory, neoplastic and connective nuclei (Kruskal test, p-value \(<0.001\) for all pairings). Sharp-LIME weights are compared to those obtained by explaining a random CNN, that is the model with randomly initialized parameters. The Sharp-LIME explanation weights for the trained and random CNN present significant differences (paired t-test, p-value\(<0.001\)), with the explanations for the latter being almost-zero values as shown by the boxplot in Fig. 3b.
Consistency. The consistency of Sharp-LIME explanations for multiple seed initialization is shown in Figs. 4a and 4b. The mean of LIME SRCC is significantly lower than that of Sharp-LIME, 0.015 against 0.18 (p-value\(<0.0001\)). As Fig. 4b shows, super-pixels with large average absolute value of the explanation weight are more consistent across re-runs of Sharp-LIME, with lower CV. We compare the SRCC of the five super-pixels with the highest ranking, obtaining average LIME explanation weights 0.029 and 0.11 for Sharp-LIME. The ICC of the most salient super-pixel in the image, i.e. first in the rankings, for different initialization seeds, further confirms the largest agreement of Sharp-LIME, with ICC 0.62 against the 0.38 of LIME. As expected, the cascading randomization of network weights shows nearly-zero SRCC in Fig. 4c. A visual example of LIME robustness to constant input shifts is given in Fig. 5a. The SRCC of LIME and Sharp-LIME is compared for original and shifted inputs with unchanged model prediction in Fig. 5b. Sharp-LIME is significantly more robust than LIME (t-test, p-value\(<0.001\)).
a) SRCC of the entire and top-5 super-pixel rankings obtained over three re-runs with changed initialization. The means of the distributions are significantly different (paired t-test, p-value\(<0.001\)); b) CV against average explanation weight for three re-runs with multiple seeds; c) SRCC of the super-pixel rankings obtained in the cascading randomization test
4 Discussion
The experiments evaluate the benefits of the Sharp-LIME approach against the standard LIME, showing improvements in the understandability and reliability of the explanations. This improvement is given by the choice of a segmentation algorithm that identifies regions with a semantic meaning in the images. Differently from standard LIME, Sharp-LIME justifies the model predictions by the relevance of image portions that are easy to understand as shown in Fig. 2. Our visualizations have higher explanation weights and show lower variability than standard LIME. The feedback from the domain-experts is encouraging (Sect. 3.2). Despite being only qualitative, it reinforces the importance of a feature often overseen in explainability development, namely considering the target of the explanations during development to provide them with intuitive and reliable tools. The quantitative results in Sect. 3.2 show the improved reliability of Sharp-LIME. Neoplastic nuclei appear more relevant than other nuclei types, aligning with clinical relevance. Since these nuclei are more frequent than other types in the data, the results are compared to a randomly initialized CNN to confirm that their importance is not due to hidden biases in the data (Fig. 3). The information contained in the background, often highlighted as relevant by LIME or Grad-CAM [6], seems to rather explain the negative class, with large and negative explanation weights on average. Large Sharp-LIME explanation weights point to relevant super-pixels with little uncertainty, shown by low variation and high consistency in Figs. 4b and 4a. The instability of LIME reported in [6] can therefore be explained by the choice of the segmentation algorithm, an observation in line with the work in [20].
The simplicity of this approach is also its strength. Our super-pixel choice of nuclei segmentation adds little complexity to the default LIME, being a standard data analysis step in various histopathology applications [8, 9]. Extensive annotations of nuclei contours are not needed since automated contouring can be learned from small amounts of labeled data [8] (Fig. 2b). Additionally, the users may directly choose the input super-pixels to compare, for example, the relevance of one image area against the background or other areas. Requiring only a few seconds to be computed, Sharp-LIME is faster than other perturbation methods that require a large number of forward passes to find representative super-pixels. For this reason, the technique represents a strong building-block to develop interactive explainability interfaces where users can visually query the network behavior and quickly receive a response.
The small number of available experts is a limitation of this study, which does not propose quantitative estimates of user confidence and satisfaction in the explanations. We will address this point in future user-evaluation studies.
5 Conclusions
This work shows important points in the development of explainability for healthcare. Optimizing existing methods to the application requirements and user satisfaction promotes the uptake and use of explainability techniques.
Our proposed visualizations are sharp, fast to compute and easy to apply to black-box histopathology classifiers by focusing the explanations on nuclei contours and background portions. Other image modalities may benefit from this approach. The relevance of the context surrounding tumor regions, for example, can be evaluated in radiomics. Further research should focus on the specific demands of the different modalities.
Notes
- 1.
camelyon17.grand-challenge.org and jgamper.github.io/PanNukeDataset.
- 2.
(github.com/maragraziani/sharp-LIME).
References
Adebayo, J., Gilmer, J., Muelly, M., Goodfellow, I., Hardt, M., Kim, B.: Sanity checks for saliency maps. In: Proceedings of the 32nd International Conference on Neural Information Processing Systems, p. 9525–9536. NIPS 2018, Curran Associates Inc., Red Hook, NY, USA (2018)
Alber, M., Let al.: Innvestigate neural networks! J. Mach. Learn. Res. 20(93), 1–8 (2019). http://jmlr.org/papers/v20/18-540.html
Fraggetta, F.: Clinical-grade computational pathology: alea iacta est. J. Pathol. Inf. 10 (2019)
Gamper, J., Alemi Koohbanani, N., Benet, K., Khuram, A., Rajpoot, N.: PanNuke: an open pan-cancer histology dataset for nuclei instance segmentation and classification. In: Reyes-Aldasoro, C.C., Janowczyk, A., Veta, M., Bankhead, P., Sirinukunwattana, K. (eds.) ECDP 2019. LNCS, vol. 11435, pp. 11–19. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-23937-4_2
Graziani, M., Andrearczyk, V., Müller, H.: Regression concept vectors for bidirectional explanations in histopathology. In: Stoyanov, D., et al. (eds.) MLCN/DLF/IMIMIC -2018. LNCS, vol. 11038, pp. 124–132. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-02628-8_14
Graziani, M., Lompech, T., Müller, H., Andrearczyk, V.: Evaluation and comparison of CNN visual explanations for histopathology. In: Explainable Agency in Artificial Intelligence at AAAI21, pp. 195–201 (2020)
He, K., Gkioxari, G., Dollar, P., Girshick, R.: Mask r-CNN. In: Proceedings of the IEEE International Conference on Computer Vision (ICCV) (2017)
Janowczyk, A., Madabhushi, A.: Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inf. 7 (2016)
Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M., Madabhushi, A.: Histoqc: an open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inf. 3, 1–7 (2019)
Jung, H., Lodhi, B., Kang, J.: An automatic nuclei segmentation method based on deep convolutional neural networks for histopathology images. BMC Biomed. Eng. 1, 1–2 (2019). https://doi.org/10.1186/s42490-019-0026-8
Kindermans, P.J., et al.: The (un) Reliability of Saliency Methods. Interpreting, Explaining and Visualizing Deep Learning, Springer International Publishing, Explainable AI (2019)
Kumar, N., et al.: A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 36(7), 1550–1560 (2017). https://doi.org/10.1109/TMI.2017.2677499
Litjens, G., et al.: 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience 7(6), giy065 (2018)
Lundberg, S.M., Lee, S.I.: A unified approach to interpreting model predictions. In: Guyon, I., et al. (eds.) Advances in Neural Information Processing Systems 30, pp. 4765–4774. Curran Associates, Inc. (2017). http://papers.nips.cc/paper/7062-a-unified-approach-to-interpreting-model-predictions.pdf
Palatnik de Sousa, I., Bernardes Rebuzzi Vellasco, M.M., Costa da Silva, E.: Evolved Explainable Classifications for Lymph Node Metastases. arXiv e-prints arXiv:2005.07229, May 2020
Reyes, M., et al.: On the interpretability of artificial intelligence in radiology: challenges and opportunities. Radiol.: Artif. Intell. 2, e190043 (2020). https://doi.org/10.1148/ryai.2020190043
Ribeiro, M.T., Singh, S., Guestrin, C.: Why Should I Trust You?: explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016, pp. 1135–1144 (2016)
Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017)
Sokol, K., Flach, P.: One explanation does not fit all. KI-Künstliche Intelligenz, pp. 1–16 (2020)
Palatnik de Sousa, I., Bernandes Rebuzzi Vellasco, M.M., Costa da Silva, E.: Local interpretable model-agnostic explanations for classification of lymph node metastases. Sensors (Basel, Switzerland) 19 (2019)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Tonekaboni, S., Joshi, S., McCradden, M.D., Goldenberg, A.: What clinicians want: contextualizing explainable machine learning for clinical end use. In: Machine Learning for Healthcare Conference, pp. 359–380. PMLR (2019)
Vedaldi, A., Soatto, S.: Quick Shift and Kernel Methods for Mode Seeking. In: Forsyth, D., Torr, P., Zisserman, A. (eds.) ECCV 2008. LNCS, vol. 5305, pp. 705–718. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-88693-8_52
Wang, D., Khosla, A., Gargeya, R., Irshad, H., Beck, A.H.: Deep learning for identifying metastatic breast cancer. arXiv preprint arXiv:1606.05718 (2016)
Acknowledgements
We thank MD. Dr. Filippo Fraggetta for his relevant feedback on our methods. We also thank Lena Kajland Wilén, Dr. Francesco Ciompi and the Swiss Digital Pathology consortium for helping us getting in contact with the pathologists that participated in the user studies. This work is supported by the European Union’s projects ExaMode (grant 825292) and AI4Media (grant 951911).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Graziani, M., Palatnik de Sousa, I., Vellasco, M.M.B.R., Costa da Silva, E., Müller, H., Andrearczyk, V. (2021). Sharpening Local Interpretable Model-Agnostic Explanations for Histopathology: Improved Understandability and Reliability. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12903. Springer, Cham. https://doi.org/10.1007/978-3-030-87199-4_51
Download citation
DOI: https://doi.org/10.1007/978-3-030-87199-4_51
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87198-7
Online ISBN: 978-3-030-87199-4
eBook Packages: Computer ScienceComputer Science (R0)