Abstract
The brain tumor segmentation is essential for diagnosis and treatment of brain diseases. However, most of current 3D deep learning technologies require large number of magnetic resonance images (MRIs). In order to make full use of small dataset like BraTS 2020, we propose a deep supervision-based 2D residual U-net for efficient and automatic brain tumor segmentation. In our network, residual blocks are used to alleviate the gradient dispersion caused by excessive depth of network, while multiple deep supervision branches are used as the regularization of the network, they can improve the training stability and enable the encoder to extract richer visual features. The CBICA’s IPP’s evaluation of the segmentation results verifies the effectiveness of our method. The average Dice of ET, WT and TC are 0.7593, 0.8726 and 0.7879 respectively.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glioma is the most common primary brain tumor, it has strong aggressiveness and high mortality rate. The average life expectancy of patients with high-grade tumors is usually no more than two years, but finding the tumor on MRI images as early as possible can improve the survival time and survival probability of patients. However, it is time-consuming and inefficient for doctors to manually label brain tumors on such huge number of MRIs, so automatic brain tumor segmentation plays an important role in assisting doctors in diagnosis, surgical planning and evaluation of postoperative recovery effects.
The Multimodal Brain Tumor Segmentation Challenge (BraTS) is currently one of the most authoritative competitions in the field of brain tumor segmentation. It aims to evaluate the performances of the latest methods in this field [1,2,3]. The BraTS 2020 training dataset [4,5,6,7,8] provides 369 multi-institutional routine clinically-acquired multimodal MRI scans of glioblastoma (GBM/HGG) and lower grade glioma (LGG). This dataset is available as NIfTI files (.nii.gz), including MRIs of native (T1), post-contrast T1-weighted (T1Gd), T2-weighted (T2), and T2 Fluid Attenuated Inversion Recovery (T2-FLAIR) volumes (Fig. 1). Our task is to segment each tumor area into enhancing tumor, the peritumoral edema, and the necrotic and non-enhancing tumor core.
2 Related Work
With the development of deep learning, the automatic segmentation of brain tumors gets excellent performance. Especially since the emergence of Convolutional Neural Networks (CNN), it greatly boosts the development of brain tumor segmentation. It is very suitable for processing image data because it can obtain local features more efficiently. Thus CNN-based brain tumor segmentation methods emerge in endlessly.
The first to mention is U-Net [9] (see Fig. 2(a)), it is extensively studied in the field of brain tumor segmentation. Its encoder-decoder structure and ship-connection became the basis of many later researches. Isensee et al. [10, 11] proposed a modified version of U-net for brain tumor segmentation task. It increased the depth of U-net by pre-activation residual blocks, so that achieved excellent results in BraTS 2017 and BraTS 2018. Specifically, the skip-connections ensured that deep feature maps can contain more low-level informations. Zhou et al. proposed a segmentation network called U-Net++ [13]. As shown in Fig. 2(b), it was developed as an improved U-net by nested and dense skip-connections. Then multi-scale features were fused through dense connections like DenseNet [14,15,16,17]. In addition, deep supervision was used to train multiple sub-networks in this method.
There are also some improvements to the encoder-decoder structure of U-net. Myronenko [18] proposed an asymmetric encoder-decoder architecture, it used multi-task learning [19, 20] method to train the network. The network contained two sub-networks, especially one of them used variational U-net structure to reconstruct original input images. The variational U-net can be regarded as regularization of the network to prevent over-fitting caused by small size of training set, also it enabled encoder to extract more features. In addition, Chen et al. [21] proposed MASSL, it used tumor masks obtained by roughly segmentation as the labels of the reconstruction task. This method encouraged model to learn differences between tumor area and background through pseudo-labels mentioned above. Thus it can avoid the network from reconstructing original images, which may lead encoder to pay much attention to original image features.
Furthermore, the first place in BraTS 2019 challenge was a two-stage cascaded U-Net [22] (see Fig. 2(c)). The first stage of this network was an asymmetric encoder-decoder architecture. The prediction results of the first stage were concatenated with original input images, then they were used as the input of the second stage. The second stage network architecture included two branches, and they perform segmentation tasks simultaneously. The purpose of two branches is to help encoder learn richer features. And the difference between the two branches lies in the decoder’s up-sampling technologies, which leads to different segmentation results.
Based on the above-mentioned development of the U-net, we propose a new network. As can be seen in Fig. 2(d), we use residual U-net as the main structure. Then we replace the skip-connection of U-net with shortcut. Also, we use different annotations as deep supervision labels to regularize the network, which enable the encoder to obtain more features and correlations between tumor regions.
Almost all segmentation methods use 2D or 3D CNN architecture. 2D CNNs use 2D image slices to train the network, they use less memory but cannot obtain the spatial context information of image slices. 3D CNNs use voxel to train the network, it use enormous memory but can obtain the spatial context information, thus can improve accuracy of segmentation. Due to the small size of training set, we use 2D CNN architecture in this experiment.
3 Methods
In this paper, our network has a main network and three sub-networks. We use U-net with the residual block as the backbone to perform different brain tumor segmentation tasks, i.e.segmentation of GD-enhancing tumor, the peritumoral edema, or the necrotic and non-enhancing tumor core. And use deep supervision method on sub-networks to realize multi-brunch architecture with different labels. We following introduce the main structure and deep supervision of the network respectively.
3.1 Main Network
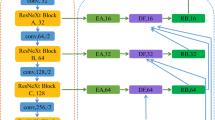
As can be seen in Fig. 3, the input of network includes four slices, which belong to four modalities respectively, so that the network can obtain richer information. The encoder includes input, four down-sampling layers and 2, 2, 2, 3 residual blocks after each down-sampling layer. Among them, each residual block contains two sub-blocks, which are composed of a layer normalization, a relu activation function and a \(3\times 3\) 2D convolutional layer. In the down-sampling stage, we use \(3\times 3\) 2D convolutional layer with strides of 2 for operations. Basically, the input slices first pass through a 2D convolutional layer with relu activation function and a dropout layer with dropout rate of 0.3, then enter followed residual blocks and down-sampling layers successively.
The decoder includes transpose blocks, add blocks and residual blocks. The transpose block is used for up-sampling. It includes a \(1\times 1\) 2D convolution and a \(3\times 3\) deconvolution layer with strides of 2. The \(1\times 1\) 2D convolution is used for dimensionality reduction, and the deconvolution layer is used to restore the size of feature map. The add block includes an add layer and a \(3\times 3\) 2D convolution layer for the fusion of low-dimensional and high-dimensional features. Every time an up-sampling and add operation is over, a residual block will be used for further feature extraction, and there are 3 residual blocks in decoder. At different stages of decoder, we match different labels for the three sub-networks to obtain different types of segmentation results. In this way, the encoder can learn more diverse features. For the main network, we set the corresponding label for the required type of segmentation, so as to we can obtain the corresponding segmentation result.
3.2 Deep Supervision Method
We use the relevance between three types of labels to reduce the mutual misjudgment between multiple tumor regions, thus we can get better segmentation result of a single label. Therefore, for three different labels, we add three additional sub-networks for segmentation. We elicit a branch before each transpose block as decoder of a sub-network. The decoder of the first sub-network includes three transpose blocks, the decoder of the second sub-network includes two transpose blocks, and the third sub-network has only one transpose block. Multiple networks share a same encoder, it can increase the demand for features of encoder and enable the encoder to obtain richer features.
For different segmentation tasks of the main network, the tasks of three sub-networks are variable. For example, when our main network task is to segment enhancing tumor, the label of the first sub-network is necrotic and non-enhancing tumor core, the label of the second sub-network is peritumoral edema, and the label of the third sub-network is GD-enhancing tumor. In this way, the task of each sub-network depends on the segmentation task of main network. We need to choose different solutions according to different tasks.
3.3 Loss Functions
Due to the imbalance between the tumor area and the background in the brain tumor segmentation task, the target tumor area only occupies a small part of the entire slice. So cross-entropy loss function always pay more attention to background area. To solve this problem, we use the combination of binary cross-entropy and dice loss [23] as our loss function.
Among them, dice loss is used to solve the problem of class imbalance, it is expressed as follows:
where y is the ground truth and \(\hat{y}\) is the prediction.
And binary cross-entropy loss function is as follows:
where n is the number of categories, \(y_i\) is the ground truth and \(\hat{y_i}\) is the prediction.
Therefore, the total loss of our network is described as follows:
where \(BCE_{weights}\) is the weight of BCE in the total loss, which is set to 0.5.
4 Experiments
In this section, we introduce our pre-processing method, post-processing method and some experimental details.
4.1 Pre-processing
The data of each patient contains slices of four modalities. On a axial, each modal has 155 slices with a size of \(240\times 240\) pixels. In the process of pre-processing, by dividing by the maximum pixel value of 155 slices, we normalize the pixel values of all slices to 0–1. Then, so as to obtain more training data, we flip the slices and therefore get the same number of data as original.
4.2 Post-processing
In order to reduce the influence of false positive on prediction maps, we identify the relative positions of GD-enhancing tumor, the necrotic and non-enhancing tumor core and peritumoral edema in the segmentation results. Then we delete those pixels that belong to the GD-enhancing tumor but are outside of the peritumoral edema. It can eliminate some theoretically impossible pixels.
4.3 Training Details
We use the Adam optimizer and train for 60 epochs. The initial learning rate is set to \(1e^{-4}\). Then it becomes \(2e^{-5}\) when reach to the 10th epoch. And when the 20th epoch is reached, the learning rate is reduced to \(1e^{-5}\). After 30 epochs, the learning rate remains at \(2e^{-6}\).
5 Results
We train our network on BraTS 2020 training set. After data enhancement, we use 520 cases as the training set, 115 cases as the validation set and 115 cases as the test set. Then we adjust the network parameters through the performance of the test set. After that we make predictions on the validation dataset provided by BraTS 2020, and finally we submit them to the online evaluation platform to evaluate the segmentation results. Finally, the average Dice we get on ET, WT, and TC are 0.7040, 0.8794 and 0.7731. And the median of our Dice scores on ET, WT and TC are 0.8350, 0.9101, and 0.8642 respectively. The performances are shown in Table 1.
The results of test set are presented in Table 2. We can see that the results of the test set are slightly higher than of the validation set.
6 Conclusion
In this paper, we propose a CNN network that uses residual blocks and deep supervision method. There are two key points in this paper. Firstly, the shortcut in the residual blocks can effectively alleviate the problem of gradient dispersion, so that the network can capture high-level visual features. Secondly, deep supervision method helps to improve the training stability of deeper networks, and enables the encoder to obtain richer features. Currently, our method is dedicated to 2D segmentation based on a single axis. However, due to the lack of spatial information in 2D data, it cannot effectively segment brain tumors with spatial correlation. So in the future work, we will apply the above method to 3D networks to improve segmentation performance.
References
McKinley, R., Rebsamen, M., Meier, R., Wiest, R.: Triplanar ensemble of 3D-to-2D CNNs with label-uncertainty for brain tumor segmentation. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 379–387. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_36
Zhao, Y.-X., Zhang, Y.-M., Liu, C.-L.: Bag of tricks for 3D MRI brain tumor segmentation. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 210–220. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_20
Zhou, C., Chen, S., Ding, C., Tao, D.: Learning contexualand attentive information for brain tumor segmentation. In: Pre-conference Proceedings of the 2018 7th MICCAI BraTS Challenge, pp. 571–578 (2018)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J.S., et al.: Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Nat. Sci. Data 4, 170117 (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629 (2018)
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017). https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive (2017). https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maierhein, K.H.: Brain tumor segmentation and radiomics survival prediction: Contribution to the BRATS 2017 challenge. In: 2017 Proceedings of the 6th MICCAI BraTS Challenge, pp. 100–107 (2017)
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maierhein, K.H.: No new-net. In: 2018 Pre-conference Proceedings of the 7th MICCAI BraTS Challenge, pp. 222–231 (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Zhou, Z.W., Siddiquee, M.M.R., Tajbakhsh, N., Liang, J.M.: UNet++: a nested U-Net architecture for medical image segmentation. In: Deep Learning in Medical Image Anylysis and Multimodal Learning for Clinical Decision Support, pp. 3–11 (2018)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: 2017 Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, pp. 2261–2269 (2017)
Zhang, X., Jian, W., Cheng, K.: 3D dense U-nets for brain tumor segmentation. In: 2018 Pre-conference Proceedings of the 7th MICCAI BraTS Challenge, pp. 562–570 (2018)
Stawiaski, J.: Leveraging a DenseNet encoder pre-trained on ImageNet for brain tumor segmentation. In: 2018 Pre-conference Proceedings of the 7th MICCAI BraTS Challenge, pp. 438–447 (2018)
Mckinley, R., Meier, R., Wiest, R.: Ensembles of densely-connected CNNs with label-uncertainty for brain tumor segmentation. In: 2018 Pre-conference Proceedings 7th MICCAI BraTS Challenge, pp. 322–330 (2018)
Myronenko, A.: 3D MRI brain tumor segmentation using autoencoder regularization. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 311–320. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_28
Weninger, L., Liu, Q., Merhof, D.: Multi-task learning for brain tumor segmentation. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 327–337. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_31
Zhou, C., Ding, C., Lu, Z., Wang, X., Tao, D.: One-pass multi-task convolutional neural networks for efficient brain tumor segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11072, pp. 637–645. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00931-1_73
Chen, S., Bortsova, G., García-Uceda Juárez, A., van Tulder, G., de Bruijne, M.: Multi-task attention-based semi-supervised learning for medical image segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 457–465. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_51
Jiang, Z., Ding, C., Liu, M., Tao, D.: Two-stage cascaded U-Net: 1st place solution to BraTS challenge 2019 segmentation task. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 231–241. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_22
Milletari, F., Navab, N., Ahmadi, S.-A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 4th International Conference on 3D Vision (3DV). IEEE (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Ma, S., Zhang, Z., Ding, J., Li, X., Tang, J., Guo, F. (2021). A Deep Supervision CNN Network for Brain Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12659. Springer, Cham. https://doi.org/10.1007/978-3-030-72087-2_14
Download citation
DOI: https://doi.org/10.1007/978-3-030-72087-2_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72086-5
Online ISBN: 978-3-030-72087-2
eBook Packages: Computer ScienceComputer Science (R0)