Abstract
Alzheimer’s disease (AD) is a progressive brain disorder that causes neurons to degenerate and die as the disease progress. AD is the most common cause of dementia, accounting for 60% to 80% of all cases, and has been recognized as a public health problem by the World Health Organization. In this study, we propose a method to aid in the diagnosis of AD that automatically extracts and classifies image features of the white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) tissues from the hippocampal regions. Our method uses the features as input to support vector machine (SVM) classifiers to perform the MR image classification in CN × AD and CN × MCI cases. For that, we preprocess all ADNI images and define the regions of interest for analysis. Then, we extract the GM, WM, and CSF tissues using an automated brain tissue segmentation method. Considering the intensities inside both hippocampal regions and each segmented tissue, we extract five statistical metrics from the voxel intensities inside each hippocampal region to use as features. Then, we train SVM classifiers with distinct kernels using a ten-fold nested cross-validation to perform the classification. From the classification experiments, the highest obtained AUC values for the CN × MCI and CN × AD classification cases were 0.814 and 0.922, respectively. It is important to emphasize that we obtained these results using an automated pipeline, with no human intervention, and a relatively small set of features.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Alzheimer’s disease (AD) is a progressive brain disorder that causes neurons to degenerate and die as the disease progress. AD is the most common cause of dementia, accounting for 60% to 80% of all cases, and has been recognized by the World Health Organization (WHO) as a public health problem. In fact, projections from the WHO estimate that 50 million people have dementia, and every year there are nearly 10 million new cases. The total number of people with dementia can reach 82 million in 2030 and as high as 152 million in 2050 [1].
Some AD studies presented in the literature have reported that early signs of this disease may be detected 10 to 20 years before symptoms arise [2,3,4], with structural brain changes that do not noticeably affect the patient. After many years of brain damage, the individuals will start to experience noticeable symptoms, such as memory loss and language problems. The observed symptoms associated with AD are caused by the damage and death of neurons (neurodegeneration), with other brain changes, including inflammation and atrophy [2].
Since there is no single test for AD, doctors use various approaches and tools to make the diagnosis. They often apply tests for assessing the individual abilities in solving problems and retrieving memories and using family history and reports from relatives or caregivers to determine changes in the individual’s skills and behavior. Magnetic resonance imaging (MRI) has played an important role in the diagnosis of AD, since this imaging modality provides images with very rich anatomical details that allow the assessment of atrophies and other brain changes resulting from the disease.
Based on the above-described scenario, it is latent the need for modern solutions to take on the challenge of diagnosing AD. There are many works proposed in the literature [5,6,7,8,9,10,11] addressing the problem of classification of MR images in one of the three diagnostic groups: cognitively normal (CN), mild cognitive impairment (MCI), and AD. The MCI group corresponds to an intermediate stage of the disease, in which the individuals present minor cognitive issues but with a high probability of evolving to AD.
In this study, we propose a computerized method to assist in the diagnosis of AD. Our method automatically extracts and classifies image features of the white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) tissues from the hippocampal regions and use them as input to support vector machine (SVM) classifiers to perform MR brain image classification in the cases CN × AD and CN × MCI.
The rest of this paper is organized as follows. Section 2 describes all the image datasets used, Sect. 3 provides a description of all methods and processes used in this work, and Sect. 4 presents the relevant experimental results. Finally, Sect. 5 presents a conclusion and summarizes future possibilities.
2 Image Datasets
In this study, we use two image datasets; the Alzheimer’s Disease Neuroimaging Initiative (ADNI) [12] and the Neuroimage Analysis Center (NAC) [13].
The ADNI dataset contains images obtained over 50 sites across the USA and CanadaFootnote 1. Due to the size and diversity of the images in this dataset, image specifications will be omitted here. However, as we will discuss in Sect. 3.1, proper standardization steps were conducted to overcome this issue. For this study, we use only images acquired using the MPRAGE sequenceFootnote 2 from patients with MMSE information and age ranging between 70 and 85 years, distributed among three diagnostic groups, i.e., CN, MCI, and mild-AD. Considering these restrictions, we randomly select a total of 762 images, one image for each subject, in which the numbers of CN, MCI, and mild-AD subjects are 302, 251, and 209, respectively.
The NAC dataset consists of 149 3-D triangular meshes of distinct brain structures; they are all spatially aligned to a T1-weighted (T1-w) image of a healthy 42-year-old male subject. This dataset provides the binary masks used to delimit the hippocampal regions in this study.
3 Methods
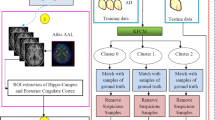
Figure 1 illustrates the general framework of the proposed method. First, we preprocessed all ADNI images and defined the regions of interest (ROI)s - the hippocampal regions in this study. Next, we extracted the GM, WM, and CSF tissues in the intracranial brain region using the fully automated brain tissue segmentation package FMRIB’s Automated Segmentation Tool (FAST). Then, considering the intensities of the voxels in the hippocampal regions of each segmented tissue, we extracted five statistical metrics, i.e., sum, mean, variance, skewness, and kurtosis to use as image features. Finally, we train SVM classifiers with distinct kernels using a ten-fold nested cross-validation to perform the brain MR image classification.
3.1 Preprocessing
In the preprocessing stage, all ADNI images were first processed for noise reduction using the Non-Local Means technique [14], following by bias field correction with the N4-ITK technique [15] and image intensity standardization using the proposed method in [16], with the T1-w NAC template image [13] as a reference. After that, the spatial alignment of all study images (ADNI) was conducted using affine transformations implemented in the Nifty-Reg image registration tool [17]. Similar to the image intensity standardization stage, the T1-w NAC template image was used as the reference image. Finally, brain extraction was performed on all images using the ROBEX [18] technique.
3.2 Hippocampal Extraction
To extract each study image’s hippocampal ROIs, we first spatially aligned (registered) the NAC T1-w template image with the study image using a deformable transformation [19]. In this case, the NAC T1-w image was used as a moving image and each study image was treated as a fixed image. Then, the obtained deformable transformation was applied to hippocampus mesh models to define the ROIs. Finally, we dilated the hippocampus binary masks (obtained from the mesh models) using the dilatation morphological operator [20, 21] with a sphere structuring element of radius four to encompass surroundings hippocampal structures.
3.3 Automatic Tissue Segmentation
The proper segmentation of the brain tissues is crucial since the segmented images will be used as sources of information for the feature extraction procedure. Therefore, we used the fully automated brain tissue segmentation software FAST [22] (version 4.1), which is considered the state-of-art on tissue segmentation [23] and is available on the FSL packageFootnote 3. The FAST implementation is based on a Hidden Markov Random Field Model, optimized using the Expectation-Maximization (EM) algorithm. A detailed description of the algorithm is available in [22]. The input image required by the FAST is a skull stripped MR image, which we obtained as a result of the preprocessing step using the ROBEX [18] technique. The outputs are an intensity non-uniformity corrected version of the input image and the segmented GM, WM, and CSF brain tissues. Figure 2 illustrates an example of an input MR image (first line) and the output image (second line) showing the segmented brain tissues. The segmentation data are color-coded as follows: WM is colored white, GM is light gray, and CSF is dark gray.
3.4 Feature Extraction
In this study, feature extraction was performed for each individual brain tissue within each hippocampal region. For that, both the resulting binary GM, WM, and CSF segmented images and the left (LH) and right hippocampal (RH) binary masks were used to define the regions to compute the features, i.e., GM ∩ LH, GM ∩ RH, WM ∩ LH, WM ∩ RH, CSF ∩ LH, and CSF ∩ RH. Then, we analyzed the voxel intensities inside each mask in order to describe their intensity distributions. Our analyses extracted the sum of the voxel intensities and the first four statistical moments, i.e., mean, variance, skewness, and kurtosis, resulting in 30 features.
3.5 MR Image Classification
For the image classification in CN × MCI and CN × AD groups, we used the extracted features (30 in total) to train SVM classifiers with four different kernels, linear, radial basis function (RBF), polynomial, and sigmoid. Furthermore, since we have a slightly unbalanced dataset, we have automatically adjusted the model weights to be inversely proportional to class frequencies in the input data using the class_weightFootnote 4 parameter from the scikit-learn python library.
We adopted four metrics for performance evaluation, including the area of the receiver operating characteristic (ROC) curve - AUC, accuracy (ACC), balanced accuracy (BACC), and F1-score. A detailed description of these metrics can be found in [24]. Also, we considered the images from the subjects with a cognitive condition (MCI or AD) as positives samples and the images of CN subjects as negative.
We conducted the classification experiments using a ten-fold nested cross-validation. For each SVM kernel, we used a coarse to fine grid search to determine the best C parameter value that maximizes the AUC. The coarse-grid search was performed within a range of values [−5; 10] and steps of 0.5, and the finer search was conducted in the neighborhood of the best coarse-grid parameter, C, with a grid range of values [2(log2C)−2; 2(log2C)+2.1] and incremental steps of 0.25.
4 Results
The highest obtained AUC values for the MR image classification experiments for the CN × MCI and CN × AD classification cases were 0.814 and 0.922, respectively. Table 1 presents the average classification results of the ten-fold nested cross-validation for the four SVM kernel functions used on the experiments mentioned above.
5 Conclusions
As presented in the previous section, the classification results for the CN × AD case were better than the CN × MCI case, presenting higher metrics for all SVM kernels. These results can be explained due to the AD continuum phases, named the preclinical (CN), the MCI, and the AD. All these phases occur sequentially in the given order; therefore, the measurable brain changes in the AD brain are more intense than MCI [2]. Thus, the brain differences among CN and AD are more distinct than CN and MCI. Our method achieved average AUC values of 0.814 and 0.922 for the CN × MCI and CN × AD classifications, respectively, and accuracy of 0.743 and 0.845 for the same cases. These results are comparable to the state-of-art accuracy results, which vary among 75–80% for the CN × MCI case and 88–92% for the CN × AD case [25,26,27,28]. It is also important to emphasize that these results were obtained from a fully automated pipeline, with no human intervention, and with a relatively simple and small set of features. In addition, the size of the dataset used in this study is larger than most published works. Another aspect of the proposed method is that it can be used alongside other methods to build a broader and more comprehensive approach, using it as a complementary tool. Despite the results, further research must be conducted to explore new features capable of classifying more difficult cases such as MCI and AD groups.
Notes
- 1.
- 2.
These MPRAGE files are considered the best in the quality ratings and have undergone preprocessing steps - https://adni.loni.usc.edu/methods/mri-tool/mrianalysis/.
- 3.
- 4.
References
World Health Organization et al (2018) Towards a dementia plan: a who guide. World Health Organization
Alzheimer’s Association (2020) Alzheimer’s disease facts and figures. Alzheimer’s Dementia 16(3):391–460
Dubois B, Hampel H, Feldman HH, Scheltens P, Aisen P, Andrieu S, Bakardjian H, Benali H, Bertram L, Blennow K et al (2016) Preclinical Alzheimer’s disease: definition, natural history, and diagnostic criteria. Alzheimer’s Dementia 12(3):292–323
Aisen PS, Cummings J, Jack CR, Morris JC, Sperling R, Frölich L, Jones RW, Dowsett SA, Matthews BR, Raskin J et al (2017) On the path to 2025: understanding the Alzheimer’s disease continuum. Alzheimer’s Res Ther 9(1):60
Ahmed OB, Mizotin M, Benois-Pineau J, Allard M, Catheline G, Amar CB, Alzheimer’s Disease Neuroimaging Initiative et al (2015) Alzheimer’s disease diagnosis on structural MR images using circular harmonic functions descriptors on hippocampus and posterior cingulate cortex. Comput Med Imaging Graph 44:13–25
Duara R, Loewenstein DA, Potter E, Appel J, Greig MT, Urs R, Shen Q, Raj A, Small B, Barker W et al (2008) Medial temporal lobe atrophy on MRI scans and the diagnosis of Alzheimer disease. Neurology 71(24):1986–1992
Aderghal K, Benois-Pineau J, Afdel K, Gwenaelle C (2017) Fuseme: classification of sMRI images by fusion of deep CNNs in 2D+ ε projections. In: International workshop on content-based multimedia indexing, Florence, Italy. ACM, pp 1–7
Chupin M, Gérardin E, Cuingnet R, Boutet C, Lemieux L, Lehéricy S, Benali H, Garnero L, Colliot O, Alzheimer’s Disease Neuroimaging Initiative et al (2009) Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus 19(6):579
Cuingnet R, Gerardin E, Tessieras J, Auzias G, Leh’ericy S, Habert M, Chupin M, Benali H, Colliot O, Alzheimer’s Disease Neuroimaging Initiative et al (2011) Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage 56(2):766–781
Poloni KM, Ferrari RJ (2018) Detection and classification of hippocampal structural changes in MR images as a biomarker for Alzheimer’s disease. In: International conference on computational science and its applications, Melbourne, Australia. Springer, pp 406–422
Poloni KM, Villa-Pinto CH, Souza BS, Ferrari RJ (2018) Construction and application of a probabilistic atlas of 3D landmark points for initialization of BTSym2020, 129, v6:’ Classification of brain MR images for the diagnosis of Alzheimer’s. . . 7 8 Chaves Cambui et al. hippocampus mesh models in brain MR images. In: International conference on computational science and its applications, Melbourne, Australia. Springer, pp 310–322
Jack CRJ, Bernstein MA, Fox NC, Thompson G, Alexander P, Harvey et al (2017) The Alzheimer’s disease neuroimaging initiative: MRI methods. J Magn Reson Imaging 27(4):685–691
Halle M, Talos IF, Jakab M, Makris N, Meier D, Wald L, Fischl B, Kikinis R (2017) Multi-modality MRI-based atlas of the brain. https://www.spl.harvard.edu/publications/item/view/2037
Buades A, Coll B, Morel J-M (2005) A review of image denoising algorithms, with a new one. Multiscale Model Simul 4(2):490–530
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Ny’ul LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 19(2):143–150
Ourselin S, Stefanescu R, Pennec X (2002) Robust registration of multi-modal images: towards real-time clinical applications. In: Medical image computing and computer-assisted intervention. Springer, Heidelberg, pp 140–147
Iglesias JE, Liu CY, Thompson PM, Tu Z (2011) Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 30(9):1617–1634
Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S (2010) Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed 98(3):278–284
Vincent L (1991) Morphological transformations of binary images with arbitrary structuring elements. Signal Process 22(1):3–23
Nikopoulos N, Pitas I (1997) An efficient algorithm for 3D binary morphological transformations with 3D structuring elements of arbitrary size and shape. In: Workshop on nonlinear signal and image processing, Michigan, USA. IEEE
Zhang Y, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20(1):45–57
Rehman HZU, Hwang H, Lee S (2020) Conventional and deep learning methods for skull stripping in brain mri. Appl Sci 10(5):1773
Kelleher JD, Mac Namee B, D’arcyA (2015) Fundamentals of machine learning for predictive data analytics: algorithms, worked examples, and case studies, 1 edn. MIT Press, Cambridge
Liu M, Zhang J, Adeli E, Shen D (2018) Landmark-based deep multi-instance learning for brain disease diagnosis. Medical Image Anal 46
Lian C, Liu M, Zhang J, Shen D (2020) Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Trans Pattern Anal Mach Intell 24(4):880–893
Liu M, Li F, Yan H, Wang K, Ma Y, Shen L, Xu M, Alzheimer’s Disease Neuroimaging Initiative et al (2020) A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. NeuroImage 208(1):116459
Zhang J, Liu M, An L, Gao Y, Shen D (2017) Alzheimer’s disease diagnosis using landmark-based features from longitudinal structural MR images. IEEE J Biomed Health Inform 21(5):1607–1616
Acknowledgments
Funding for ADNI can be found at https://adni.loni.usc.edu/about/#fund-container.
Funding
The authors would like to thank the São Paulo Research Foundation (FAPESP) (grant numbers 2018/08826-9 and 2018/06049-5) and the National Council for Scientific and Technological Development (CNPq) (grant number 166082/2019-8 - PIBITI) for the financial support of this research.
Author information
Authors and Affiliations
Consortia
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Cambui, V.H.C., Poloni, K.M., Ferrari, R.J., for the Alzheimer’s Disease Neuroimaging Initiative. (2021). Classification of Brain MR Images for the Diagnosis of Alzheimer’s Disease Based on Features Extracted from the Three Main Brain Tissues. In: Iano, Y., Saotome, O., Kemper, G., Mendes de Seixas, A.C., Gomes de Oliveira, G. (eds) Proceedings of the 6th Brazilian Technology Symposium (BTSym’20). BTSym 2020. Smart Innovation, Systems and Technologies, vol 233. Springer, Cham. https://doi.org/10.1007/978-3-030-75680-2_25
Download citation
DOI: https://doi.org/10.1007/978-3-030-75680-2_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-75679-6
Online ISBN: 978-3-030-75680-2
eBook Packages: EngineeringEngineering (R0)