Abstract
Phylum Planctomycetes are unique, atypical, and distinct of the bacteria species resembling that of eukaryotes. The organisms have unusual features such as cystolic compartmentalization. They also possess characteristics such as endocytosis-like macromolecule uptake. These peculiar bacteria are equally important environmentally, biologically, and biotechnologically. The organization of cells of the phylum planctomycetes species shall be examined by thin ultrastructure sections of the cryosubstituted cell. Studies revealed cell architecture of the phylum is a gram-negative bacterium. This chapter in detail provides the knowledge of cell organization of planctomycetes, structural significance, and its importance in cell biology and control mechanism of intracellular compartmentalization.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Planctomycetes is a bacteria phylum initially described by a Hungarian biologist Nandor Gimesi in Lake Lagymanyos, Budapest (Gimesi 1924). When cultured, they seem to be atypical microcolonies. He named the species “Planctomycetes bekefi” in the name of a Hungarian abbot, Remigius Bekefi, assuming the species be a planktonic fungus. The organism showed unique morphology with several circular cells, pear-shaped, ellipsoid, or club-shaped, each in its noncellular stalk with a holdfast at its tip giving the shape as microcolonies rosette. A wide range of species has originated in different regions all over the world. Few such species are Planctomyces guttaeformis (Hortobágyi 1965) and Planctomyces stranskae, Rhodopirellula, and Thermogutta. The bacteria reproduce by budding are named as Blastocaulis sphaerica for the stalked form and Blastobacter for the non-stalked (Henrici and Johnson 1935).
Taxonomically, the phylum Planctomycetes and superphylum PVC divides into two classes and orders. Grouping the class Phycisphaerae into different orders is based on the cultured and uncultured strains such as Phycisphaerales and Algisphaera, Phycisphera and Tepidisphaera, respectively (Fukunaga et al. 2009; Kovaleva et al. 2015; Yoon et al. 2014). While the orders Planctomycetales and Brocadiales (anammox planctomycetes) descend under the class Planctomycetia (Schlesner and Stackebrandt 1986; Jetten et al. 2015; Ward 2015). The 16S rRNA sequencing suggested diverse microbial community and much larger taxonomy dividing into ten classes, sixteen orders, and 43 families and much genetic diversity (Yilmaz et al. 2016).
Studies earlier considered that the planctomycetes cell wall does not constitute peptidoglycan where almost all the bacterial species have (König et al. 1984; Liesack et al. 1986), the same was noted in Chlamydia also. But the latest studies on re-examination found out peptidoglycan polymer in both Chlamydia and planctomycetes; however, the function and its localization are not that apparent (Jeske et al. 2015; van Teeseling et al. 2015; Pilhofer et al. 2013). Most planctomycetes are multicellular while, few form biofilms, for example, Planctomyces limnophilus (Giovannoni et al. 1987). Planctomycetes have a complex life cycle relating to both sessile or swimmer stage (Jogler et al. 2011). With the aid of fimbriae, the sessile cells anchor on biofilms and take part in the dwelling of microbial communities (DeLong et al. 1993).
The planctomycetes, namely Gemmata obscuriglobus, Rhodopirellula baltica, and Planctomyces limnophilus, played a crucial role as genetic models for developing genetic tools (Jogler and Jogler 2013). The physiology speaks diverse functions. Growth requirements of planctomycetes reported many species generally belong to chemoautotrophic aerobes such as Blastopirellula, Gemmata, Pirellula, Planctopirus, Rhodopirellula, and Gimesia, although at times oligotrophic example, Isosphaera pallida and slow-growing Obscuriglobus and Maris species. Acidophilic species like Zavarzinella and Singulisphaera are detailed. Taking the temperature requirement, most of the planctomycetes belong to mesophiles and few be thermophilic, for example Isosphaera pallid grows at 55 °C at maximum (Giovannoni et al. 1987). The electron acceptor is sulfur-giving sulfides besides, nitrate, mono, di, and polysaccharides (Slobodkina et al. 2016).
2 Structural Organization of Prokaryotes and Eukaryotes
Ahead of understanding more about cell organization of planctomycetes, it is indispensable to know the different cell plan compartmentalization of prokaryotes and eukaryotes. Distinguishing the cellular life of prokaryotes and eukaryotes is usually based on the cell organization. Earlier in the nineteenth century, the dichotomy organization was explained by the presence of a nucleus of cells as plants using a light microscope; however, later in the twentieth century, Edouard Chatton proposed prokaryotes as non-nucleated cells (Chatton 1925). Studies using an electron microscope revealed a cavernous difference in the two cell types; the presence of membrane-bound cell organelles such as the mitochondria, nucleus, and the chloroplast of archetypal eukaryote and the absence of double-membrane organelles as prokaryotes, the bacterial species (Chatton 1938). Another distinct feature is the presence of the nuclear envelope in eukaryotic cells and lack of enveloped membrane in prokaryotes (Stanier and Van Niel 1962). The three-domain system proposal was accepted with the establishment of Archaea.
For a eukaryotic organism, the presence of a nucleus, its nuclear envelope, and the nuclear pore complex is the main cell plan. The nuclear pores entrenched within it, and the structural components that transport amidst the nucleus and cytoplasm and the proteins from the nuclear pore complex. Outwardly, the nuclear envelope is deemed double membrane but, it is not continuous. At the junction of nuclear pores, the loop link is formed between the membranes of the double-layered sections, such that the envelope is believed to be as a single folded membrane (Martin 1999). This characteristic is noteworthy to consider the origin of the nucleus for the viability of models. Few models that cite symbiotic fusion of two phylogenetic diverge cells mean that the nuclear envelope has no continuity of inner and outer membranes (Poole and Penny 2001; Rotte and Martin 2001).
The traditional prokaryote cell plan entails a typically naked genomic DNA with no membrane. In models of E. coli and B. subtilis, the DNA is shown as a corralled nucleoid, expands throughout the cytoplasm, and into tiny ribosome-free spaces occupying a large area of the cell. This arrangement is vital for co-translational protein secretion during coupled transcription and translation processes. This process of coupled transcription and translation excludes the separation of genomic DNA from the cytoplasmic region and the ribosomal bound regions and is dependable on short-lived mRNA. This unique feature is selective to prokaryotes. Yet, studies are still fragmentary whether this process occurs in eukaryotes (Iborra et al. 2004). Prokaryotic cells do comprise various internal membrane-bound organelles or intracytoplasmic membrane (ICM) development though, in general, is studied as prokaryotes lack intracellular compartmentalization (Table 1) (Stolz 1998); however, these do not constitute prokaryotic cell plan as they are not concerned for compartments of genomic DNA.
3 Why Planctomycete is an Exception to the Prokaryotic Cell Plan?
From the above context of prokaryotes cell organization, it is definite that they lack the nuclear membrane. However, planctomycetes being a typical phylum of bacteria has a structural arrangement contradictory to prokaryotes, as the cells are separated into compartments by ICM into either single or two or more than two are dependent on the genus. So, these variations of cell compartmentalization confer doubt whether all genera of planctomycete show cell compartmentalization and if they do exhibit, are there characteristics helpful in knowing the basis or source and mechanism?
4 Compartmentation of Planctomycetes Cell
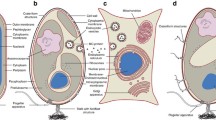
As discussed earlier, all planctomycetes contribute to a typical cell plan relating to internal compartments; within this arrangement there lays additional multifaceted disparity concerning more compartments and cell membranes (Fig. 1). The freshwater planctomycetes, Gemmata obscuriglobus, when cultured and examined revealed a new characteristic not found in any other bacteria. The feature is the presence of three membranes: condensed nucleoid bound by two intracytoplasmic membranes (ICM) and the pirellulosome enclosing the nuclear body (Fuerst and Webb 1991). In anammox planctomycetes, namely Kuenenia, a single membrane-bound anammoxosome is seen apart with double membrane ICM. In Pirellula marina and Pirellula staleyi (Lindsay et al. 1997), Planctomyces limnophilus (Jogler et al. 2011) and Schlesneria paludicola (Kulichevskaya et al. 2007) have two compartments, the paryphoplasm separating a ribosome-free outer region from major compartment the pirellulosome enclosing the ribosome and nucleoid; however, the species exhibits cellular compartmentalization (Lindsay et al. 1997).
Current studies on planctomycetes suggested the organism executes protein uptake (endocytosis) via putative or integral membrane protein that looks like eukaryotic protein clathrins (Lonhienne et al. 2010; Santarella-Mellwig et al. 2010). This feature of endocytosis plays a significant role in eukaryogenesis lineage giving an interlink bridge to prokaryotes and eukaryotes (Devos and Reynaud 2010; Forterre and Gribaldo 2010; Fuerst and Sagulenko 2011).
5 Components of Planctomycetes Cell
5.1 Cell Wall and Cell Surface Proteins
The principal component we would discuss is the cell wall. The chemical analysis for planctomycetes reported the absence of muramic acid an important component typical for peptidoglycan polymer which has a key for structural integrity and potency. This unique feature is contradictory to other bacterial cell walls (Liesack et al. 1986). While in few planctomycetes and ancestral PVC superphylum, the operon gene “dcw” is observed which is essential in synthesizing the peptidoglycan in bacteria (Pilhofer et al. 2008). Homologous to the “dcw” gene, planctomycetes have genes such as the FtsZ gene. The cell wall is composed mainly of proteins and made up of amino acids in abundance (Table 2). The amino acid cystine is predominant in most planctomycetes, wherein few low contents of cystine are reported, for example G. obscuriglobus. The presence of cystine shows an integral cell wall due to the presence of disulfide cross-links (Liesack et al. 1986). This feature gives the cell shape, strength, and integrity similar to that of peptidoglycan.
5.2 Cateriform
A special characteristic is spotted on the cell surface of planctomycete, the appearance of pit-like structures called crateriform. The allocation of crateriform varies down the taxonomical position as well as by the size. The nature of crateriform is not understandable. Indeed, it can withstand its uniformity after SDS treatment. The distribution might be either polar buds or uniform. The size varies from 12 nm at least to 7–5 nm in mature buds (Tekniepe et al. 1981). The larger is close to the mother cell while the smaller is present at the distal end to that of the mother cell. The change in diameter to 30 nm is noticed in the mature mother cells, where the smaller structures turn out to be invisible.
5.3 Paryphoplasm
The region paryphoplasm is present amidst the cytoplasmic and intracytoplasmic membrane. The region is ribosome free. The term “paryphe” means border woven along with a robe. Based on the electron density, it varies from species to species; for example, G. obscuriglobus and Candidatus Kuenenia stuttgartiensis where the former shows relatively denser while the latter is transparent (van Niftrik et al. 2009). It is arranged mostly in the region of the cell border or occupies larger space within the cell bound to the intracytoplasmic membrane. In Pirellula group species, the paryphoplasm is dense at polar region than the opposite (Fig. 2b), while in Rhodopirellula baltica, vesicles-like structures similar to pirellulosome is present but with no nucleoid (Schlesner et al. 2004). The cell plan in Planctomyces limnophilus is identical to that in Blastopirellula, but the paryphoplasm shape changes with different cells along the z-axis and is symmetrically nonrotational (Jogler et al. 2011). In Blastopirellula, the pirellulosome and the paryphoplasm are separated by a thin single ICM (Fig. 2a). Studies have reported contradictory outcomes replacing the term pireplasm in place of paryphoplasm. In species Gemmata obscuriglobus, invagination of the cytoplasmic membrane into the paryphoplasmic space from vesicles topologically alike to the pireplasm (Lonhienne et al. 2010), another argument is the asymmetrical distribution of paryphoplasm in species of Blastopirellula and Pirellula. However, analysis using cytochemical methods on anammox planctomycetes is reliable with the theory of paryphoplasm be the actual cytoplasmic compartment.
Transmission electron micrograph view shows cell compartmentalization of parypholasm (P) and pirellulosome (PI) separated by ICM with condensed nucleoid (N) dispersed in the pirellulosome (a) Blastopirellula marina species (b) of Pirellula staleyi (Source: Fuerst 2005)
Proteins such as non-FTsZ kustd1438 (Candidatus Kuenenia) and YTV protein accumulates in the paryphoplasm. Staining Blastopirellula and Pirellula indicates the presence of glycoprotein and polysaccharide in the paryphoplasm, RNA in Pirellula group, and G. obscuriglobus species (Lindsay et al. 1997, 2001).
5.4 Intracytoplasmic Membrane
The ICM present below the paryphoplasm is an exclusive trilaminar membrane. The electron micrographs sections of ICM is visible than the cytoplasmic membrane. In species Planctomyces limnophilus, ICM is bilayered and is of 6 nm wide as same as the cytoplasmic membrane (Jogler et al. 2011). The ICM is never intact with the cytoplasmic membrane, as other bacterial cell membranes appear. Due to the invagination of the cytoplasmic membrane at endocytotic protein intake, ICM form vesicles inside the paryphoplasm (Lonhienne et al. 2010), one such example is Isophaera pallida where 60% of the cell interior is occupied forming large inclusion due to infolding of ICM (Fig. 1b). Through these vesicles in the cytoplasmic membrane, there is the molecular trafficking of components, which is not evident. On the other hand, in groups of planctomycetes such as G. obscuriglobus and Anammox species, an extra compartment within the ICM-bound pirellulosome giving an arrangement of two membranes enfolding the nucleoid, while in anammox group, a single membrane component anammoxosome with no nucleoid is present. A common fundamental model of the pirellulosome partition bound by the ICM and a paryphoplasm bound by the ICM within the cell observes this cell plan in almost all planctomycetes.
5.5 Pirellulosome
Below the ICM, the major cell component of planctomycetes, the pirellulosome with nucleoid DNA and ribosome’s is arranged. In species of G. obscuriglobus, an outer membrane is formed due to infolding ICM into the pirellulosome. But, in Pirellula group, the ICM at all times outlines an unremitting frontier to the ribosome and nucleoid, enclosing pirellulosome compartment (Fig. 1a and c) (Lindsay et al. 1997). While in Rhodopirellula baltica and other planctomycetes, the cell plan appears to have a small adaptation of pirellulosomes roofed within the paryphoplasm, most likely surrounded with ribosome’s, but it seems lacking nucleoid (Schlesner et al. 2004); however, this need to be reassessed by tomography to rule out the 3D structure of a distinct pirellulosome. The same cell plan is seen in Planctomyces limnophilus but with different forms and organization (Jogler et al. 2011). A rare example of pirellulosome forming falcate shape at the cell margins are shown in Isosphaera pallida and Planctomyces maris when the ICM infolds forming an outsized lobe of paryophoplasm. Protein synthesis by the co-translation process is suggested in Pirellula group as ribosomes of the pirellulosome are noticed in few regions lining the boundary of the ICM. In G. obscuriglobus, the ribosomes are observed arranged linearly all along the outer and inner membranes of the nuclear envelope looking like that of the eukaryotic endoplasmic reticulum. The significant role played by the pirellulosome in cell biology is the metabolic activities such as glycolysis, transcription, and translation, because of the occurrence of DNA and ribosomes in almost all sites.
5.6 Nucleoid
Section of plantomycetes cells visualized under a transmission electron microscope shows dense fibrillar nucleoid existing within the pirellulosome. In species of G. obscuriglobus, the membrane-bound nuclear region, both fibrillar nucleoid and ribosome-free particle reside; depending on this, both DNA and RNA at the same time are imaged by immuno-gold labeling techniques. This presence of the invagination of fibrils within the nucleoid signifies a highly dense concentration similar to that of E. coli nucleoid where a coralline-shaped nucleoid expands all through the cell space. During cell division in G. obscuriglobus species, the nucleoid shows to stay in the condensed form all through the cell cycle. Yet, at some point of the passageway that is the nucleoid passage from the bud neck into the new bud, the fibril unfolds to some amount but remains connected leaving gaps (Fig. 3a and b). These gaps signify that the outer and inner membranes of the membrane envelope combine to form a continuous membrane giving an impression of a single membrane nuclear envelop. Within these gaps, there is a chance of protein transport as similar to that of the nuclear pore. A liquid crystalline cholesteric organization such as DNA filaments being arranged in a sequence such that each one takes turns virtually in helical model resulting in the orderly arrangement of DNA in a series of the nested arch as analogous to eukaryotic dinoflagellates (Yee 2012). This condensation of the nucleoid in planctomycetes gives inference on how the replication and transcriptional processes are planned in these organisms. Locating the RNA polymerases might assist in clarifying the cell plan. Lieber et al. (2009) suggested that the condensation of the nucleoid in G. obscuriglobus species might be due to its obvious resistance toward ultraviolet rays and gamma radiation (Lieber et al. 2009). Still, the statement has no proper evidence whether all other planctomycetes do exhibit such resistance activities.
Diagram of G. obscuriglobus (a) showing the membrane-bound nuclear body, nucleoid, ICM, and paryphoplasm (b) enlarged section nuclear envelope showing as single membrane and gap due to infolding (Source: Fuerst 2005)
5.7 Anammoxosome
Anammox planctomycetes are usually coccoid-shaped bacteria with a long generation time (10–30 days). Species, namely Candidatus Brocadia anammoxidans, Candidatus Scalindua brodae, Candidatus Scalindua sorokinii, Candidatus Kuenenia stuttgartiensis, and Candidatutus scalindua, belong to anammox planctomycetes. They are capable enough to convert ammonium to dinitrogen donating an electron to nitrite with hydroxylamine and hydrazine as transitional compounds (Jetten et al. 2003; Liesack and Stackebrandt 1992; Strous et al. 2002; van Niftrik et al. 2004). The capacity of producing hydrazine from hydroxylamine is a key feature of anammox planctomycetes, similar to the occurrence of free hydrazine in nitrogen metabolism of microbes (Strous and Jetten 2004). Localization of anammox is detected by the presence of a key enzyme, a hydrazine-oxidizing enzyme.
In species of Anammox planctomycetes, a distinctive feature is the presence of a unique organelle named as anammoxosome and exceptional lipids molecule lining the anammoxosome membrane pictured as linear concatenated cyclobutane rings (ladderane) (Sinninghe Damste et al. 2002). rRNA studies reported anammox planctomycetes to appear deeply branched with 16sRNA having a distinctive helical structure at position 9 giving a secondary structure not seen in any other planctomycetes (Schmid et al. 2001). The organelle lies within the riboplasm region but is separated by a thin-layered membrane, these planctomycetes do have a nucleoid present in proximity with the anammoxosome membrane (Lindsay et al. 2001). The cell plan of anammox suggests a single-membrane envelope nucleoid DNA associated with anammoxosome membrane (Fig. 4). An array of tubules seen within the anammoxosome membrane suggests a major theory on the anammoxosome focusing its role in catabolism, that is the oxidation of ammonium anaerobically. The organelle signifies a lively function, i.e., the cell division or the anammoxosome movement.
Cell plan of anammox species (Brocadia anammoxidans) (Source: Fuerst 2017)
Anammox planctomycetes play an important role on the global cycling of nitrogen in the environment, especially within the marine ecosystems, where they are a major microbial component of the world’s oceanic oxygen minimum zones (OMZs) and marine sediments (Francis et al. 2007; Kuypers et al. 2005; Lam et al. 2009; Op den Camp et al. 2006; Schmid et al. 2007). OMZs are expected to increase with global warming. So, understanding their implications for nutrient cycling in the oceans is important for modeling climate change effects (Lam et al. 2009). Marine anammox planctomycetes comprise a previously unsuspected important missing link in the global nitrogen cycle. It has been estimated that 50% of the molecular dinitrogen in the atmosphere originating from the oceans may have its origin in the ammonium-oxidizing activities of anammox planctomycetes (Dietl et al. 2015).
5.8 Atypical Components of Planctomycetes
5.8.1 Sterols and Hopanoids
Recent research and discovery on Gemmata group reported sterols in its internal membrane. The presence of sterols in lanosterol form and its isomer parkeol in the original form suggest the occurrence of the metabolic sterol biosynthetic pathway (Pearson et al. 2003). Planctomycetes also can produce hopanoids (a substitute for sterols in eukaryotes) that are helpful in coding squalene hopene cyclase genes for flexibility, smoothing of membranes implicated for compartmentalization (Sinninghe Damste et al. 2004).
5.8.2 Ladderane
Ladderane is a unique lipid in the form of a staircase-like arrangement with fused moieties of cyclobutane. Ladderane is of three types: one be the ester-linked a heptyl chain, the second be ether linkage with glycerol unit, the final type constitutes both one and two linked together (Sinninghe Damste et al. 2002). The main function of ladderane lipid is to avert toxic intermediate hydrazine from dripping into the riboplasm region. However, the ladderane does not materialize to form a perfect barrier, as the intermediates are evident on the outer surface of the cells (Kartal et al. 2013). Application of ladderane is suggested in optoelectronics—for a model, transfer of electrons by chromophores via pie-pie interactions, in materials science for long-range electron transfer by alpha bonds, as molecular rods or stiff spacers among functional groups (Nouri and Tantillo 2006). Ladderane is also used to treat wastes in industries by providing an economical source.
5.8.3 Hydrocarbons
Gimesia maris once classified as Planctomycetes maris reported to generate olefinic long chain hydrocarbon with nine double bonds and also has a set of “ole” genes for synthesis (Sukovich et al. 2010). Besides G. maris, P. limnophilus is capable of synthesizing long-chain fatty acids through the FAS/PKS mechanism, and also offers precursors for biosynthesis of olefinic hydrocarbon (Shulse and Allen 2011).
5.8.4 Sulfatases
Of all varied planctomycetes, marine planctomycetes have genes for sulfatases associated with the habituate with algae-producing sulfated polysaccharides but studies are not evident regarding the function. However, studies with Rhodopirellula baltica (110 genes) and other species of Rhodopirellula reported having genes for sulfates (Glöckner et al. 2003; Wegner et al. 2013). Correlation of marine metagenomes with marine planctomycete genomes for sulfatases gene is recommended (Woebken et al. 2007). Of the 110 genes, 82 are similar to the prokaryotes, while 28 are alike eukaryotic sulfatases (Glöckner et al. 2003). Studies later instituted more than 1000 ORF annotated with sulfatases. Few of these sulfatases are useful in stereochemical transformation in chemical industries, for example, R. baltica (Gadler et al. 2006). Deracemization of compounds by R. balticais is a unique feature in this planctomycete where it undergoes enantioselective hydrolysis of alkyl sulfate esters retaining the configuration (Gadler and Faber 2007). An experimental model to yield maximum product by deracemization approach was attained by the sulfatase in mixture with a stereo-complementary inverting enzyme (Wallner et al. 2005).
5.8.5 Polyketide Genes
The ease of having either a partial genome sequence or complete suggests the possibility of assessing biological and chemical assays of novel secondary metabolites generated by the planctomycetes (Jeske et al. 2013). From these analyses, it is found to have the possibility for synthesis of bacteriocins, non-ribosomal peptides, and polyketide genes in planctomycetes such as Blastopirellula marina, R. baltica, Schelsneria paludicola, Singulisphaera acidiphila, and G. obscuriglobus having highest gene clusters 11, 10, 13, 10, and 12 respectively, while anammox planctomycete having smallest genome and lowest be Phycisphaera mikurensis.
5.8.6 Esterases
In thermophilic planctomycetes example, Thermogutta, a carboxylesterase is found which has a major role in applied perspective (Sayer et al. 2015). The enzyme is active at 70 °C with a unique active site for α-β hydrolase lineage, most of the “cap” area is missing at the active site exposing to solvent. This phenomenon explains the relationships of esterases linking substrate predilection and structural pocket that is the active site. Gene coding for esterase synthesis has been recovered from a sludge metagenome and instituted closest association (33% identical amino acids) to G. obscuriglobus esterase/lipase protein (Zhang et al. 2009).
5.8.7 Polysaccharide Lyase Enzymes
Polysaccharide lyase is one of the main sources of R. baltica (Dabin et al. 2008). The most common debasing polysaccharide enzyme is the extracellular glycoside hydrolase 10 endo-β-1,4-xylanases present in almost all planctomycetes, as well as Isosphaera-Singulisphaera group (Naumoff et al. 2014). Studies in soils using ecological radioisotope probing specified that uncultured planctomycetes perhaps be the prime degraders of composite heteropolysaccharide and consequently may be a primary contender for investigating the role and function of polysaccharide lyase enzyme and the enzyme intermediates in the cloning of metagenomes (Wang et al. 2015).
5.8.8 Ornithine Lipids
Low pH habitat planctomycetes such as Telmatocola sphagniphila, Singulisphaera rosea, and Singulisphaera acidiphila have methylated ornithine lipids (OL) in the form of mono, di, and tri at the epsilon nitrogen region. The hypothesis endows with a different mechanism for membrane stability at low pH conditions and extremely little phosphate conditions. Studies for ecology or lipid analysis of Sphagrum wetland, the habitat for acidophilic planctomycetes, reported trimethyl OL at the interface of oxic/noxic region and abundant rRNA sequence (Moore et al. 2015). An enzyme for N-methylation of OL remained unclear, but later a study in Northern Russia identified and characterized OL N-methyltransferase OlsG gene/enzyme from planctomycetes Singulisphaera acidiphila (Escobedo-Hinojosa et al. 2015).
6 Cell Division of Planctomycetes
The cell cycle of planctomycetes and the division is budding, where an identical smaller daughter cell is formed from the larger mother cell. Budding is observed in almost all planctomycetes such as Pirellula, Rhodopirellula, Aquisphaera, Isosphaera, Planctomyces, Gemmata, and Blastopirellula, but in Isosphaera where intercalary buds are produced, all along the filament of cells and are not separated from the mother cell as seen in the usually budding process (Giovannoni et al. 1987). Alternatively, fission takes place in anammox planctomycete Candidatus Kuenenia stuttgartiensis and Phycisphaera (van Niftrik et al. 2008; Fukunaga et al. 2009). In anammox planctomycetes, it is not just the cell limits division at the midpoint but also the organelle anammoxosome divides within the cell. A protein ring is seen in the paryphoplasm region unusual to the FSTZ ring seen in the cytoplasm of most bacteria (van Niftrik et al. 2009). Consequently, even where budding is not seen in a planctomycete, cell division occurs, which is a unique feature.
In Pirellula staleyi (belongs to Blastocaulis-Planctomyces group), the cell division has an exclusive element that indulge in the motility of the cell known as “swarmer cell phase” (Tekniepe et al. 1981). After swarmer phase, the cell matures into the mother cell and undergoes budding forming a daughter cell about half to that of the mother cell. The duration is nearly 3 h. A flagellum is synthesized at the time of daughter cell formation (attachment phase) and consequently the motile swarmer is released as soon as the daughter cell is detached from the mother cell. The same cycle is shown in Planctomyces limnophilus and Rhodopirellula baltica (Gade et al. 2005; Jogler et al. 2011; Wecker et al. 2010). Nevertheless, its studies are unclear and no evidence of all planctomycetes having the swarmer phase. During the cell cycle, we come across significant phases: the lag phase or resting phase and the synthesized phase or S phase where DNA replication occurs.
In Isosphaera pallida, the motility is of gliding type rather than swimming. In Rhodopirellula baltica, the cells are nonmotile but the polar association is seen (crateriform structures exhibit the polar distribution where fimbriae start at the border cell pole). In Gemmata species both the mother cell and the swarmer cells have flagella (Gade et al. 2005). Rosette-type structure occurs at nonreproductive cell pole through holdfast components. Budding in Gemmata overall takes 12 h with new bud detaching from the matured mother cell and that reportedly undergo fission (Fig. 5).
Diagrammatic representation of cell division in planctomycetes. (a and g) are Mother Cells; Stage b shows bud formation from mother cell; In stage c, the double membraned nuclear envelope opens releasing nucleoid and the nucleoid is freely passed via the neck and enters the newly formed bud. Stage d shows free nucleoid with envelope be absent. (e) shows the formation of a nuclear envelope formed in continuous with the ICM of the mother cell. (f) Membrane fusion and pinching off results in two cells; however, incomplete separation is observed in the micrographic image. (g) shows the complete formation of two identical cells, one of which initiates the next budding cycle while the second mother cell starts a new budding cycle at a lag phase duration of 3–5.5 h [Source: Lee et al. 2009 modified)
7 Synthetic Biology and Compartmentalized Planctomycetes Cell
The purpose of Synthetic biology is that it utilizes components and modules of an existing organism to produce modified or novel types of organisms with definite roles. Earlier, for synthetic organism generation, genes and regulatory elements of gene group have been in use. Yet, recent studies reported the use of organelles such as microcomponents, pirellulosome, and anammoxosome unique to planctomycetes (Giessen and Silver 2016) and magnetosome (Borg et al. 2015) are considered as a possible element for engineering synthetic organisms.
Giving a detailed account of organelle function in synthetic biology: the magnetosome element in arrangement with nano trap system transmits protein localization in Magnetotactic bacterium altering the taxonomical activities of the bacterium. Microcompartments enclosed by proteins anchorage enzymes and such encapsulated enzymes be capable of constructing new catalytic functions being a module. One such example of microcompartments degrading enzyme (1-fucose and 1-rhamnose) useful in encapsulation is seen in planctomycetes that degrade sugars from algal and plant cell walls (Erbilgin et al. 2014). Similar way, the anammoxosome organelle of planctomycetes may be used to disclose innovative ways and mechanisms to compartmentalize other bacteria for particular functions and transfer of substances within the cell. But due to limiting cause in understanding about the formation of compartments, it is not evident how such arrangement is formed, but if awareness can be attained through synthetic cell biology research on planctomycetes, then that may lead to getting a more or increased yield of the desired compounds by cellular metabolism (Giessen and Silver 2016). The very initial step for such a process is the expansion of a genetic system for planctomycetes which is still up and coming. Over and above, advanced research on the isolation of compartments and organelles might be useful directly in the field of biotechnology (Neumann et al. 2014).
8 Conclusion
For understanding the cell organization, the cell plan of Planctomycetes has proven to present novel models and imminent new ways of living earlier indefinite for bacteria or certainly for any other life form. Due to their varied compartmentalization, exclusive enzymatic and chemical mechanisms, the planctomycetes have previously provided evidence as a functional microbial source. Planctomycetes as a model organism have variety or continuum of structural organization and interrelated molecular properties among eukaryotes and prokaryotes; however, studies at the genomic level may further help in understanding the fundamental molecular cell biology of their cell plan and their evolutionary impact for the basis of eukaryote cell biology.
References
Borg S, Popp F, Hofmann J et al (2015) An intracellular nanotrap redirects proteins and organelles in live bacteria. M Bio 6(1):e02117-14
Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA (2013) Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155(5):1131–1140
Chatton E (1925) Pansporella perplexa. Reflexions sur la biologie et la phylogenie des protozoaires. Ann Sci Nat Zool 10e:1–84
Chatton E (1938) Titres et Travaux Scientifiques (1906–1937). S’ete, France: Sottano
Dabin J, Jam M, Czjzek M, Michel G (2008) Expression, purification, crystallization and preliminary X-ray analysis of the polysaccharide lyase RB5312 from the marine planctomycete Rhodopirellula baltica. Acta Crystallogr Sect F: Struct Biol Cryst Commun 64(3):224–227
DeLong EF, Franks DG, Alldredge AL (1993) Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr 38(5):924–934
Devos DP, Reynaud EG (2010) Evolution. Intermediate Steps Sci 330:1187–1188
Dietl A, Ferousi C, Maalcke WJ et al (2015) The inner workings of the hydrazine synthase multiprotein complex. Nature 527(7578):394–397
Erbilgin O, McDonald KL, Kerfeld CA (2014) Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. App Environ Microbiol 80(7):2193–2205
Escobedo-Hinojosa WI, Vences-Guzmán MÁ, Schubotz F et al (2015) OlsG (Sinac_1600) is an ornithine lipid N-methyltransferase from the planctomycete Singulisphaera acidiphila. J Biol Chem 290(24):15102–15111
Forterre P, Gribaldo S (2010) Bacteria with a eukaryotic touch: a glimpse of ancient evolution? Proc Natl Acad Sci U S A 107:12739–12740
Francis CA, Beman JM, Kuypers MM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1(1):19–27
Fuerst JA (2005) Intracellular compartmentation in planctomycetes. Annu Rev Microbiol 9:299–328
Fuerst JA (2017) Planctomycetes—new models for microbial cells and activities. In: Microbial resources. Academic Press, pp 1–27
Fuerst JA, Sagulenko E (2011) Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413
Fuerst JA, Webb RI (1991) Membrane-bounded nucleoid in the eubacterium Gemmatata obscuriglobus. Proc Natl Acad Sci U S A 88(18):8184–8188
Fukunaga Y, Kurahashi M, Sakiyama Y et al (2009) Phycisphaera mikurensis gen. Nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. Nov., Phycisphaerales Ord. Nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J Gen Appl Microbiol 55(4):267–275
Gade D, Stuhrmann T, Reinhardt R, Rabus R (2005) Growth phase dependent regulation of protein composition in Rhodopirellula baltica. Environ Microbiol 7:1074–1084
Gadler P, Faber K (2007) New enzymes for biotransformations: microbial alkyl sulfatases displaying stereo-and enantioselectivity. Trends Biotechnol 25(2):83–88
Gadler P, Glueck SM et al (2006) Biocatalytic approaches for the quantitative production of single stereoisomers from racemates. Biochem Soc Trans 34(2):296–300
Giessen TW, Silver PA (2016) Encapsulation as a strategy for the design of biological compartmentalization. J Mol Biol 428(5):916–927
Gimesi N (1924) Hydrobiologiai Tanulmányok [Hydrobiological studies]. I. Planctomyces Bekefi I Gim. Nov. gen. Et sp. [Hungarian with German translation]. Kiadja a Magyar Ciszterci rend, Budapest, 1–8
Giovannoni SJ, Schabtach E, Castenholz RW (1987) Isosphaera pallida, gen. And comb. nov., a gliding, budding eubacterium from hot springs. Arch Microbiol 147(3):276–284
Glöckner FO, Kube M, Bauer M et al (2003) Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci U S A 100(14):8298–8303
Henrici AT, Johnson DE (1935) Studies of fresh water bacteria. II. Stalked bacteria, a new order of Schizomycetes. J Bacteriol 30:61–93
Hortobágyi T (1965) Neue Planctomyces—Arten. Botaniki Kozlemenyek 52:111–119
Iborra FJ, Jackson DA, Cook PR (2004) The case for nuclear translation. J Cell Sci 117:5713–5720
Jeske O, Jogler M, Petersen J et al (2013) From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 104(4):551–567
Jeske O, Schüler M, Schumann P et al (2015) Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6(1):1–7
Jetten MSM, Sliekers O, Kuypers M et al (2003) Anaerobic ammonium oxidation by marine and freshwater planctomycete-like bacteria. Appl Microbiol Biotechnol 63:107–114
Jetten MS, den Camp HJ, Gijs Kuenen J et al (2015) “Candidatus Brocadiales” Ord. Nov. Bergey’s Manual Sys Archaea Bacteria 17:1
Jogler M, Jogler C (2013) Toward the development of genetic tools for Planctomycetes. In: Planctomycetes: cell structure, origins and biology. Humana Press, Totowa, NJ, pp 141–164
Jogler C, Glockner FO, Kolter R (2011) Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl Environ Microbiol 77:5826–5829
Kartal B, de Almeida NM, Maalcke WJ et al (2013) How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev 37(3):428–461
Komeili A, Vali H, Beveridge TJ, Newman DK (2004) Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A 101:3839–3844
König E, Schlesner H, Hirsch P (1984) Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol 138(3):200–205
Kovaleva OL, Merkel AY, Novikov AA et al (2015) Tepidisphaera mucosa gen. Nov., sp. nov., a moderately thermophilic member of the class Phycisphaerae in the phylum Planctomycetes, and proposal of a new family, Tepidisphaeraceae fam. Nov., and a new order, Tepidisphaerales Ord. Nov. Int J Syst Evol Microbiol 65(2):549–555
Kulichevskaya IS, Ivanova AO, Belova SE et al (2007) Schlesneria paludicola gen. Nov., sp. nov., the fi rst acidophilic member of the order Planctomycetales, from sphagnum-dominated boreal wetlands. Int J Syst Evol Microbiol 57:2680–2687
Kuypers MM, Lavik G, Woebken D et al (2005) Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci U S A 102(18):6478–6483
Lam P, Lavik G, Jensen MM et al (2009) Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci U S A 106(12):4752–4757
Lee KC, Webb RI, Fuerst JA (2009) The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol 10:4
Lieber A, Leis A, Kushmaro A et al (2009) Chromatin organization and radio resistance in the bacterium Gemmata obscuriglobus. J Bacteriol 191:1439–1445
Liesack W, Stackebrandt E (1992) Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol 174:5072–5078
Liesack W, Konig H, Schlesner H, Hirsch P (1986) Chemical composition of the peptidoglycanfree cell envelopes of budding bacteria of the Pirella / Planctomyces group. Arch Microbiol 145:361–366
Lindsay MR, Webb RI, Fuerst JA (1997) Pirellulosomes: a new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiol 143:739–748
Lindsay MR, Webb RI, Strous M et al (2001) Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch Microbiol 175(6):413–429
Lonhienne TG, Sagulenko E, Webb RI et al (2010) Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A 107:12883–12888
Martin W (1999) A briefly argued case that mitochondria and plastids are descendants of endosymbionts, but that the nuclear compartment is not. Proc R Soc London Ser B 266:1387–1395
Moore EK, Villanueva L, Hopmans EC et al (2015) Abundant trimethylornithine lipids and specific gene sequences are indicative of planctomycete importance at the oxic/anoxic interface in sphagnum-dominated northern wetlands. App Environ Microbiol 81(18):6333–6344
Murat D, Byrne M, Komeili A (2010) Cell biology of prokaryotic organelles. Cold Spring Harbor Percept Biol 2(10):a000422
Naumoff DG, Ivanova AA, Dedysh SN (2014) Phylogeny of β-xylanases from Planctomycetes. Mol Biol 48(3):439–447
Neumann S, Wessels HJ, Rijpstra WI et al (2014) Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium K uenenia stuttgartiensis. Mol Microbiol 94(4):794–802
Nouri DH, Tantillo DJ (2006) They came from the deep: syntheses, applications, and biology of ladderanes. Curr Org Chem 10(16):2055–2074
Op den Camp HJ, Kartal B, Guven D et al (2006) Global impact and application of the anaerobic ammonium-oxidizing (anammox) bacteria. Biochem Soc Trans 34(1):174–178
Pearson A, Budin M, Brocks JJ (2003) Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A 100:15352–15357
Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G (2008) Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol 190:3192–3202
Pilhofer M, Aistleitner K, Biboy J et al (2013) Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4(1):1–7
Poole A, Penny D (2001) Does endosymbiosis explain the origin of the nucleus? Nat Cell Biol 3:E173–E174
Rotte C, Martin W (2001) Does endosymbiosis explain the origin of the nucleus? Nat Cell Biol 3:E173–E174
Santarella-Mellwig R, Franke J, Jaedicke A et al (2010) The compartmentalized bacteria of the Planctomycetes -Verrucomicrobia – Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol 8:e1000281
Sayer C, Isupov MN, Bonch-Osmolovskaya E, Littlechild JA (2015) Structural studies of a thermophilic esterase from a new Planctomycetes species, Thermogutta terrifontis. FEBS J 282(15):2846–2857
Schlesner H, Stackebrandt E (1986) Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. Nov. and description of the order Planctomycetales Ord. Nov. Syst Appl Microbiol 8(3):174–176
Schlesner H, Rensmann C, Tindall BJ et al (2004) Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. Nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. Nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 54:1567–1580
Schmid M, Schmitz-Esser S, Jetten M, Wagner M (2001) 16S-23SrDNAintergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ Microbiol 3:450–459
Schmid MC, Risgaard-Petersen N, Van De Vossenberg J et al (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9(6):1476–1484
Seufferheld M, Vieira MC, Ruiz FA et al (2003) Identification of organelles in bacteria similar to acidocalcisomes of unicellular eukaryotes. J Biol Chem 278(32):29971–29978
Seufferheld M, Lea CR, Vieira M, Oldfield E, Docampo R (2004) The H+-pyrophosphatase of Rhodospirillum rubrum is predominantly located in polyphosphate-rich acidocalcisomes. J Biol Chem 279(49):51193–51202
Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL et al (1998) Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes—polyhedral bodies. Can J Biol 76:906–916
Shulse CN, Allen EE (2011) Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One 6(5):e20146
Sinninghe Damste JS, Strous M, Rijpstra WI et al (2002) Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nat 419:708–712
Sinninghe Damste JS, Rijpstra WI, Strous M et al (2004) A mixed ladderane/n-alkyl glycerol diether membrane lipid in an anaerobic ammonium-oxidizing bacterium. Chem Commun 22:2590–2591
Slobodkina GB, Panteleeva AN et al (2016) Thermostilla marina gen. Nov., sp. nov., a thermophilic, facultatively anaerobic planctomycete isolated from a shallow submarine hydrothermal vent. Int J Syst Evol Microbiol 66(2):633–638
Stackebrandt E, Wehmeyer U, Liesack W (1986) 16S ribosomal RNA-and cell wall analysis of Gemmata obscuriglobus, a new member of the order Planctomycetales. FEMS Microbiol Lett 37(3):289–292
Stanier RY, Van Niel CB (1962) The concept of a bacterium. Arch Microbiol 42:17–35
Stolz JF (1998) Bacterial intracellular membranes. In: Mitchell A, Trapnell J, Hadfield S, Kerguelen V, Richmond F (eds) Nature encyclopedia of life sciences. Wiley, Chichester, UK
Strous M, Jetten MS (2004) Anaerobic oxidation of methane and ammonium. Annu Rev Microbiol 58:99–117
Strous M, Kuenen JG, Fuerst JA et al (2002) The anammox case: a new experimental manifesto for microbiological eco-physiology. Antonie Van Leeuwenhoek 81:693–702
Sukovich DJ, Seffernick JL, Richman JE et al (2010) Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. App Enviorn Microbiol 76(12):3850–3862
Tekniepe BL, Schmidt JM, Starr MP (1981) Life-cycle of a budding and appendaged bacterium belonging to Morphotype-IV of the Blastocaulis – Planctomyces group. Curr Microbiol 5:1–6
van Niftrik LA, Fuerst JA, Sinninghe Damste JS et al (2004) The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol Lett 233:7–13
van Niftrik L, Geerts WJ, van Donselaar EG et al (2008) Combined structural and chemical analysis of the anammoxosome: a membrane- bounded intracytoplasmic compartment in anammox bacteria. J Struct Biol 161:401–410
van Niftrik L, Geerts WJ, van Donselaar EG et al (2009) Cell division ring, a new cell division protein and vertical inheritance of a bacterial organelle in anammox planctomycetes. Mol Microbiol 73:1009–1019
Van Teeseling MC, Mesman RJ, Kuru E et al (2015) Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun 6:6878
Wallner SR, Bauer M, Würdemann C et al (2005) Highly enantioselective sec-alkyl sulfatase activity of the marine planctomycete Rhodopirellula baltica shows retention of configuration. Angew Chem 117(39):6539–6542
Wang X, Sharp CE, Jones GM et al (2015) Stable-isotope probing identifies uncultured Planctomycetes as primary degraders of a complex heteropolysaccharide in soil. App Environ Microbiol 81(14):4607–4615
Ward NL (2015) Planctomycetia class. Nov. Bergey’s Manual of Systematics of Archaea and Bacteria 17:1
Wecker P, Klockow C, Schuler M, Dabin J, Michel G, Glockner FO (2010) Life cycle analysis of the model organism Rhodopirellula baltica SH 1(T) by transcriptome studies. Microb Biotechnol 3:583–594
Wegner CE, Richter-Heitmann T, Klindworth A et al (2013) Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar Genomics 9:51–61
Woebken D, Teeling H, Wecker P et al (2007) Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J 1(5):419^–435
Yee B (2012) The diversity and cell biology of Planctomycetes. School of Chemistry and Molecular Biosciences, The University of Queensland, St. Lucia, Queensland
Yilmaz P, Yarza P, Rapp JZ, Glöckner FO (2016) Expanding the world of marine bacterial and archaeal clades. Front Microbiol 6:1524
Yoon J, Jang JH, Kasai H (2014) Algisphaera agarilytica gen. Nov., sp. nov., a novel representative of the class Phycisphaerae within the phylum Planctomycetes isolated from a marine alga. Antonie Van Leeuwenhoek 105(2):317–324
Zhang T, Han WJ, Liu ZP (2009) Gene cloning and characterization of a novel esterase from activated sludge metagenome. Microb Cell Factories 8(1):67
Acknowledgements
The authors Pavani Sanapala and Sudhakar Pola are grateful to authorities of Andhra University Visakhapatnam, India for providing necessary facilities to carry out the research work and for extending constant support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Sanapala, P., Pola, S. (2021). Understanding the Cell Organization in Planctomycetes. In: Villa, T.G., de Miguel Bouzas, T. (eds) Developmental Biology in Prokaryotes and Lower Eukaryotes. Springer, Cham. https://doi.org/10.1007/978-3-030-77595-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-77595-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-77594-0
Online ISBN: 978-3-030-77595-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)