Abstract
In this work we present a coupled electromechanical model of the heart for patient-specific simulations, and in particular cardiac resynchronisation therapy. To this end, we propose a fast fully autonomous and flexible pipeline to generate and optimise the data required to run the mechanical simulation. After the meshing of the biventricular segmentation image and the construction of the associated fibres arrangement, we compute the electrical potential propagation in the myocardial tissue from selected onset points on the endocardium. We generate a 12-lead electrocardiogram corresponding to the latter activation map by extrapolating the electrical potential on a virtual torso. This electrical activation is coupled to a mechanical model, featuring a small set of interpretable parameters. We also propose an efficient algorithm to optimise the model parameters, based on patient data. The whole pipeline including a cardiac cycle is computed in 30 min, enabling to use this digital twin for diagnosis and therapy planning.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Building patient-specific 3D models of the heart can help improving diagnosis and therapy selection for various cardiac diseases. They can for instance help interventional cardiologists in choosing the best pacing method and anticipate the patient response based on the chosen pacing sites [12]. However, personalisation, i.e. adapting a generic cardiac model to a specific patient, is still challenging due to theoretical (identifiability) and practical (computation time) issues.
The model presented here combines different modelling approaches and tools that are well suited for personalisation, and results in a computational time compatible with clinical constraints. In this manuscript, we detail the different elements, how we can adjust their parameters, and present simulation results to evaluate their feasibility. The computational cost of each step is also detailed.
This pipeline is based on various previous studies that have been combined to propose for the first time in the team a complete and fast method for running a personalised patient-specific simulation of a beating heart, including:
-
a local anatomical correction tool,
-
a realistic approach to simulate the Purkinje activation and electrical propagation,
-
algorithms to quickly generate body surface electrocardiograms,
-
a ready-to-use mechanical model,
-
a sensitivity analysis to assess the model’s observability and
-
algorithms to personalise the models’ parameters based on patient data.
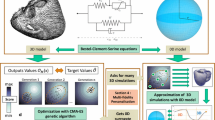
Pipeline for patient-specific simulation. a. Initial and modified geometries. b. Fibre directions. c. Activation points and activation times on the endocardial surfaces. d. Activation map. e. Leads location on a virtual torso. f. 12-lead ECG. This pipeline generates all the data required to run the mechanical simulation
2 Anatomical Model
We suppose here that the patient biventricular myocardium was already segmented from 3D images. This is more and more available thanks to deep learning approaches. Note that the atria are not included in our 3D model, but their interactions with the ventricles are modelled in different ways: a delay is introduced in the electrical model to simulate the propagation of the electrical signal from the sino-atrial to the auriculo-ventricular node; the atrial pressure is modelled as a sigmoid curve; and the atrial contraction is taken into account in the boundary conditions, as a spring force on the basal tetrahedra.
2.1 Meshing and Labelling
The binary mask from the segmentation (Fig. 2a) is resampled into \(1\times 1\times 1mm^3\) voxels and contains the labels for left and right ventricles. The labels for endocardium, myocardium and epicardium are built with a ray-tracing method, emerging from each ventricle barycentre. A tetrahedral mesh is built using the remeshing software MMG [6] (Fig. 2b), resulting in a mesh of approximately 90k tetrahedra and 20k vertices.
In addition to the generated mesh, tools were developed to virtually modify the anatomy, in order to increase the available number of healthy and pathological cases, and to correct any inaccuracies from the segmentation method. For instance, on Fig. 1a, the right ventricle volume has been reduced by 20%. Several pathological cases could be generated similarly from a healthy heart geometry, for example dilated or hypertrophic cardiomyopathies.
2.2 Cardiac Fibres
The spatial orientation of the muscle fibres plays a major role in the excitation and contraction of the heart [13]. We suppose that the fibre helix angle varies from \(-\alpha \) to \(+\alpha \) across the myocardial wall and remains constant throughout the cardiac cycle. We neglect the angle variations in any other direction.
To compute the fibres direction, we need to build the local coordinate system in each voxel. To this end, we first compute the tissue thickness using a simple diffusion equation on the endocardium and epicardium as in [14]. The resultant vector field gives the radial direction. The other directions are then easy to obtain. The fibre directions are given by the circumferential basis vector, rotated by an angle \(\alpha _f\in \left[ -\alpha ,+\alpha \right] \) along the radial direction (Fig. 1b). In this framework, the angle variation is chosen to linearly increase from the endocardium \((\alpha _f=-80^o)\) to the epicardium \((\alpha _f=80^o)\). Note that the septum is considered as left ventricle for the fibres generation.
2.3 AHA Regions
The 17 American Heart Association (AHA) segments are defined as a standardised segmentation of the left ventricle. We can extend this to 29 regions (Fig. 2b) for the whole myocardial tissue. These segments are mostly used in the mechanical simulation. They can also give a good insight on regional properties such as stress and strain, thus help in assessing intraventricular dyssynchrony.
To build these regions for each ventricle, we let an ellipsoid fit the ventricle mesh vertices and use the spherical coordinates to generate the regions on the ellipsoid. Finally, the segments are projected onto the tetrahedral mesh [3]. At the base of the mesh, a stiffer and less conductive region is introduced to model the valves fibrous tissue (beige region at the top of the mesh in Fig. 2b).
3 Electrophysiology Simulation
3.1 Electrical Activation
At each cardiac cycle, the mechanical contraction of the heart is driven by the electrical activation. The electrical signal emerging from the auriculo-ventricular node is conducted along the bundle of His and the Purkinje network to the endocardium (Fig. 1c). The numerous termination points of the Purkinje fibres on the endocardium are reduced to a dozen of points (Fig. 1d) and selected matching a real endocardial map (Fig. 3). To account for the fast potential propagation due to the Purkinje fibres, a thin endocardial layer is assigned a higher isotropic conductivity. These onset points are activated between 0 and 21ms, \(t=0\) marking the beginning of the QRS complex. In the remaining tissue, the potential propagation is computed using a fast marching method and solving the anisotropic Eikonal equation [10] for each mesh vertex:
After a short period of time (duration of the QRS), all the myocardial cells are activated and contract until the repolarisation wave arrives. The action potential duration (APD) is linearly interpolated from the endocardium to the epicardium (shorter on the epicardium), using a wall depth map.
3.2 ECG Generation
The ECG records the bio-electric activity of the cardiac cells by measuring the electrical potential at the 6 precordial leads placed on the patient’s torso and 3 (or 4) limb leads (Fig. 1e). In order to simulate this electrical activity, we consider each voxel of the image as a dipole of current density \(\mathbf{j} _{eq} = -\sigma \nabla v\) where \(\nabla v\) is the spacial gradient of the potential v [4]. By using the chain rule, we obtain:
where \(\sigma \) is the local conductivity, \(\nabla T\) the gradient of the activation map and \(\frac{\partial v}{\partial T}\) is given by solving the Mitchell-Schaeffer model [11] using a forward Euler scheme. We suppose here that the body is a homogeneous material with constant conductivity \(\sigma _T\). The electrical potential at a distance r from the source is developed in [8] and is given by:
We finally derive the potential contribution of each voxel with the Einthoven triangle to obtain the augmented leads and plot the electrocardiogram Fig. 1f.
4 Cardiac Mechanics
After the electrical depolarisation, the muscle cells in the heart release ions that lead to sarcomere shortening and active muscle contraction. These cardiomyocites are surrounded by an extracellular matrix that accounts for the passive material. We use here the Bestel-Clement-Sorine [1] model, further improved by [5], based on a multi-scale physiological description of the myocardial muscle function.
4.1 Bestel-Clement-Sorine Model
In this model, the heart is described as a passive isotropic Mooney-Rivlin material with 3 parameters, which accounts for the elasticity and friction in the cardiac extracellular matrix (mainly collagen) surrounding the fibres.
The electrical stimulation is derived from the Eikonal activation map computed beforehand and is coupled to the active orthotropic contraction part, which has 7 main parameters and accounts for the active stress (Fig. 4) along the cardiac fibres and the elasticity between sarcomeres and Z-discs. Note that combining the Mooney-Rivlin isotropic material with the elasticity of the Z-discs, the material is globally considered transversely isotropic.
The cardiac cycle is decomposed into four phases: filling, isovolumetric contraction, ejection and isovolumetric relaxation (Fig. 5 and Fig. 6). We couple a haemodynamic model implementing those phases and use the four-element Windkessel model to compute the arterial pressures.
Moreover, we performed a sensivity analysis (Table 1) on the mechanical model using a Latin Hypercube Sampling method, coupled with the computation of the Partial Ranked Correlation Coefficient [9] for a collection of relevant model outputs, selected from [2].
The four first parameters evaluated are the four elements of the Windkessel model and thus are not, or poorly, correlated with non-ejection indexes such as LPEI or the isovolumetric contraction time. Since they control the shape of the arterial and ventricular pressures during the ejection, a strong correlation on the ejection time and ejection fraction was expected. The closing time of the aortic and pulmonary valves is also depending on those pressures, thus the correlation observed on the isovolumetric relaxation time. We can notice that the relaxation rate, the myocardial passive stiffness and the fibres extremum angle do not seem to impact these model outputs. Finally, the contractility is strongly correlated with all the considered indexes.
5 Personalisation Method
This model can be customised with only a few set of parameters and is therefore adapted to personalisation. To this end, we use the Covariance Matrix Adaptation Evolution Strategy (CMA-ES), an evolutionary algorithm for difficult non-linear non-convex black-box optimisation problems [7]. The parameters are sampled according to a multi-variate normal distribution. The covariance matrix of this distribution is iteratively updated such as the new set of parameters minimises the cost function. The latter is built such that the output of the model with the current parameters fits the patient data. Moreover, this personalisation method is embarrassingly parallel, as all simulations are independent within an iteration.
Preliminary results on the mechanical model are presented in Fig. 7. For these results, only the Windkessel and the contractility parameters have been included in the CMA-ES framework, thus spanning a five-dimensional parameter space.
5.1 Anatomy
Depending on the available patient data, it is possible to generate a mesh that meets several constraints: axis length, volumes, wall thickness, etc. If the anatomy of the heart is poorly defined, it is possible to adjust the geometry of mesh based on the patient’s cardiomyopathy.
5.2 Electrophysiology
In order to match the QRS axis and the QRS duration of each patient, the following parameters are being optimised:
-
depolarisation delay of the Purkinje breakthrough points (one parameter for all termination points or one per point),
-
position on the endocardium of these points (two parameters per point) and
-
endocardial and myocardial conductivities (two parameters).
5.3 Mechanics
All the indexes presented in Table 1 (columns of the table) will be used for the mechanical personalisation. Again, the following parameters (rows of the table) are being optimised:
-
four elements of the Windkessel model. Note that the parameters for the right ventricle are adjusted according to those of the left ventricle and are not counted here (four parameters),
-
contraction and relaxation parameters (two parameters),
-
Mooney-Rivlin material parameters (three parameters) and
-
active parameters, sarcomere contractility and viscosity, Z-discs elasticity (three parameters).
5.4 Discussion
Figure 7 presents some personalisation results regarding two and four patient indexes, on the mechanical model, optimised with five parameters. In both cases the algorithm converges toward the set of parameters that minimises the global error. With four indexes, the algorithm only achieved a global relative error of 25% compared to the user-defined target values. Either the global minimum of the error function was found, or the algorithm has fallen into a local minimum, and the global minimum has been excluded from the parameter space due to overly restrictive bounds.
This personalisation method has shown great promises in our preliminary tests, in terms of computational cost and global error. We are currently implementing the CMA-ES method for both the electrophysiological and mechanical frameworks on a computer cluster.
This algorithm converges in only a few iterations, for example 30 iterations if the population size is ten times the dimension of the parameter space. This space is spanned by a user-defined range of realistic values for each parameter.
With this configuration, the electrophysiological model can be personalised in less than two hours and the mechanical model in less than twelve hours on a computer cluster.
6 Conclusion
In this manuscript, we have presented a fast (see Table 2) pipeline for patient-specific electromechanical simulations, using a model relying on a small set of interpretable parameters, allowing for extensive personalisation. This pipeline is built upon three submodels: an anatomical one, allowing local corrections, as well as mesh and fibres generation; an electrophysiological one, generating ECGs and simulating the complex activation and propagation of the electrical potential; and a mechanical submodel. We also lead a sensitivity analysis to assess the model’s observability, and developed methods to efficiently fit the model output to patient data.
Such approach opens up possibilities in order to use cardiac modelling within clinical applications.
References
Bestel, J., Clément, F., Sorine, M.: A biomechanical model of muscle contraction. In: Niessen, W.J., Viergever, M.A. (eds.) MICCAI 2001. LNCS, vol. 2208, pp. 1159–1161. Springer, Heidelberg (2001). https://doi.org/10.1007/3-540-45468-3_143
Cazeau, S., Toulemont, M., Ritter, P., Reygner, J.: Statistical ranking of electromechanical dyssynchrony parameters for CRT. Open Heart 6(1), e000933 (2019)
Cedilnik, N., et al.: Fast personalized electrophysiological models from computed tomography images for ventricular tachycardia ablation planning. EP Europace 20(suppl_3), iii94–iii101 (2018)
Cedilnik, N., et al.: Fully automated electrophysiological model personalisation framework from CT imaging. FIMH 2019, 325–333 (2019)
Chapelle, D., Le Tallec, P., Moireau, P., Sorine, M.: An energy-preserving muscle tissue model: formulation and compatible discretizations. Int. J. Multiscale Comput. Eng. 10(2), 189–211 (2012)
Dapogny, C., Dobrzynski, C., Frey, P.: Three-dimensional adaptive domain remeshing, implicit domain meshing, and applications to free and moving boundary problems. J. Comput. Phys. 262, 358–378 (2014)
Hansen, N.: The CMA evolution strategy: a comparing review. In: Lozano, J.A., Larrañaga, P., Inza, I., Bengoetxea, E. (eds.) Towards a New Evolutionary Computation. Studies in Fuzziness and Soft Computing, vol. 192, pp. 75–102. Springer, Berlin https://doi.org/10.1007/3-540-32494-1_4
Malmivuo, J., Plonsey, R.: Bioelectromagnetism - Principles and Applications of Bioelectric and Biomagnetic Fields (10 1995)
Marino, S., Hogue, I.B., Ray, C.J., Kirschner, D.E.: A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 254(1), 178–196 (2008)
Sermesant, M., et al.: An anisotropic multi-front fast marching method for real-time simulation of cardiac electrophysiology. In: Sachse, F.B., Seemann, G. (eds.) FIMH 2007. LNCS, vol. 4466, pp. 160–169. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-72907-5_17
Mitchell, C.: A two-current model for the dynamics of cardiac membrane. Bull. Math. Biol. 65(5), 767–793 (2003)
Sermesant, M., et al.: Patient-specific electromechanical models of the heart for the prediction of pacing acute effects in CRT: a preliminary clinical validation. Med. Image Anal. 16(1), 201–215 (2012)
Streeter, D.D., Spotnitz, H.M., Patel, D.P., Ross, J., Sonnenblick, E.H.: Fiber orientation in the canine left ventricle during diastole and systole. Circ. Res. 24(3), 339–347 (1969)
Yezzi, A.J., Prince, J.L.: An eulerian pde approach for computing tissue thickness. IEEE Trans. Med. Imaging 22(10), 1332–1339 (2003)
Acknowledgments
This work has been supported by the French government through the National Research Agency (ANR) Investments in the Future 3IA Côte d’Azur (ANR-19-P3IA-000) and by Microport CRM funding.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Desrues, G., Feuerstein, D., Legay, T., Cazeau, S., Sermesant, M. (2021). Personal-by-Design: A 3D Electromechanical Model of the Heart Tailored for Personalisation. In: Ennis, D.B., Perotti, L.E., Wang, V.Y. (eds) Functional Imaging and Modeling of the Heart. FIMH 2021. Lecture Notes in Computer Science(), vol 12738. Springer, Cham. https://doi.org/10.1007/978-3-030-78710-3_43
Download citation
DOI: https://doi.org/10.1007/978-3-030-78710-3_43
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-78709-7
Online ISBN: 978-3-030-78710-3
eBook Packages: Computer ScienceComputer Science (R0)