Abstract
Satellite repeats make up a large fraction of the genomes of many higher eukaryotes. Until recently these sequences were viewed as molecular parasites with few functions. Drosophila melanogaster and related species have a wealth of diverse satellite repeats. Comparative studies of Drosophilids have been instrumental in understanding how these rapidly evolving sequences change and move. Remarkably, satellite repeats have been found to modulate gene expression and mediate genetic conflicts between chromosomes and between closely related fly species. This suggests that satellites play a key role in speciation. We have taken advantage of the depth of research on satellite repeats in flies to review the known functions of these sequences and consider their central role in evolution and gene expression.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1.1 Introduction
Repetitive DNA makes up a large portion of the genomes of higher eukaryotes. Satellite DNA, composed of tandem repeats that assemble into constitutive heterochromatin, was first described when mouse DNA was subjected to density gradient centrifugation and “satellite bands” of different densities formed above or below the bulk of the genome (Kit 1961; reviewed in Garrido-Ramos 2017). Fifty years ago the pioneering technique of in situ hybridization to mitotic chromosomes demonstrated that mouse satellite DNA was strikingly localized around the centromere (Pardue and Gall 1970). Although satellites propagate and may be mobile, expansion and movement are passive, relying on processes such as replication slippage, unequal crossing-over, or gene conversion to expand and move. This distinguishes satellites from transposable elements that typically insert as monomers and encode genes necessary for mobilization. However, this dichotomy is not clean. Some satellite repeats may be derived from transposable elements (Dias et al. 2015; Meštrović et al. 2015). Both transposable elements and satellite repeats are enriched in heterochromatic regions, are subject to silencing by heterochromatin formation, and are often grouped with transposable elements for the purpose of analysis and discussion.
In spite of their abundance, satellite repeats are typically thought of as having few cellular functions besides contributing to the formation of heterochromatin, centromeres, and telomeres. But satellite DNA and RNA participate in a number of diverse processes, including gene regulation, stress response, and nuclear organization in Drosophila melanogaster and many other organisms. The mutability of satellites makes them prominent actors in the evolution of genomes. In accord with this, satellite repeats are a potent and adaptable weapon in genomic conflicts between species, and between chromosomes within a species. This review will focus on the functions of satellite repeats in Drosophila with particular attention to the properties that make satellites a versatile and powerful force in nuclear organization, gene regulation and evolution.
1.2 Seeing the Dark Matter of the Genome
Much of eukaryotic genomes are comprised of vast, uncharted blocks of heterochromatin surrounding centromeres and telomeres. These regions, made up of satellite repeats and transposable elements, resist cloning and have posed an insurmountable barrier to traditional methods of genome sequencing and assembly. But advances in long-read sequencing of unamplified DNA have allowed the most challenging regions of genomes to be assembled. Nanopore sequencing of high molecular weight DNA was used to complete human centromeres (Miga et al. 2020). PacBio sequencing has similarly enabled Drosophila centromeres to be assembled (Chang et al. 2019). These methods avoid bias in library preparation but have high error rates, making assembly of repetitive regions challenging. The performance of correction and assembly methods must consequently be validated before being used to reconstruct repetitive regions (Khost et al. 2017). At present, sequencing and assembly of major repetitive regions remains labor intensive and technically challenging. In contrast, the diversity and abundance of different types satellite repeats in the genome can be determined by sequencing of unamplified genomic DNA (Lower et al. 2018). This approach revealed differences in satellite composition between strains of Drosophila melanogaster, supporting the idea that satellites are a rapidly evolving portion of the genome (Wei et al. 2014). Interestingly, the satellite composition of different chromosomes is often distinct. This is observed in humans, where the variants of the α-satellite arrays that make up centromeres are chromosome specific (Rudd et al. 2006). It is also the rule in flies, where distinctive combinations of satellite repeats make up the pericentric heterochromatin of different chromosomes (Lohe et al. 1993; Blattes et al. 2006; Jagannathan et al. 2017; Chang et al. 2019).
1.3 Biophysical Properties of Satellites
Many satellites have interesting biophysical properties that are often commented on. How these contribute to function remains unclear in most instances. With the exception of Dodeca, D. melanogaster satellites are notably AT rich (Table 1.1). Indeed, AT richness is common in satellite DNA and may contribute to a curving of the duplex that enhances nucleosome stability (Fitzgerald et al. 1994; reviewed in Palomeque and Lorite 2008). Also suggestive is the observation that monomers of longer and more complex satellites often approximate the length of mono-, di-, or tri-nucleosomes, suggesting the potential for nucleosome phasing (Henikoff et al. 2001). For example, α-satellite repeats of human centromeres (171 bp) and the 359 bp satellite family of D. melanogaster suggest mono- and di-nucleosomes, respectively (Table 1.1). Nucleosomes are phased over the centromeric satellites of multiple species (reviewed in Heslop-Harrison and Schwarzacher 2013). The human centromere protein CENP-B enforces phasing by binding a 17 bp sequence in α-satellite repeats (Ando et al. 2002). The Responder (Rsp) repeats of D. melanogaster, composed of two similar 120 bp units, also enforce phasing as demonstrated by an extended nucleosome periodicity of 240 bp (Doshi et al. 1991). The Rsp locus is known for its role as the target of meiotic drive, but the potential role of nucleosome phasing in this process is unknown. Taken together, these observations suggest that satellites display intrinsic features that are expected to contribute to nucleosome stability and influence the biophysical properties of chromatin.
1.4 How Do Satellites Expand, Move and Change?
D. melanogaster is rich in satellites, having approximately twice the diversity as humans (Shatskikh et al. 2020). The most abundant of these are 12 bp or less. As satellites generally lack coding potential their movement is nonautonomous. In spite of this limitation, they have been extraordinarily successful. Expansion and mobilization of satellite DNA reflects the propensities of replication and repair systems. For example, short tandem repeats are intrinsically unstable as they expand and contract by replication slippage (Fig. 1.1a) (Tautz et al. 1986; Bzymek and Lovett 2001; reviewed in Richards and Sutherland 1994; Levinson and Gutman 1987). Satellites also expand and contract by unequal crossing over during replication or repair (Fig. 1.1b). As longer, more complex monomers pose less of a challenge to the replication machinery, unequal crossing over is presumed to be a major factor variation of long repeats (Cabot et al. 1993; Southern 1975). A relevant question is why the expansion of noncoding sequence is tolerated. Some organisms have a considerably lower accumulation of satellite DNA, suggesting differences in susceptibility to slippage and unequal crossing over or tolerance of repetitive sequence. A comparison of related organisms with dramatic differences in genome size supports the idea that tolerance of additional genetic material is species specific (Petrov et al. 2000; Hartl 2000).
The repetitive structure of satellite repeats facilitates movement and change. (a) Replication stalling at repeats allows mispairing and template slippage. This produces contraction (top) or expansion (bottom) of tandem repeats. (b) Unequal crossing over leads to the expansion and contraction of tandem arrays. Cycles of unequal crossing over homogenize repeats at the center of an array. (c) Extrachromosomal loops generated by recombination within a tandem array can insert at a new site. (d) Gene conversion occurs when a related sequence serves as a template for recombination or repair
The movement of satellites may also occur by the formation of extrachromosomal loops that occur by recombination within an array. rDNA and noncoding tandem repeats are recovered as extrachromosomal loops in Drosophila and mammalian cells (Kiyama et al. 1986, 1987; Pont et al. 1987; Cohen et al. 2003, 2006; reviewed in Cohen and Segal 2009). This suggests a simple mechanism for movement to new sites. Extrachromosomal loops could undergo recombination with similar sequences or insert at random (Fig. 1.1c). The risk of extrachromosomal loops to genome integrity is moderated by the assembly of satellite repeats into heterochromatin. In accord with this idea, the loss of heterochromatin factors elevates the level of extrachromosomal loops and increases genomic damage (Larson et al. 2012; Peng and Karpen 2007). The erosion of heterochromatic silencing that is observed in aging and cancer is presumed to lead to the increase in extrachromosomal loops and contribute to genome instability in these cells (Sinclair and Guarente 1997; Larson et al. 2012; Turner et al. 2017; deCarvalho et al. 2018; Kim et al. 2020).
Tandem arrays are also subject to gene conversion by recombination within a cluster or with similar sequences from elsewhere in the genome (Fig. 1.1d). This general process is of interest for its role in the evolutionary divergence of duplicated genes (Osada and Innan 2008). The extreme abundance of satellite repeats that could be used as templates favors this process but raises the potential for large, damaging genome rearrangements. The idea that protection of repetitive DNA from inappropriate recombination is one of the functions of heterochromatin is supported by the behavior of repair foci in heterochromatic regions (Caridi et al. 2018). These foci move out of the nuclear territory occupied by heterochromatin before completion of the repair, supporting the idea that recombination and repair of repetitive DNA are potentially dangerous and under tight control.
1.5 Evolution of Satellite Repeats Is Rapid and Driven
Closely related species often display striking variations in satellite repeat composition and abundance (Bosco et al. 2007; Jagannathan et al. 2017; Lohe and Brutlag 1987). A comparative study of the satellite composition of four closely related species, D. melanogaster, D. sechellia, D. simulans, and D. mauritiana used hybridization to mitotic chromosomes to compare satellite composition and localization (Jagannathan et al. 2017). Some classes of satellites undergo complete replacement in closely related species. For example, Prodsat (AATAACATAG, Table 1.1) makes up 2% of the D. melanogaster genome but is not detected in the other three species (Török et al. 2000; Jagannathan et al. 2017). One caveat of this approach is that sites with low copy numbers of repeats are below the detection limit on mitotic chromosomes. For example, several megabases of 359 bp satellites in pericentromeric heterochromatin on the D. melanogaster X are detected by this method, but hundreds of closely related satellites dispersed throughout X euchromatin are not.
Dispersed satellites in euchromatin have also been subject to rapid, widespread changes in sequence, position, and abundance (Sproul et al. 2020). Although striking, this wholesale replacement is the natural outcome of two mutagenic processes with vastly different speeds. Point mutations diversify sequence, and the accumulation of mutations will eventually destroy the identity between sequences derived from the same progenitor. In contrast, gene conversion occurs orders of magnitude much more rapidly and acts to homogenize repeats within an array, and between arrays at different sites in the genome (Ohta and Dover 1984). The outcome of these competing mutational processes is the replacement and homogenization of satellites throughout the genome, termed molecular drive (Dover 1982). This is particularly dramatic when comparing closely related species, such as the Drosophilids (Jagannathan et al. 2017; Sproul et al. 2020; de Lima et al. 2020; Larracuente 2014).
1.6 Satellites, Silencing, and Organization of Chromatin
One of the most prominent features of satellite repeats is their role in heterochromatin formation. Large arrays of tandem repeats trigger silencing through heterochromatin formation that is largely sequence independent (Henikoff 1998). This has bedeviled mouse genetic studies because random transgene insertions produce tandem arrays subject to silencing. Using Cre/LoxP to excise extra copies from a mouse transgene array, Garrick et al. (1998) demonstrated that chromatin compaction and transgene silencing was not an intrinsic feature of the insertion site or transgene sequence, but was instead induced by multicopy arrays. Silencing of tandem transgenes is also observed in flies and plants, indicative of a common strategy for inactivating repetitive DNA (Dorer and Henikoff 1994). As most repetitive sequences are potential threats to genome integrity, the recognition and silencing of repeats represent a triumph of genome defense.
RNA derived from transposable elements and satellites direct the chromatin modifications that initiate heterochromatin formation in fission yeast and this serves as a useful model for the process (reviewed by Grewal and Elgin 2007). Transcription through repeats generates RNAs that are processed into siRNAs and loaded onto Argonaut effector complexes (Höck and Meister 2008). Nascent RNA from cognate regions of the genome is bound by these complexes, which recruit a histone methyltransferase that places the H3K9me mark. The heterochromatin protein Swi6, and a number of small RNA processing factors, are recruited by H3K9me to ensure maintenance of silencing (Zhang et al. 2008). While Drosophila heterochromatin is heterogeneous, evidence suggests that small RNA pathways also contribute to chromatin regulation in flies (Swenson et al. 2016; Cernilogar et al. 2011). Mislocalization of heterochromatin proteins and break down of silencing have been observed when Argonaut effectors or the genes necessary to produce small RNAs are inactivated (Fagegaltier et al. 2009). A genetically distinct silencing system in the germ line controls transposable elements by message destruction and transcriptional silencing (Khurana et al. 2010). This is directed by Piwi RNAs (piRNAs), generated from transposon sequences archived in piRNA clusters and expressed in the germ line (Brennecke et al. 2007). The resulting piRNAs enable Piwi, a germ line-specific Argonaut protein, to identify and bind nascent transcripts from mobile elements. Piwi recruits an H3K9 methyltransferase through an adapter protein to establish silencing (Sienski et al. 2015). Components of the Piwi system are also involved in chromatin compaction and silencing at later developmental stages. Maternal depletion of Piwi impairs heterochromatic silencing in the adult, a long-lasting effect that is observed by reduction of Position Effect Variegation (PEV) (Gu and Elgin 2013). PEV occurs when transgenes in repressive environments are silenced in some cells (Elgin and Reuter 2013). The majority of piRNAs have the identity to transposons, consistent with their vital role in the repression of mobile elements (Brennecke et al. 2007). But satellite piRNA are also present and may direct heterochromatin compaction of some repeats in the early embryo. Maternally deposited cues, possibly small RNA, direct the formation of zygotic heterochromatin over a cluster of 359 bp satellites on the X chromosome (Ferree and Barbash 2009; Yuan and O’Farrell 2016). The 359 bp satellites are notable for their role in hybrid incompatibility between closely related species, discussed in a following section.
Heterochromatin itself displays remarkable biophysical properties. Visualization of D. melanogaster heterochromatin reveals a subnuclear compartment that is distinct from euchromatin and which may consolidate the major heterochromatic regions of all chromosomes (see Caridi et al. 2018). The discovery that fly and human HP1 phase separate in vitro, and that heterochromatin itself displays the properties of phase separation in cells, suggested a biophysical explanation for how segregation is achieved (Strom et al. 2017; Larson et al. 2017). Phase separation of subcellular bodies occurs by the self-association of disordered proteins (reviewed in Hall et al. 2019). Separation is favored by multivalent interactions, protein crowding, and assembly with a polymer, such as RNA or chromatin. HP1 has disordered domains, interacts with a large number of proteins and also binds RNA (Alekseyenko et al. 2014; Muchardt et al. 2002; Roach et al. 2020). A functional role for RNA in HP1 localization is suggested by the finding that HP1a is released from mouse nuclei by RNase and the association of fly HP1a with chromatin is also RNA dependent (Maison et al. 2002; Piacentini et al. 2003).
Many chromatin proteins in addition to HP1 have RNA-binding domains or interact with RNA-binding proteins, in a manner that suggests a structural role for RNA in chromatin organization. One of these, Decondensation factor 31 (Df31), a small, hydrophobic, and highly disordered RNA binding protein, also boasts a large protein–protein interaction network. The general distribution of Df31 in the nucleus suggests a role in maintaining chromosome territories (Rohrbaugh et al. 2013). In cultured Drosophila cells association of Df31 with RNA is necessary for accessible chromatin (Schubert et al. 2012). In vitro assays found that Df31 association with chromatin was RNA dependent and RNase treatment collapsed chromatin into a nuclease-resistant state. While Df31 shows hallmarks of a protein involved in phase separation, it is enriched in euchromatic regions.
Scaffold Attachment Factor A (SAF-A, HNRNPU in humans) and SAF-B have DNA binding domains that recognize AT-rich matrix or scaffold attachment sites (Fackelmayer et al. 1994; Göhring and Fackelmayer 1997; Nozawa et al. 2017; Fan et al. 2018). These similar proteins also have RNA binding domains and large disordered regions. Loss of these proteins disrupts chromatin structure and DNA accessibility, as does RNase digestion (Nickerson et al. 1989; Nozawa et al. 2017; Fan et al. 2018). Mouse SAF-B binds a variety of long noncoding RNAs, but transcripts from pericentric satellite repeats are its predominant partners (Huo et al. 2020). Depletion of mouse SAF-B allowed heterochromatin bodies in the nucleus to expand and make interchromosomal contacts. Imaging reveals that SAF-B coats the exterior of H3K9me3-rich heterochromatin bodies, suggesting a SAF-B shell that prevents inappropriate mingling of phase-separated heterochromatin domains from different chromosomes (Huo et al. 2020). Drosophila SAF-B binds chromatin and is also visualized as an extrachromosomal network (Alfonso-Parra and Maggert 2010). The Association of fly SAF-B with chromatin responds to transcription and is differentially affected by mutation of its DNA binding domain and RNase treatment, but whether or not fly SAF-B interacts with specific RNAs is unknown.
Responses to heat shock and stress suggest that satellite RNA is situated in an interconnected web of RNA-binding proteins that organize chromatin and coordinate mRNA processing. When mammalian cells are subjected to stress, transcription of Sat III RNA is dramatically upregulated (Rizzi et al. 2004). SAF-B, and many RNA-binding factors involved in message processing, are recruited to nuclear stress bodies that form at sites of Sat III transcription (Valgardsdottir et al. 2008). Knockdown of Sat III RNA partially reversed the transcriptional repression induced by heat shock, suggesting a mechanism for rapidly restructuring chromatin and RNA processing pathways during stress (Goenka et al. 2016). In flies the Heat shock RNA omega (Hsrω) RNA serves a similar function. This noncoding transcript orchestrates stress response by sequestering splicing and RNA processing factors (reviewed by Jolly and Lakhotia 2006). D. melanogaster Hsrω includes 20 kb of AT-rich, 280 bp tandem repeats, thus conforming to the pattern of AT-rich satellites with a repeat length corresponding to multiples of nucleosome length.
1.7 Tandem Repeats in Euchromatin Modulate Nearby Genes
The role of satellite repeats in nucleating heterochromatin formation is well known, but tandem repeats of all types, including satellites, play interesting and surprising roles in gene regulation in euchromatin. A portion of satellite DNA is distributed throughout the euchromatic genome in tandem arrays. Changes in the number of repeats have created a wealth of genetic variation that has been exploited in forensic analysis, population genetics, and conservation. Although often considered neutral, microsatellites are highly represented in the promoters of human genes (Sawaya et al. 2013; Tomilin 2008). Dinucleotide repeats are enriched in fly enhancers, where they contribute to normal expression levels (Yanez-Cuna et al. 2014). These authors concluded that the association of short tandem repeats with regulatory regions is broadly conserved. Roughly 25% of the promoters in baker’s yeast, Saccharomyces cerevisiae, also contain tandem repeats (Vinces et al. 2009). These increase gene expression as the length of the repeats expanded. Tandem repeats also mediate repression. In the beetle Tribolium castaneum a major satellite DNA family near euchromatic genes maintains repression after heat stress (Feliciello et al. 2015). The mutability of short tandem repeats suggests a potential source of phenotypic variation. In accord with this idea, variation in repetitive DNA has been linked to expression differences in plants and insects (Ranathunge et al. 2018; Brajković et al. 2012). Repeat length variations in developmental genes, coupled with selection by breeders, are responsible for rapid phenotypic evolution in dogs (Fondon and Garner 2004). Social behavior in voles is influenced by satellite polymorphisms in a vasopressin receptor and length variants of repeats in the period (per) gene of D. melanogaster determine the male courtship song rhythm (Yu et al. 1987; Hammock and Young 2005). In addition to providing a source of genetic diversity, satellite repeats have been recruited to wage genomic conflicts and enable sex chromosome dosage compensation, described in the following sections. Their usefulness in these contexts owes to the properties described above: mobility, rapid evolution, and multifaceted roles in the structure and regulation of chromatin.
1.8 Chromosome Identification During Dosage Compensation
Organisms with highly differentiated sex chromosomes, such as humans and Drosophila, must address the problem of sex chromosome gene dosage. Males are functionally hemizygous for X-linked genes. Flies meet this challenge by increasing expression from virtually every gene on the single male X chromosome to match that of the two female X chromosomes. The Male-Specific Lethal (MSL) complex, composed of five proteins and one of two redundant RNAs, is essential for this process (reviewed in Kuroda et al. 2016). The MSL complex is selectively recruited to actively expressed X-linked genes (Alekseyenko et al. 2006; Bell et al. 2008; Sural et al. 2008). One of the MSL proteins, Males absent on the first (Mof), is a histone acetyltransferase that deposits the H4K16ac mark within the gene body (Kind et al. 2008; Copur et al. 2018). Histone acetylation increases the likelihood that initiated transcripts will be completed, raising the level of transcripts approximately twofold (Larschan et al. 2011). A long noncoding RNA, roX1 or roX2, must be part of the complex for proper X localization (Meller and Rattner 2002). Severe roX1 roX2 mutants are male lethal, the expression of X-linked genes is reduced and MSL proteins localize to ectopic autosomal sites (Deng and Meller 2006). How the MSL complex identifies the X chromosome with the required selectivity is still unknown. Studies in a number of laboratories characterized Chromatin Entry Sites (CES) on the X chromosome that bind an adapter protein and recruit the MSL complex directly (Alekseyenko et al. 2008; Straub et al. 2008; Soruco et al. 2013). However, the adapter protein binds related sites on all chromosome arms but only recruits the MSL complex in the context of X-linked sites. This suggests the presence of additional X identity elements.
The striking enrichment of a clade of 359 bp repeats, termed the 1.688X repeats (Table 1.1) in X euchromatin pointed to a potential role in an X chromosome-specific process such as dosage compensation (Hsieh and Brutlag 1979; Waring and Pollack 1987; Dibartolomeis et al. 1992). The 1.688X repeats are enriched near genes, including promoters and introns, leading to the suggestion that they could modulate expression (Kuhn et al. 2012). Autosomal insertions of short clusters of these repeats induced recruitment of the MSL complex and partial compensation of genes as much as 140 kb away (Joshi and Meller 2017; Deshpande and Meller 2018). A clue to how these repeats function came from the discovery that mutations in the siRNA pathway enhanced the lethality of males with partial loss of function roX1 and roX2 chromosomes (Menon and Meller 2012). Furthermore, ectopic expression of siRNA from one 1.688X repeat partially restored MSL localization and rescued roX1 roX2 males (Menon et al. 2014). Taken together, these studies reveal that the 1.688X satellite repeats are X identify elements and suggest that the siRNA pathway mediates their function. Interestingly, other Drosophilid X chromosomes are highly enriched for chromosome-specific repeats, although the sequence of these repeats is not highly conserved (Gallach 2014). Particularly striking is the rapid acquisition of repeats by neo-X chromosomes that arise by the fusion of an autosome to a sex chromosome. These fusions also produce a neo-Y, fated to degenerate as it passes exclusively through males without recombination (reviewed by Wei and Barbash 2015). Degeneration of the neo-Y necessitates compensation of genes on the neo-X. The relative mobility of satellite repeats makes them well suited for marking a young X chromosome to enable it to capture the dosage compensation machinery.
The manner in which the 1.688X satellites identify the X chromosome remains unknown, but there are clues that the mechanism is very different than that of the CES. The 1.688X satellites on the X chromosome are not generally enriched for the MSL proteins, suggesting that they do not recruit directly but mark the X chromosome in some fashion (Deshpande and Meller 2018). Ectopic expression of 1.688X siRNA increased the repressive H3K9me2 mark on autosomal 1.688X satellite insertions (Deshpande and Meller 2018). Contrary to conventional expectations for a repressive mark, this increased the expression of nearby genes in males. The finding that HP1 is modestly enriched on the male X chromosome, and that mutations in several heterochromatin factors selectively disrupt the structure of the polytenized male X, support the idea that repressive marks are in some way linked to fly dosage compensation (Spierer et al. 2005, 2008; De Wit et al. 2005). How satellite repeats and repressive marks might accomplish this is speculative, but one possibility is through influencing chromatin organization in the nucleus. Nuclear organization is a factor in X chromosome compensation in other organisms. For example, the single male X chromosome of C. elegans is located at the periphery of the nucleus, but the two female X chromosomes are centrally located (Sharma et al. 2014). Interaction with nuclear pore proteins may elevate the expression of X-linked genes in this species. A role for nuclear pore proteins in MSL loading and activation has also been proposed in flies (Mendjan et al. 2006). More generally, the ability of X-linked genes to acquire dosage compensation during development is attributed to the three-dimensional organization of X chromosomes in the nucleus of mammals and flies (Engreitz et al. 2013; Schauer et al. 2017; Ramírez et al. 2015). A role of the location or organization of the X chromosome is one way that 1.688X satellites might promote X recognition.
1.9 Satellites and Centromeres
The most widely appreciated function of satellite DNA is at centromeres. Human centromeres contain α-satellite arrays harboring a motif that interacts with the centromeric H3 variant CENP-A, suggesting determination by sequence (Masumoto et al. 1989; reviewed by McNulty et al. 2017; Willard 1985; Schueler et al. 2001). However, human centromeres occupy only part of the array and satellites are absent from some neo-centromeres, challenging the idea that sequence is the primary centromere determinant. Fly centromeres also form within extensive arrays of satellite repeats, but the centromere itself assembles at “islands” of transposons embedded in this sea of satellites (Chang et al. 2019). Fly centromeres are defined by the incorporation of an H3 variant called Cid. The importance of epigenetic information in specifying centromeres in flies is demonstrated by the fact that the transposons at the fly centromere are by no means limited to the centromere, appearing in both heterochromatic and euchromatic contexts throughout the genome (Chang et al. 2019). It is also reflected in the persistence of a functional centromere following transient anchoring of the centromere-specific chaperone CAL-1, which is capable of loading Cid at ectopic sites (Chen et al. 2014). These findings indicate that both fly and humans centromeres are specified by a combination of DNA sequence, genomic context, and epigenetic marking. Centromeres are surrounded by heterochromatin that contributes to their function. A large deletion of heterochromatin flanking a fly centromere produced mitotic instability and premature sister chromatid separation (Wines and Henikoff 1992). This is consistent with the enrichment of cohesin in heterochromatin and suggests that the mitotic machinery is tuned to a certain arrangement of heterochromatin surrounding the centromere (Bernard and Allshire 2002).
RNA from satellites has also been found to localize to centromeres. Transcripts from a large block of pericentric 359 bp satellites on the D. melanogaster X chromosome bind in cis to the centromeric region (Bobkov et al. 2018). RNA from the mammalian α-satellite also binds to centromeric proteins and localizes in cis at centromeres (Wong et al. 2007; reviewed in Ideue and Tani 2020; McNulty et al. 2017). This RNA is also necessary for characteristic localization of centromeric proteins CENPC1 and INCENP, and so may function to recruit or stabilize components of the centromere.
1.10 Satellites Are the Ammunition of Genomic Conflicts
Chromosomes are fundamental units of inheritance and take an active role in biasing their own transmission to the next generation. Meiotic drive, when a genetic element manipulates reproduction to favor its own transmission and overthrow Mendel’s rules, is the outcome. A selfish chromosome able to accomplish this will increase in the population. Evolutionary theory posits that systems of meiotic drive emerge frequently and sweep through the population, driving enrichment of one chromosome and limiting genetic variation. In addition, unfavorable, genetically linked alleles are allowed to proliferate (Courret et al. 2019). This extracts a cost in fitness that enables suppressors of drive to emerge and restore Mendelian segregation. A history of recurring cycles of drive and suppression is revealed when wild-caught flies are outcrossed and suppressed drivers emerge (Hartl and Hartung 1975). These conflicts are often mediated through satellite repeats and the outcome shapes the genome.
Sex differences in meiosis ensure that a strategy for biasing chromosome transmission can only function in one sex. While all products of male meiosis have the potential to develop into sperm, only one of the four products of female meiosis will become the egg. To gain an advantage in female meiosis the critical point is the alignment of homologs on the spindle at the first division (Fig. 1.2a). A centromere that attaches to the egg pole will escape elimination in the polar body (Rosin and Mellone 2017; Kursel and Malik 2018). To take advantage of this requires asymmetry in the meiotic spindle and a centromere able to exploit the asymmetry. The extraordinary reproductive advantage that a stronger centromere holds is thought to fuel an evolutionary race that drives rapid changes in centromeric DNA and proteins (Malik 2009). Predictions of this model are fulfilled in mice where an expansion of satellite repeats has produced a large centromere with an advantage over a homolog with fewer satellites (Iwata-Otsubo et al. 2017). Interestingly, the kinetochores of the larger centromere detach more frequently from cortical spindle fibers, providing an opportunity to reattach to the egg pole (Akera et al. 2019). Suppressors of centromere drive would benefit a population in which a chromosome had begun to cheat. The observation that the centromeric variants Cid and CEN-A are remarkably fast-evolving and divergent from other histones suggests their involvement (Black et al. 2004). Amino acid changes in Cid are concentrated in a region that interacts with H4 and an extended loop that contacts DNA (Vermaak et al. 2002). The rapid evolution of centromeres and the proteins that bind them seems at odds with the very conservative function of centromeres but is in accord with the idea that these structures are the site of an evolutionary battle (Malik 2009).
Meiotic drive in female and male germlines. (a) Stronger centromeres (larger dot) gain an advantage in the female germline by avoiding the cortical spindle and becoming an egg nucleus more than 50% of the time. (b) Chromosomes that achieve drive in males do so by sabotaging their homolog. This may be direct, as implied in the cartoon, or indirect by the establishment of an environment that is toxic to cells inheriting the susceptible homolog. Left) The Paris sex ratio X chromosome produces a factor that blocks segregation of the Y at the second meiotic division. Failure to form Y-bearing sperm ensures predominantly female broods. Right) The Sd and Winters drivers sabotage the maturation of sperm carrying susceptible homologs. In both systems, arrest occurs before chromatin compaction and these malformed cells are eliminated (center). Sperm carrying the driver (right) develop normally
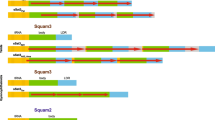
All products of male meiosis have the opportunity to become sperm. To gain an advantage in the male germ line a chromosome must exert a negative effect on its homolog (Fig. 1.2b). Several examples of this are well known, including Segregation distorter/Responder (Sd/Rsp) in D. melanogaster. Rsp, the target of drive, is an array of two very similar 120 bp repeats present in dozens to thousands of copies (Khost et al. 2017). Larger arrays confer increased sensitivity to Sd (Larracuente and Presgraves 2012; Moschetti et al. 1996). Sd is a truncated but enzymatically active duplication of RanGAP that mislocalizes to the interior of the nucleus (Kusano et al. 2001). When a male has Sd on one homolog and a sensitive Rsp allele (RspS) on the other, maturation of sperm carrying RspS is arrested and these cells are eliminated. The sperm carrying Sd develop normally and are responsible for most or all fertilizations. The Sd phenotype is enhanced by a number of modifiers, all genetically linked to Sd on the second chromosome (reviewed by Larracuente and Presgraves 2012). Of course, Sd chromosomes must themselves carry insensitive Rsp arrays in order to escape elimination. Although the precise molecular defect that causes arrest is unclear, abnormal localization of mutant RanGAP is thought to disrupt the RanGTP/GDP gradient across the nuclear envelope and this may interfere with transport in and out of the nucleus (Kusano et al. 2003). In this environment, the expanded repeats on RspS chromosome precipitate failure of sperm maturation. The process that is affected must be unique to sperm development as Sd/RspS females produce normal offspring ratios. During sperm maturation chromatin is remodeled by the replacement of histones, a process requiring the import of protamine. This step is male-limited and appropriate to the stage of arrest, but it is unclear how expanded RspS arrays and defects in protamine levels would induce arrest. Disruption of small RNA import leading to a defect in repackaging Rsp chromatin is also possible. Small RNAs from Rsp have been identified in the germ line and mutations in aubegine (aub), an argonaut family protein that participates in piRNA production, enhances distortion by Sd (Gell and Reenan 2013; Nagao et al. 2010). This finding suggests that an additional role of germ line small RNA systems is to defend against meiotic drive.
Rsp provides an excellent example of satellite turnover. Two families of repeats, the Rsp and Rsp-like family and the 359 bp family, which includes an extensive array of pericentromeric 359 bp repeats in X heterochromatin and the euchromatic 1.688X satellites, were found to occupy overlapping sites in related species (Sproul et al. 2020). Both the Rsp and 359 families are AT-rich, but the 359 bp repeats are widespread, older, and more diversified in related species. Examination of satellite repeats in several Drosophila species revealed that 1.688X and Rsp-like satellites occupy many of the same euchromatic sites (Sproul et al. 2020). Sites in which Rsp-like repeats have been inserted in an existing 1.688X array, possibly in the process of replacing it, were identified in D. simulans and D. mauritiana. This suggests a model in which young Rsp-like repeats use homology with existing 1.688X repeats to enter these sites, a process that may be facilitated by long-range interactions in the nucleus. Extrachromosomal circular DNAs are also a potential mechanism for movement. The correlation between the abundance of one of the repetitive elements and extrachromosomal DNA also suggests a role in Rsp-like invasion (Sproul et al. 2020).
D. simulans has at least three meiotic drive systems that bias sex chromosome inheritance and thus distort the sex ratio. All of these involve drivers on the X chromosome. In the Winters system, named for the location where the flies were collected, the X chromosome distorter prevents Y-bearing gametes from completing maturation. Failure occurs during condensation of the haploid nucleus, a timing that is similar to that observed in the D. melanogaster Sd/Rsp system (Tao et al. 2007b). The driver, Distorter on the X chromosome (Dox), is a partial duplication that produces an RNA with limited coding potential (Tao et al. 2007a). The mechanism of Dox action is unknown but suppressors of Dox on the second chromosome generate siRNAs that reduce levels of the Dox transcript (Lin et al. 2018). As the Y chromosome is primarily composed of satellite repeats and transposons, it is likely that the toxic effect of Dox depends on the unique sequence and chromatin composition of this chromosome. A second D. simulans sex ratio distortion system, Paris, induces anaphase bridges and failure of Y chromosome disjunction during the second meiotic division (Fig. 1.2b, Cazemajor et al. 2000). One component of the X-linked driver was discovered to be a loss of function mutation in a rapidly evolving member of the HP1 family, HP1D2 (Helleu et al. 2016). Intriguingly, the HP1D2 protein is specifically enriched on the Y chromosome, suggesting that a defect in the organization or compaction of this chromosome prevents segregation. Sex ratio distortion leads to populations with unbalanced ratios of males and females and creates a strong selective advantage for an individual with a novel suppressor of drive. In accord with this, Y chromosomes that are resistant to the Paris or Winters driver have been discovered (Branco et al. 2013; Helleu et al. 2019). As the coding potential on the Y is limited, it is quite possible that these suppressors are changes in satellite or transposon content that make them insensitive to the X-linked driver.
1.11 Satellite Repeats Mediate Conflict Between Species
Hybrid incompatibilities enforce the reproductive isolation that defines species (Castillo and Barbash 2017). The rapid evolution of heterochromatin DNA and proteins is a potential source of incompatibilities that produce lethality or infertility upon hybridization of closely related Drosophilids (Presgraves 2010; Ferree and Barbash 2009; Gatti et al. 1976; Yunis and Yasmineh 1971; reviewed in Ferree and Prasad 2012). For example, when D. melanogaster males are mated to D. simulans females, male offspring emerge as sterile adults but females die as embryos. Early female lethality is attributable to the D. melanogaster X chromosome in D. simulans cytoplasm. Specifically, the large array of 359 bp repeats at the base of the D. melanogaster X chromosome fails to compact and X chromatids become entangled in anaphase bridges (Ferree and Barbash 2009). But when D. melanogaster males transmitted a Zygotic hybrid rescue (Zhr) chromosome that was deleted for the pericentric 359 bp repeats, female offspring survived (Sawamura et al. 1993). Small RNAs from the 359 bp satellite are present in oocytes from D. melanogaster females, and it is plausible that these direct heterochromatin formation over the 359 bp satellites in fertilized embryos (Ferree and Barbash 2009). The X chromosome of D. simulans lacks 359 bp repeats and the relevant class of siRNA is not present in D. simulans eggs. These ideas are supported by a study demonstrating that heterochromatin formation at the 359 bp satellites occurred with different timing than that of another large satellite array and required maternal factors missing from D. simulans ooplasm (Yuan and O’Farrell 2016).
The reciprocal mating, D. melanogaster females mated to D. simulans males, produced sterile female adults but no male offspring. The toxic interaction producing male lethality can be traced to heterochromatin proteins, Lethal hybrid rescue (Lhr, D. simulans) and Hybrid male rescue (Hmr, D. melanogaster) (Maheshwari and Barbash 2012). Higher expression of Lhr from the D. simulans chromosome is the basis of hybrid lethality. Loss of D. simulans Lhr rescues hybrid lethality, but loss of D. melanogaster Lhr, which is expressed at only half the rate as D. simulans, does not achieve rescue. Lhr encodes a rapidly evolving heterochromatin protein that interacts with HP1 (Brideau et al. 2006; Brideau and Barbash 2011; Thomae et al. 2013). Loss of Lhr results in bloated polytene chromosomes in D. simulans, a phenotype associated with a loss of chromosome structure (Pal Bhadra et al. 2006). The Hmr gene is also rapidly evolving and encodes a DNA binding protein that localizes to heterochromatin in a complex with Lhr and HP1a (Satyaki et al. 2014; Alekseyenko et al. 2014). Hybrid males, which die as larvae, display poorly condensed chromosomes and anaphase bridges between sister chromatids, consistent with the idea that heterochromatin assembly and compaction is the primary defect (Blum et al. 2017).
The conflicts between genetic elements within a species that produce meiotic drive, and between species that lead to hybrid incompatibility, rely on an overlapping cast of characters. In accord with this, it has been suggested that meiotic drive contributes to the genetic divergence that produces hybrid incompatibility (McDermott and Noor 2010). This notion is supported by the discovery that a single gene, Overdrive (Ovd), appears responsible for both meiotic drive and hybrid incompatibility in D. pseudoobscura (Phadnis and Orr 2009). Males from a mating between subspecies are sterile when young but become weakly fertile and produce almost exclusively daughters when aged. Although the molecular mechanisms at play are currently unknown, the finding that one gene is involved in both phenomena supports the idea that meiotic drive and hybrid incompatibility are produced by similar genetic conflicts.
Summary
Satellite repeats appeared both troublesome and singularly unpromising at the dawn of the genomics era. The typical concentration of satellites in vast, unclonable blocks of heterochromatin was an additional deterrent to their study. But the ability of satellites to move, expand, and undergo relatively rapid genome-wide replacement enables them to shape genomes and respond to evolutionary pressures. Satellite DNA, and small RNA pathways capable of directing modifications to chromatin, are a powerful combination that can be adapted to novel roles. This can be appreciated by the dispersed, euchromatic 1.688X satellites that recruit dosage compensation while very similar heterochromatic 359 bp satellites mediate hybrid incompatibility at a different life stage and in different sex. In spite of the stark differences in the roles of these satellites, it is likely that small RNA normally directs chromatin modifications to both, and that these modifications are essential for normal function. When heterochromatin is compromised satellites become unstable and devastating disruptions of nuclear organization result. This intrinsic risk can be appreciated by the destruction unleased by 359 bp satellites in a hybrid environment. The remarkable ability of heterochromatin to assemble satellite DNA into a nondestructive form can be credited with enabling satellite repeats to expand to their current position of prominence in higher eukaryotes. All of the properties described above, including the mutability of satellites and their intrinsic danger, put repetitive sequences at the leading edge of evolution.
References
Abad JP, Carmena M, Baars S, Saunders RD, Glover DM, Ludeña P, Sentis C, Tyler-Smith C, Villasante A (1992) Dodeca satellite: a conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. PNAS 89:4663–4667. https://doi.org/10.1073/pnas.89.10.4663
Akera T, Trimm E, Lampson MA (2019) Molecular strategies of meiotic cheating by selfish centromeres. Cell 178(5):1132–1144.e10. https://doi.org/10.1016/j.cell.2019.07.001
Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI (2006) High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20(7):848–857. https://doi.org/10.1101/gad.1400206
Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, Kuroda MI (2008) A sequence Motif within chromatin entry sites directs MSL establishment on the drosophila X chromosome. Cell 134(4):599–609. https://doi.org/10.1016/j.cell.2008.06.033
Alekseyenko AA, Gorchakov AA, Zee BM, Fuchs SM, Kharchenko PV, Kuroda MI (2014) Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev 28(13):1445–1460. https://doi.org/10.1101/gad.241950.114
Alfonso-Parra C, Maggert KA (2010) Drosophila SAF-B links the nuclear matrix, chromosomes, and transcriptional activity. PLoS One 5(4). https://doi.org/10.1371/journal.pone.0010248
Ando S, Yang H, Nozaki N, Okazaki T, Yoda K (2002) CENP-A, -B, and -C chromatin complex that contains the I-type α-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol 22(7):2229–2241. https://doi.org/10.1128/mcb.22.7.2229-2241.2002
Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schübeler D (2008) Transcription-coupled methylation of histone H3 at Lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol 28(10):3401–3409. https://doi.org/10.1128/mcb.00006-08
Bernard P, Allshire RC (2002) Centromeres become unstuck without heterochromatin. Trends Cell Biol 12(9):419–424. https://doi.org/10.1016/S0962-8924(02)02344-9
Black BE, Foliz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW (2004) Structural determinants for generating centromeric chromatin. Nature 430(6999):578–582. https://doi.org/10.1038/nature02766
Blattes R, Monod C, Susbielle G, Cuvier O, Wu JH, Hsieh TS, Laemmli UK, Käs E (2006) Displacement of D1, HP1 and topoisomerase II from satellite heterochromatin by a specific polyamide. EMBO J 25(11):2397–2408. https://doi.org/10.1038/sj.emboj.7601125
Blum JA, Bonaccorsi S, Marzullo M, Palumbo V, Yamashita YM, Barbash DA, Gatti M (2017) The hybrid incompatibility genes Lhr and Hmr are required for sister chromatid detachment during anaphase but not for centromere function. Genetics 207(4):1457–1472. https://doi.org/10.1534/genetics.117.300390
Bobkov GOM, Gilbert N, Heun P (2018) Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J Cell Biol 217:1957–1972
Bosco G, Campbell P, Leiva-Neto JT, Markow TA (2007) Analysis of drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics 177(3):1277–1290. https://doi.org/10.1534/genetics.107.075069
Brajković J, Feliciello I, Bruvo-Madarić B, Ugarković D (2012) Satellite DNA-like elements associated with genes within euchromatin of the beetle Tribolium castaneum. G3 Genes Genomes Genetics 2(8):931–941. https://doi.org/10.1534/g3.112.003467
Branco AT, Tao Y, Hartl DL, Lemos B (2013) Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis-specific gene expression in Drosophila simulans. Heredity 111(1):8–15. https://doi.org/10.1038/hdy.2013.5
Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128(6):1089–1103. https://doi.org/10.1016/j.cell.2007.01.043
Brideau NJ, Barbash DA (2011) Functional conservation of the Drosophila hybrid incompatibility gene Lhr. BMC Evol Biol 11(1):57. https://doi.org/10.1186/1471-2148-11-57
Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA (2006) Two Dobzhansky-Muller genes interact to cause hybrid lethality in drosophila. Science 314(5803):1292–1295. https://doi.org/10.1126/science.1133953
Bzymek M, Lovett ST (2001) Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci USA 98(15):8319–8325. https://doi.org/10.1073/pnas.111008398
Cabot EL, Doshi P, Wu ML, Wu CI (1993) Population genetics of tandem repeats in centromeric heterochromatin: unequal crossing over and chromosomal divergence at the responder locus of Drosophila melanogaster. Genetics 135(2):477–487
Caridi CP, D’agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I (2018) Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559(7712):54–60. https://doi.org/10.1038/s41586-018-0242-8
Castillo DM, Barbash DA (2017) Moving speciation genetics forward: modern techniques build on foundational studies in drosophila. Genetics 207(3):825–842. https://doi.org/10.1534/genetics.116.187120
Cazemajor M, Joly D, Montchamp-Moreau C (2000) Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics 154(1):229–236
Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Sardo FL, Saxena A, Miyoshi K, Siomi H, Siomi MC, Carninci P, Gilmour DS, Corona DFV, Orlando V (2011) Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480(7377):391–395. https://doi.org/10.1038/nature10492
Chang CH, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen CC, Erceg J, Beliveau BJ, Wu CT, Larracuente AM, Mellone BG (2019) Islands of retroelements are major components of Drosophila centromeres. PLoS Biol 17(5). https://doi.org/10.1371/journal.pbio.3000241
Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG (2014) CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol 204(3):313–329. https://doi.org/10.1083/jcb.201305036
Cohen S, Segal D (2009) Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res 124(3–4):327–338. https://doi.org/10.1159/000218136
Cohen S, Yacobi K, Segal D (2003) Extrachromosomal circular DNA of tandemly repeated genomic sequences in drosophila. Genome Res 13(6):1133–1145. https://doi.org/10.1101/gr.907603
Cohen Z, Bacharach E, Lavi S (2006) Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway. Oncogene 25(33):4515–4524. https://doi.org/10.1038/sj.onc.1209485
Copur Ö, Gorchakov A, Finkl K, Kuroda MI, Müller J (2018) Sex-specific phenotypes of histone H4 point mutants establish dosage compensation as the critical function of H4K16 acetylation in Drosophila. Proc Natl Acad Sci USA 115(52):13336–13341. https://doi.org/10.1073/pnas.1817274115
Courret C, Chang CH, Wei KHC, Montchamp-Moreau C, Larracuente AM (2019) Meiotic drive mechanisms: lessons from Drosophila. Proc R Soc B Biol Sci 286(1913). https://doi.org/10.1098/rspb.2019.1430
de Lima LG, Hanlon SL, Gerton JL (2020) Origins and evolutionary patterns of the 1.688 satellite DNA family in drosophila phylogeny. G3 Genes Genomes Genetics 10(11):4129–4146. https://doi.org/10.1534/g3.120.401727
De Wit E, Greil F, Van Steensel B (2005) Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res 15(9):1265–1273. https://doi.org/10.1101/gr.3198905
deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, Koeman J, Seth S, Protopopov A, Felicella M, Zheng S, Multani A, Jiang Y, Zhang J, Nam D-H, Petricoin EF, Chin L, Mikkelsen T, Verhaak RGW (2018) Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 50(5):708–717. https://doi.org/10.1038/s41588-018-0105-0
Deng X, Meller VH (2006) roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 174(4):1859–1866. https://doi.org/10.1534/genetics.106.064568
Deshpande N, Meller VH (2018) Chromatin that guides dosage compensation is modulated by the siRNA pathway in Drosophila melanogaster. Genetics 209(4):1085–1097. https://doi.org/10.1534/genetics.118.301173
Dias GB, Heringer P, Svartman M, Kuhn GCS (2015) Helitrons shaping the genomic architecture of Drosophila: enrichment of DINE-TR1 in α- and β-heterochromatin, satellite DNA emergence, and piRNA expression. Chromosom Res 23(3):597–613. https://doi.org/10.1007/s10577-015-9480-x
Dibartolomeis SM, Tartof KD, Rob Jackson F (1992) A superfamily of Drosophila satellite related (SR) DNA repeats restricted to the X chromosome euchromatin. Nucleic Acids Res 20(5):1113–1116. https://doi.org/10.1093/nar/20.5.1113
Dorer DR, Henikoff S (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77(7):993–1002. https://doi.org/10.1016/0092-8674(94)90439-1
Doshi P, Kaushal S, Benyajati C, Wu C-I (1991) Molecular analysis of the responder satellite DNA in Drosophila melanogaster: DNA bending, nucleosome stucture, and Rsp-binding proteins. Mol Biol Evol 8(5):721–741
Dover G (1982) Molecular drive: a cohesive mode of species evolution. Nature 299:111–117
Elgin SCR, Reuter G (2013) Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5(8):1–26. https://doi.org/10.1101/cshperspect.a017780
Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341(6147):1–13. https://doi.org/10.1126/science.1237973
Fackelmayer FO, Dahm K, Renz A, Ramsperger U, Richter A (1994) Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem 221(2):749–757. https://doi.org/10.1111/j.1432-1033.1994.tb18788.x
Fagegaltier D, Bougé AL, Berry B, Poisot É, Sismeiro O, Coppée JY, Théodore L, Voinnet O, Antoniewski C (2009) The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci USA 106(50):21258–21263. https://doi.org/10.1073/pnas.0809208105
Fan H, Lv P, Huo X, Wu J, Wang Q, Cheng L, Liu Y, Tang Q-Q, Zhang L, Zhang F, Zheng X, Wu H, Wen B (2018) The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res 28(2):192–202. https://doi.org/10.1101/gr.224576.117
Feliciello I, Akrap I, Ugarković Đ (2015) Satellite DNA modulates gene expression in the Beetle Tribolium castaneum after heat stress. PLoS Genet 11(8):1–18. https://doi.org/10.1371/journal.pgen.1005466
Ferree PM, Barbash DA (2009) Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol 7(10). https://doi.org/10.1371/journal.pbio.1000234
Ferree PM, Prasad S (2012) How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. Genetics Res Int 2012:1–11. https://doi.org/10.1155/2012/430136
Fitzgerald DJ, Dryden GL, Bronson EC, Williams JS, Anderson JN (1994) Conserved patterns of bending in satellite and nucleosome positioning DNA. J Biol Chem 269(33):21303–21314. https://doi.org/10.1016/S0021-9258(17)31963-4
Fondon JW, Garner HR (2004) Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA 101(52):18058–18063. https://doi.org/10.1073/pnas.0408118101
Gallach M (2014) Recurrent turnover of chromosome-specific satellites in Drosophila. Genome Biol Evol 6(6):1279–1286. https://doi.org/10.1093/gbe/evu104
Garrick D, Fiering S, Martin DIK, Whitelaw E (1998) Repeat-induced gene silencing in mammals. Nat Genet 18(1):56–59. https://doi.org/10.1038/ng0198-56
Garrido-Ramos MA (2017) Satellite DNA: an evolving topic. Genes 8(9). https://doi.org/10.3390/genes8090230
Gatti M, Pimpinelli S, Santini G (1976) Characterization of Drosophila heterochromatin. Chromosoma 57(4):351–375. https://doi.org/10.1007/BF00332160
Gell SL, Reenan RA (2013) Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster. Genetics 193(3):771–784. https://doi.org/10.1534/genetics.112.147561
Goenka A, Sengupta S, Pandey R, Parihar R, Mohanta GC, Mukerji M, Ganesh S (2016) Human satellite-III non-coding RNAs modulate heat-shockinduced transcriptional repression. J Cell Sci 129(19):3541–3552. https://doi.org/10.1242/jcs.189803
Göhring F, Fackelmayer FO (1997) The Scaffold/Matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry 36(27):8276–8283. https://doi.org/10.1021/bi970480f
Grewal SIS, Elgin SCR (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447(7143):399–406. https://doi.org/10.1038/nature05914
Gu T, Elgin SCR (2013) Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in drosophila. PLoS Genet 9(9). https://doi.org/10.1371/journal.pgen.1003780
Hall AC, Ostrowski LA, Mekhail K (2019) Phase separation as a melting pot for DNA repeats. Trends Genet 35(8):589–600. https://doi.org/10.1016/j.tig.2019.05.001
Hammock EAD, Young LJ (2005) Genetics: microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308(5728):1630–1634. https://doi.org/10.1126/science.1111427
Hartl DL (2000) Molecular melodies in high and low C. Nat Rev Genet 1(2):145–149. https://doi.org/10.1038/35038580
Hartl DL, Hartung N (1975) High frequency of one element of segregation distorter in natural populations of Drosophila melanogaster. Evolution 29(3):512–518
Helleu Q, Gérard PR, Dubruille R, Ogereau D, Prud’homme B, Loppin B, Montchamp-Moreau C (2016) Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proc Natl Acad Sci USA 113(15):4110–4115. https://doi.org/10.1073/pnas.1519332113
Helleu Q, Courret C, Ogereau D, Burnham KL, Chaminade N, Chakir M, Aulard S, Montchamp-Moreau C (2019) Sex-ratio meiotic drive shapes the evolution of the y chromosome in Drosophila simulans. Mol Biol Evol 36(12):2668–2681. https://doi.org/10.1093/molbev/msz160
Henikoff S (1998) Conspiracy of silence among repeated transgenes. BioEssays 20(7):532–535. https://doi.org/10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. https://doi.org/10.1126/science.1062939
Heslop-Harrison JS, Schwarzacher T (2013) Nucleosomes and centromeric DNA packaging. Proc Natl Acad Sci USA 110(50):19974–19975. https://doi.org/10.1073/pnas.1319945110
Höck J, Meister G (2008) The Argonaute protein family. Genome Biol 9(2). https://doi.org/10.1186/gb-2008-9-2-210
Hsieh T-S, Brutlag D (1979) Sequence and sequence variation within the 1.688 g/cm3 satellite DNA of Drosophila melanogaster. J Mol Biol 135(2):465–481. https://doi.org/10.1016/0022-2836(79)90447-9
Huertas D, Cortes A, Casanova J, Azorin F (2004) Drosophila DDP1, a multi-KH-domain protein, contributes to centromeric silencing and chromosome segregation. Curr Biol 14:1611–1620
Huo X, Ji L, Zhang Y, Lv P, Cao X, Wang Q, Yan Z, Dong S, Du D, Zhang F, Wei G, Liu Y, Wen B (2020) The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol Cell 77(2):368–383.e7. https://doi.org/10.1016/j.molcel.2019.10.001
Ideue T, Tani T (2020) Centromeric non-coding RNAs: conservation and diversity in function. Non-coding RNA 6(1). https://doi.org/10.3390/ncrna6010004
Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE (2017) Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol 27(15):2365–2373.e8. https://doi.org/10.1016/j.cub.2017.06.069
Jagannathan M, Warsinger-Pepe N, Watase GJ, Yamashita YM (2017) Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 Genes Genomes Genetics 7(2):693–704. https://doi.org/10.1534/g3.116.035352
Jagannathan M, Cummings R, Yamashita YM (2019) The modular mechanism of chromocenter formation in Drosophila. BioRxiv:1–16. https://doi.org/10.1101/481820
Jolly C, Lakhotia SC (2006) Human sat III and Drosophila hsrω transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res 34(19):5508–5514. https://doi.org/10.1093/nar/gkl711
Joshi S, Meller VH (2017) Satellite repeats idenitfy X chromatin for dosage compesation in Drosophila melanogaster males. Curr Biol 176(1):139–148. https://doi.org/10.1016/j.physbeh.2017.03.040
Khost DE, Eickbush DG, Larracuente AM (2017) Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila. Genome Res 27(5):709–721. https://doi.org/10.1101/gr.213512.116
Khurana JS, Xu J, Weng Z, Theurkauf WE (2010) Distinct functions for the Drosophila piRNA pathway in genome maintenance and telomere protection. PLoS Genet 6:e1001246
Kim H, Nguyen N-P, Turner K, Wu S, Gujar AD, Luebeck J, Liu J, Deshpande V, Rajkumar U, Namburi S, Amin SB, Yi E, Menghi F, Schulte JH, Henssen AG, Chang HY, Beck CR, Mischel PS, Bafna V, Verhaak RGW (2020) Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet 52(9):891–897. https://doi.org/10.1038/s41588-020-0678-2
Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A (2008) Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in drosophila. Cell 133(5):813–828. https://doi.org/10.1016/j.cell.2008.04.036
Kit S (1961) Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol 3(6):711–716. https://doi.org/10.1016/S0022-2836(61)80075-2
Kiyama R, Matsui H, Oishi M (1986) A repetitive DNA family (Sau3A family) in human chromosomes: extrachromosomal DNA and DNA polymorphism. Proc Natl Acad Sci 83(13):4665–4669. https://doi.org/10.1073/pnas.83.13.4665
Kiyama R, Matsui H, Okumura K, Oishi M (1987) A group of repetitive human DNA families that is characterized by extrachromosomal oligomers and restriction-fragment length polymorphism. J Mol Biol 193(4):591–597. https://doi.org/10.1016/0022-2836(87)90342-1
Kuhn GCS, Küttler H, Moreira-Filho O, Heslop-Harrison JS (2012) The 1.688 repetitive DNA of drosophila: concerted evolution at different genomic scales and association with genes. Mol Biol Evol 29(1):7–11. https://doi.org/10.1093/molbev/msr173
Kuroda MI, Hilfiker A, Lucchesi JC (2016) Dosage compensation in Drosophila—a model for the coordinate regulation of transcription. Genetics 204(2):435–450. https://doi.org/10.1534/genetics.115.185108
Kursel LE, Malik HS (2018) The cellular mechanisms and consequences of centromere drive. Curr Opin Cell Biol 52(206):58–65. https://doi.org/10.1016/j.ceb.2018.01.011
Kusano A, Staber C, Ganetzky B (2001) Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev Cell 1(3):351–361. https://doi.org/10.1016/S1534-5807(01)00042-9
Kusano A, Staber C, Chan HYE, Ganetzky B (2003) Closing the (Ran)GAP on segregation distortion in Drosophila. BioEssays 25(2):108–115. https://doi.org/10.1002/bies.10222
Larracuente AM (2014) The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol Biol 14(1):1–12. https://doi.org/10.1186/s12862-014-0233-9
Larracuente AM, Presgraves DC (2012) The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192(1):33–53. https://doi.org/10.1534/genetics.112.141390
Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI (2011) X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471(7336):115–118. https://doi.org/10.1038/nature09757
Larson K, Yan S-J, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX (2012) Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 8(1):e1002473. https://doi.org/10.1371/journal.pgen.1002473
Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547(7662):236–240. https://doi.org/10.1038/nature22822
Levinson G, Gutman G (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. https://doi.org/10.1093/oxfordjournals.molbev.a040442
Lin CJ, Hu F, Dubruille R, Vedanayagam J, Wen J, Smibert P, Loppin B, Lai EC (2018) The hpRNA/RNAi pathway is essential to resolve intragenomic conflict in the drosophila male germline. Dev Cell 46(3):316–326.e5. https://doi.org/10.1016/j.devcel.2018.07.004
Lohe AR, Brutlag DL (1987) Identical satellite DNA sequences in sibling species of drosophila. J Mol Biol 194:161–170
Lohe AR, Hilliker AJ, Roberts PA (1993) Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134(4):1149–1174. https://doi.org/10.1016/0168-9525(93)90135-5
Lower SS, McGurk MP, Clark A, Barbash DA (2018) Satellite DNA evolution: old ideas, new approaches. Curr Opin Genet Dev 49:70–78. https://doi.org/10.1016/j.gde.2018.03.003
Maheshwari S, Barbash DA (2012) Cis-by-trans regulatory divergence causes the asymmetric lethal effects of an ancestral hybrid incompatibility gene. PLoS Genet 8(3):e1002597. https://doi.org/10.1371/journal.pgen.1002597
Maison C, Bailly D, Peters AHFM, Quivy J-P, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30(3):329–334. https://doi.org/10.1038/ng843
Malik HS (2009) The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol 48(June):33–52. https://doi.org/10.1007/978-3-642-00182-6_2
Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109(5):1963–1973. https://doi.org/10.1083/jcb.109.5.1963
McDermott SR, Noor MAF (2010) The role of meiotic drive in hybrid male sterility. Philos Trans R Soc B Biol Sci 365(1544):1265–1272. https://doi.org/10.1098/rstb.2009.0264
McNulty SM, Sullivan LL, Sullivan BA (2017) Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev Cell 42(3):226–240.e6. https://doi.org/10.1016/j.devcel.2017.07.001
Meller VH, Rattner BP (2002) The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21(5):1084–1091. https://doi.org/10.1093/emboj/21.5.1084
Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A (2006) Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21(6):811–823. https://doi.org/10.1016/j.molcel.2006.02.007
Menon DU, Meller VH (2012) A role for siRNA in X-chromosome dosage compensation in Drosophila melanogaster. Genetics 191(3):1023–1028. https://doi.org/10.1534/genetics.112.140236
Menon DU, Coarfa C, Xiao W, Gunaratne PH, Meller VH (2014) siRNAs from an X-linked satellite repeat promote X-chromosome recognition in Drosophila melanogaster. Proc Natl Acad Sci USA 111(46):16460–16465. https://doi.org/10.1073/pnas.1410534111
Meštrović N, Mravinac B, Pavlek M, Vojvoda-Zeljko T, Šatović E, Plohl M (2015) Structural and functional liaisons between transposable elements and satellite DNAs. Chromosom Res 23(3):583–596. https://doi.org/10.1007/s10577-015-9483-7
Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA, Schneider VA, Potapova T, Wood J, Chow W, Armstrong J, Fredrickson J, Pak E, Tigyi K, Kremitzki M et al (2020) Telomere-to-telomere assembly of a complete human X chromosome. Nature 585(7823):79–84. https://doi.org/10.1038/s41586-020-2547-7
Moschetti R, Caizzi R, Pimpinellit S (1996) Segregation distortion in drosophila rnelanogaster: genomic organization of responder sequences. Genetics 144:1665–1671
Muchardt C, Guillemé M, Seeler J, Trouche D, Dejean A, Yaniv M (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1 α. EMBO Rep 3(10):975–981. https://doi.org/10.1093/embo-reports/kvf194
Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H (2010) Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16(12):2503–2515. https://doi.org/10.1261/rna.2270710
Nickerson JA, Krochmalnic G, Wan KM, Penman S (1989) Chromatin architecture and nuclear RNA. Proc Natl Acad Sci 86(1):177–181. https://doi.org/10.1073/pnas.86.1.177
Nozawa R-S, Boteva L, Soares DC, Naughton C, Dun AR, Buckle A, Ramsahoye B, Bruton PC, Saleeb RS, Arnedo M, Hill B, Duncan RR, Maciver SK, Gilbert N (2017) SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell 169(7):1214–1227.e18. https://doi.org/10.1016/j.cell.2017.05.029
Ohta T, Dover GA (1984) The cohesive population genetics of molecular drive. Genetics 108(2):501–521
Osada N, Innan H (2008) Duplication and gene conversion in the Drosophila melanogaster genome. PLoS Genet 4(12). https://doi.org/10.1371/journal.pgen.1000305
Pal Bhadra M, Bhadra U, Birchler JA (2006) Misregulation of sex-lethal and disruption of male-specific lethal complex localization in drosophila species hybrids. Genetics 174(3):1151–1159. https://doi.org/10.1534/genetics.106.060541
Palomeque T, Lorite P (2008) Satellite DNA in insects: a review. Heredity 100(6):564–573. https://doi.org/10.1038/hdy.2008.24
Pardue ML, Gall JG (1970) Chromosomal localization of mouse satellite DNA. Science 168(3937):1356–1358. https://doi.org/10.1126/science.168.3937.1356
Peng JC, Karpen GH (2007) H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9(1):25–35. https://doi.org/10.1038/ncb1514
Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL (2000) Evidence for DNA loss as a determinant of genome size. Science 287(5455):1060–1062. https://doi.org/10.1126/science.287.5455.1060
Phadnis N, Orr HA (2009) Sterility and segregation distortion in Drosophila hybrids. Science 323(January):376–379
Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S (2003) Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol 161(4):707–714. https://doi.org/10.1083/jcb.200303012
Pont G, Degroote F, Picard G (1987) Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes. J Mol Biol 195(2):447–451. https://doi.org/10.1016/0022-2836(87)90665-6
Presgraves DC (2010) The molecular evolutionary basis of species formation. Nat Rev Genet 11(3):175–180. https://doi.org/10.1038/nrg2718
Ramírez F, Lingg T, Toscano S, Lam KC, Georgiev P, Chung HR, Lajoie BR, de Wit E, Zhan Y, de Laat W, Dekker J, Manke T, Akhtar A (2015) High-affinity sites form an interaction network to facilitate spreading of the MSL complex across the X chromosome in drosophila. Mol Cell 60(1):146–162. https://doi.org/10.1016/j.molcel.2015.08.024
Ranathunge C, Wheeler GL, Chimahusky ME, Kennedy MM, Morrison JI, Baldwin BS, Perkins AD, Welch ME (2018) Transcriptome profiles of sunflower reveal the potential role of microsatellites in gene expression divergence. Mol Ecol 27(5):1188–1199. https://doi.org/10.1111/mec.14522
Richards RI, Sutherland GR (1994) Simple repeat DNA is not replicated simply. Nat Genet 6(2):114–116. https://doi.org/10.1038/ng0294-114
Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G (2004) Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell 15(2):543–551. https://doi.org/10.1091/mbc.e03-07-0487
Roach RJ, Garavís M, González C, Jameson GB, Filichev VV, Hale TK (2020) Heterochromatin protein 1α interacts with parallel RNA and DNA G-quadruplexes. Nucleic Acids Res 48(2):682–693. https://doi.org/10.1093/nar/gkz1138
Rohrbaugh M, Clore A, Davis J, Johnson S, Jones B, Jones K, Kim J, Kithuka B, Lunsford K, Mitchell J, Mott B, Ramos E, Tchedou MR, Acosta G, Araujo M, Cushing S, Duffy G, Graves F, Griffin K et al (2013) Identification and characterization of proteins involved in nuclear organization using drosophila GFP protein trap lines. PLoS One 8(1). https://doi.org/10.1371/journal.pone.0053091
Rosin LF, Mellone BG (2017) Centromeres drive a hard bargain. Trends Genet 33(2):101–117. https://doi.org/10.1016/j.tig.2016.12.001
Rudd MK, Wray GA, Willard HF (2006) The evolutionary dynamics of α-satellite. Genome Res 16(1):88–96. https://doi.org/10.1101/gr.3810906
Satyaki PRV, Cuykendall TN, Wei KH-C, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA (2014) The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet 10(3):e1004240. https://doi.org/10.1371/journal.pgen.1004240
Sawamura K, Yamamoto MT, Watanabe TK (1993) Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 133(2):307–313
Sawaya S, Bagshaw A, Buschiazzo E, Kumar P, Chowdhury S, Black MA, Gemmell N (2013) Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One 8(2). https://doi.org/10.1371/journal.pone.0054710
Schauer T, Ghavi-Helm Y, Sexton T, Albig C, Regnard C, Cavalli G, Furlong EE, Becker PB (2017) Chromosome topology guides the drosophila dosage compensation complex for target gene activation. EMBO Rep 18(10):1854–1868. https://doi.org/10.15252/embr.201744292
Schubert T, Pusch MC, Diermeier S, Benes V, Kremmer E, Imhof A, Längst G (2012) Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol Cell 48(3):434–444. https://doi.org/10.1016/j.molcel.2012.08.021
Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF (2001) Genomic and genetic definition of a functional human centromere. Science 294(5540):109–115. https://doi.org/10.1126/science.1065042
Sharma R, Jost D, Kind J, Gómez-Saldivar G, van Steensel B, Askjaer P, Vaillant C, Meister P (2014) Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev 28(23):2591–2596. https://doi.org/10.1101/gad.248864.114
Shatskikh AS, Kotov AA, Adashev VE, Bazylev SS, Olenina LV (2020) Functional significance of satellite DNAs: insights from drosophila. Front Cell Dev Biol 8(May):1–19. https://doi.org/10.3389/fcell.2020.00312
Sienski G, Batki J, Senti KA, Dönertas D, Tirian L, Meixner K, Brennecke J (2015) Silencio/CG9754 connects the piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev 29(21):2258–2271. https://doi.org/10.1101/gad.271908.115
Sinclair DA, Guarente L (1997) Extrachromosomal rDNA Circles—a cause of aging in yeast. Cell 91(7):1033–1042. https://doi.org/10.1016/S0092-8674(00)80493-6
Soruco MML, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, Vedenko A, Bulyk ML, Park PJ, Larschan E (2013) The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev 27(14):1551–1556. https://doi.org/10.1101/gad.214585.113
Southern EM (1975) Long range periodicities in mouse satellite DNA. J Mol Biol 94(1):51–69. https://doi.org/10.1016/0022-2836(75)90404-0
Spierer A, Seum C, Delattre M, Spierer P (2005) Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci 118(21):5047–5057. https://doi.org/10.1242/jcs.02623
Spierer A, Begeot F, Spierer P, Delattre M (2008) SU(VAR)3-7 links heterochromatin and dosage compensation in drosophila. PLoS Genet 4(5):3–7. https://doi.org/10.1371/journal.pgen.1000066
Sproul JS, Khost DE, Eickbush DG, Negm S, Wei X, Wong I, Larracuente AM (2020) Dynamic evolution of euchromatic satellites on the X chromosome in Drosophila melanogaster and the simulans Clade. Mol Biol Evol 37(8):2241–2256. https://doi.org/10.1093/molbev/msaa078
Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB (2008) The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4(12):1–14. https://doi.org/10.1371/journal.pgen.1000302
Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH (2017) Phase separation drives heterochromatin domain formation. Nature 547(7662):241–245. https://doi.org/10.1038/nature22989
Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI (2008) The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol 15(12):1318–1325. https://doi.org/10.1038/nsmb.1520
Swenson JM, Colmenares SU, Strom AR, Costes SV, Karpen GH (2016) The composition and organization of Drosophila heterochromatin are heterogeneous and dynamic. eLife 5:1–37. https://doi.org/10.7554/elife.16096
Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL (2007a) A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol 5(11):2576–2588. https://doi.org/10.1371/journal.pbio.0050293
Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL (2007b) A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol 5(11):2560–2575. https://doi.org/10.1371/journal.pbio.0050292
Tautz D, Trick M, Dover G (1986) Cryptic simplicity in DNA is a major source of genetic variation. Nature 322:652–656
Thomae AW, Schade GOM, Padeken J, Borath M, Vetter I, Kremmer E, Heun P, Imhof A (2013) A pair of centromeric proteins mediates reproductive isolation in drosophila species. Dev Cell 27(4):412–424. https://doi.org/10.1016/j.devcel.2013.10.001
Tomilin NV (2008) Regulation of mammalian gene expression by retroelements and non-coding tandem repeats. BioEssays 30(4):338–348. https://doi.org/10.1002/bies.20741
Török T, Gorjánácz M, Bryant PJ, Kiss I (2000) Prod is a novel DNA-binding protein that binds to the 1.686 g/cm3 10 bp satellite repeat of Drosophila melanogaster. Nucleic Acids Res 28(18):3551–3557. https://doi.org/10.1093/nar/28.18.3551
Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, Kornblum HI, Taylor MD, Kaushal S, Cavenee WK, Wechsler-Reya R, Furnari FB, Vandenberg SR, Rao PN, Wahl GM et al (2017) Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543(7643):122–125. https://doi.org/10.1038/nature21356
Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G (2008) Transcription of satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 36(2):423–434. https://doi.org/10.1093/nar/gkm1056
Vermaak D, Hayden HS, Henikoff S (2002) Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol 22(21):7553–7561. https://doi.org/10.1128/mcb.22.21.7553-7561.2002
Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ (2009) Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324(5931):1213–1216. https://doi.org/10.1126/science.1170097
Waring GL, Pollack JC (1987) Cloning and characterization of a dispersed, multicopy, X chromosome sequence in Drosophila melanogaster. Proc Natl Acad Sci USA 84(9):2843–2847. https://doi.org/10.1073/pnas.84.9.2843
Wei KHC, Barbash DA (2015) Never settling down: frequent changes in sex chromosomes. PLoS Biol 13(4):1–6. https://doi.org/10.1371/journal.pbio.1002077
Wei KHC, Grenier JK, Barbash DA, Clark AG (2014) Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc Natl Acad Sci USA 111(52):18793–18798. https://doi.org/10.1073/pnas.1421951112
Willard HF (1985) Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet 37(3):524–532
Wines DR, Henikoff S (1992) Somatic instability of a drosophila chromosome. Genetics 131(3):683–691
Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KHA (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17(8):1146–1160. https://doi.org/10.1101/gr.6022807
Wu CI, Lyttle TW, Wu ML, Lin GF (1988) Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster. Cell 54:179–189. https://doi.org/10.1016/0092-8674(88)90550-8
Yanez-Cuna JO, Arnold CD, Stampfel G, Boryń LM, Gerlach D, Rath M, Stark A (2014) Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res 24(7):1147–1156. https://doi.org/10.1101/gr.169243.113
Yu Q, Colot HV, Kyriacou CP, Hall JC, Rosbash M (1987) Behavior modification by in vitro mutagenesis of a variable region within the period gene of Drosophila. Nature 326:765–769
Yuan K, O’Farrell PH (2016) TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev 30(5):579–593. https://doi.org/10.1101/gad.272237.115
Yunis JJ, Yasmineh WG (1971) Heterochromatin, satellite DNA, and cell function. Science 174(4015):1200–1209. https://doi.org/10.1126/science.174.4015.1200
Zhang K, Mosch K, Fischle W, Grewal SIS (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15(4):381–388. https://doi.org/10.1038/nsmb.1406
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Lauria Sneideman, M.P., Meller, V.H. (2021). Drosophila Satellite Repeats at the Intersection of Chromatin, Gene Regulation and Evolution. In: Ugarković, Ð. (eds) Satellite DNAs in Physiology and Evolution. Progress in Molecular and Subcellular Biology, vol 60. Springer, Cham. https://doi.org/10.1007/978-3-030-74889-0_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-74889-0_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-74888-3
Online ISBN: 978-3-030-74889-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)