Abstract
Machine learning (ML) integrated with medical imaging has introduced new perspectives in precision diagnostics of GBM tumors, through radiomics and radiogenomics. This has raised hopes for developing non-invasive and in-vivo biomarkers for prediction of patient survival, tumor recurrence, or molecular characterization, and therefore, encouraging treatments tailored to individualized needs. Characterization of tumor infiltration based on pre-operative multi-parametric magnetic resonance imaging (MP-MRI) scans would help in predicting the loci of future tumor recurrence, and thereby aiding in planning the course of treatment for the patients, such as increasing the resection or escalating the dose of radiation. Specifying molecular properties of GBM tumors and prediction of their changes over time and with treatment would help characterize the molecular heterogeneity of a tumor, and potentially use a respective combination treatment. In this article, we will provide examples of our work on radiomics and radiogenomics, aiming to offer personalized treatments to patients with GBM tumors.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glioblastoma (GBM) is the most common and fatal primary brain tumor. The current standard of care for treatment of patients with GBM tumors involves maximal safe tumor resection followed by radiotherapy and adjuvant temozolomide (TMZ) chemotherapy, and maintenance TMZ therapy for 6–12 months. The standard treatment at the very best provides the patients with a median progression-free survival (PFS) of 6.2–7.5 months and an overall survival (OS) of around 14.6–16.7 months [1, 2]. The patients who tolerate TMZ treatment and do not show tumor progression, can be prescribed to receive tumor-treating fields (TTFields), improving the prognosis only to a median OS of 20.9 months [1, 2]. One of the main reasons for the failure of treatments in GBM patients is marked intra-tumor heterogeneity of GBM tumors, diffuse and immense infiltration of tumor cells in the adjacent brain parenchyma which mainly remain untreated, and resistance of tumor subpopulations to the given therapies.
Medical imaging, and specifically magnetic resonance imaging (MRI), has evolved into an indispensable diagnostic tool in neuro-oncology. It can contribute to personalized patient management by offering patient prognosis, treatment guidance, and monitoring the response of a patient to the therapy, based on specific characteristics of the tumor, manifested with different phenotypes on MRI scans [3]. A mounting body of literature over the past decade has shown that subvisual aspects of GBM tumor heterogeneity can be captured by integrating characteristics that relate to cellular density, neo-angiogenesis, water content, etc. from multi-parametric MRI (MP-MRI) scans [4, 5].
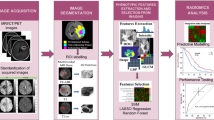
Radiomics is an emerging computational method that combines diverse imaging features through machine learning (ML) modeling into distinctive imaging signatures [6,7,8]. It can reveal patterns underlying the tumor’s progression, response to standard, adjuvant, or novel therapies, and can help to achieve a more personalized medicine for the GBM patients. Imaging phenotypes discovered by radiomics have shown promise in risk stratification, prediction of overall and progression-free survival, disease follow-up (discrimination of true vs pseudo-progression of the disease), characterization of tumor genomics [9], and upfront prediction of the response to treatment.
In this article, we will briefly review the proposed techniques for characterization, prognostication, and treatment planning of the patients with GBM.
2 Patient Prognosis
Upfront prediction of PFS and OS could potentially identify high-risk patients, who are suspected to have a short OS, e.g. of less than 6 months, and enroll them to alternative therapies or palliative care, depending on personal choices. While OS is a primary endpoint for determination of the efficacy of treatment strategies in clinical trials, PFS could serve as a surrogate for OS to overcome several limitations of OS, such as long trial times [10]. Moreover, as patients with GBM have a poor prognosis, with PFS as an early marker of OS, the course of the prescribed treatments for the patients can be modified or changed. In this regard, radiomics has shown potential in stratification of high- and low-risk patients, and prediction of OS or PFS.
In our 2016 study, quantitative imaging features were extracted from pre-operative MP-MRI scans, including pre- and post-contrast T1-weighted images (T1, T1-Gd), T2-weighted (T2), T2 fluid attenuated inversion recovery (FLAIR), dynamic susceptibility contrast-enhanced (DSC)-MRI, and diffusion tensor imaging (DTI). Features of intensity from the MP-MRI scans, volume, location, and growth model parameters were combined by support vector machine (SVM) classification algorithm for stratification of patients with GBM tumors into groups of short, medium, and short survivors. The results suggested that an overall 3-way classification into short/medium/long survivor groups was around 79% in the prospective cohort [11].
As DSC-MRI and DTI sequences are not frequently included with the routine pre-surgical brain tumor protocols at many imaging centers, in a later study, we investigated the accuracy of a predictive model built based on augmented radiomics feature panel (ARFP), including morphology and textural descriptors, extracted only from basic MP-MRI (Bas-mpMRI, comprising of T1, T1-Gd, T2, FLAIR) for stratification of OS risk groups [12]. This predictive model, generated with ARFP from Bas-mpMRI yielded a comparable accuracy to the previous model [11] that was generated with advanced MP- MRI (Adv-mpMRI) scans without using ARFP [12]. It was proposed that Bas-mpMRI and advanced radiomics features can compensate for the lack of Adv-mpMRI [12]. This approach also showed generalizability across different scanners in a multi-center study [13].
In a recent multi-center study, we investigated the role of radiomics in prediction of PFS based on MP-MRI scans, acquired prior to the primary surgery in patients with GBM [14, 15]. A prognostic model was generated with a rich panel of quantitative features of intensity, first-order histogram, texture, morphology, and volume, through an SVM classifier, resulting an AUC = 0.82 for the data from two institutions. This radiomics study was carried out using the publicly available and open-source Cancer Imaging Phenomics Toolkit (CaPTk) software [14].
A few studies have reported the added value of radiomics models to clinical and molecular predictors for risk stratification of patients with GBM tumors. A radiomics study of GBM patients showed a Concordance index (C-index) = 0.696 for prediction of OS by integrating radiomics and clinical variables, i.e. age and Karnofsky performance score (KPS), compared to a C-index = 0.640 with only clinical variables [16]. Similarly, on a larger cohort of patients, it was suggested that a combination of key clinical characteristics, i.e. age, extent of resection, and KPS, with molecular diagnosis, i.e. MGMT methylation status, yielding integrated Brier scores (IBS) of 0.119 and 0.098 for prediction of OS and PFS, respectively, improved to 0.103 and 0.0809 when integrated with radiomics variables. For prediction of PFS, [17]. Another study investigated the improvement of survival prediction using radiomics integrated with clinical and molecular profiles [18]. They found an improvement of risk stratification of GBM patients into low and high survivor groups when radiomics was combined with clinical and molecular variables, denoting an area under the curve (AUC) = 0.78 compared to an AUC = 0.70 for a model based only on clinical and molecular variables [18].
3 Intratumor Heterogeneity and Tumor Recurrence
A hallmark characteristic of GBM is diffuse infiltration into the surrounding brain tissue, extending beyond the hyperintense regions visible on T1-Gd MRI scans into the peritumoral edema, and leading to tumor recurrence. Mapping peritumoral infiltration would augment precision treatment through escalating the radiation therapy dose in densely infiltrated regions, and potentially prolonging survival of the patients [11]. Pattern analysis of MP-MRI scans can reveal infiltration of tumor cells in the peritumoral edema by quantification of spatial heterogeneity in terms of the changes in regional microvasculature, microstructure, and water content [4]. We have developed an imaging signature of tumor infiltration that serves as an early biomarker of the likely location of tumor recurrence [19]. Quantitative features estimated from MP-MRI scans (T1, T1-Gd, T2, FLAIR, DTI, and DSC-MRI) within peritumoral edema were combined using an ML approach to generate predictive maps of tumor recurrence, with an odds ratio of 9.29, AUC of 0.84, sensitivity of 91%, and specificity of 93%. Figure 1 shows examples of infiltration maps generated using pre-operative MP-MRI scans of two patients with GBM tumors and the location of tumor recurrence in the patients.
An illustration of two examples of generating infiltration maps from pre-operative MP-MRI scans. The images on the right side indicate the corresponding recurrence scans of the same patients. As it can be inferred from the images, the predicted infiltration maps are highly predictive of future recurrence.
A common dilemma in evaluating the response of GBM tumors to therapy is differentiation of progressive disease, or true progression (TP), from pseudo-progression (PsP). Radiomics signatures distinguishing between TP and PsP in patients with GBM tumors have been reported including a recent study of our group, on a cohort of GBM patients who underwent second resection due to progressive radiographic changes suspicious for recurrence [20]. A multivariate analysis of deep learning and conventional features from multi-parametric MRI scans was performed, showing an accuracy of 87% for predicting PsP and 84% for predicting TP that compared with similar categories blindly defined by board‐certified neuropathologists [20]. In another recent study on a cohort of patients with GBM tumors, a feature learning method based on deep convolutional generative adversarial networks (DCGAN) and AlexNet was implemented to discriminate between PsP and TP, which showed a performance of AUC > 0.90 [21].
4 Radiogenomics
Advances in genomic profiling of tissue specimens in a variety of diseases, especially cancers, has encouraged development of treatments targeted at the genetic makeup of the tumor and paved the way towards personalized treatments and precision medicine [22]. However, several factors, including tumor heterogeneity, sampling error during biopsy, insufficient tissue quality for sequencing, limitations of the sequencing methods, etc., may hinder characterization of tumor genomics. Radiographic imaging phenotypes have shown strong associations with the underlying biology of GBM tumors. Through an ML approach, radiogenomics studies aim to bridge the gap between the two disciplines by generating imaging signatures that represent genetic characteristics or heterogeneity of the tumor. Thereby, radiogenomics signatures can serve as noninvasive biomarkers for tumor genomics or as complementary data for predicting patient prognosis [9].
In our radiogenomics study for generating a signature of EGFRvIII mutation in GBM tumors, we integrated quantitative features derived from MP-MRI through an ML approach that predicted the EGFRvIII mutation status with an accuracy of 87% in a replication cohort [23]. The results suggested that the tumors with EGFRvIII mutations had a propensity to occur in the frontal and temporal regions of the brain, and were associated with higher neovascularization and cell density compared to the wildtype tumors [23]. Figure 2 shows the descriptive characteristics of GBM tumors with EGFRvIII mutation. We further found that heterogeneity in hemodynamic patterns within peritumoral edema, quantified by pattern analysis of perfusion MRI scans, is strongly linked to EGFRvIII mutation status [24]. EGFRvIII mutant tumors displayed a highly infiltrative-migratory phenotype while the wildtype tumors had a confined vascularization within their peritumoral area [24].
This image illustrates the characteristics of GBM tumors with mutation in EGFRvIII (EGFRvIII(+)). As indicated, EGFRvIII(+) patients have a fronto-parietal propensity, and show increased neovascularization and cell density [23].
In a study on exploring the synergies between imaging and genomics, we identified three distinct and reproducible imaging subtypes, including rim-enhancing, irregular, and solid, which exhibit clearly different clinical outcome and molecular characteristics, including IDH1, MGMT methylation, EGFRvIII, and transcriptomic molecular subtypes, i.e. classical, mesenchymal, neural, proneural [25]. Our findings signify the importance of precision diagnostics and personalized therapies for patients with GBM tumors.
There are many other notable radiogenomics studies of GBM tumors, which overviewing them is out of scope of this paper. We refer the interested readers to the relevant review papers [9, 22, 26,27,28].
5 Current Challenges and Future Directions
Despite the promises that radiomics and radiogenomics offer for achieving precision diagnostics in management of GBM tumors, challenges of reproducibility and generalizability of the proposed methods have yet to be tackled for these methods to be translated into clinical applications. First, radiomics is not directly related to biological characteristics, and reproducibility of the features mainly depends on the imaging process, from acquisition, to post-processing and feature extraction. Most radiomics studies are retrospective, therefore, not all aspects of reproducibility such as standardization of image acquisition protocol, intra-patient test-retest repeatability, and across scanner reproducibility can be addressed [29]. This issue complicates generalizability of the radiomics models across different institutions. The clinical trials follow standardized image acquisition guidelines, although the number of data is usually limited and data sharing is restricted due to ownership concerns [9].
Another impedance to reproducible radiomics research is variability in analysis methods, i.e. different results can be achieved for the same data depending on the feature extraction, feature selection and modeling approaches. Lack of clear and comprehensive description of research methodology, including data processing and parametrization of the computational algorithms, and unwillingness or hesitance of the researchers to share their methods with the community mainly due to an understandable reason of intellectual property, further poses a challenge to research reproducibility [9]. In this regard, multiple open-source software toolkits have been developed and made publicly available. To this end, Cancer Imaging Phenomics Toolkit (CaPTk), an imaging analytics suite of open-source software algorithms, has been implemented to facilitate estimation of extensive panels of quantitative imaging phenomics (CIPh) features and integrating them with prognostic models to support a course of tailored treatment strategies for the patients [30]. CaPTk has a three-level functionality design for radiomics analysis. The users can build their image preprocessing pipelines, composed of algorithms for conversion of image format, registration of the scans, segmentation of the desired regions, artifact reduction, and intensity normalization with the tools provided in the first-level functionality. The second level provides general-purpose applications, including extensive set of radiomics features with adjustable parameters for feature extraction, feature selection, and ML model construction. Numerous features representing morphology, volume, intensity, and regional heterogeneity, compliant with the guidelines provided by the Image Biomarker Standardization Initiative (IBSI) [31] to ensure reproducibility and comparability, can be extracted. In the third level, the methods in the first level features have been synthesized into a smaller and meaningful subset of features, combined with second level ML algorithms for specialized applications that aim to support specific clinical applications, including risk stratification of patients, stratification of patients according to their transcriptomic molecular subtypes, predicting genomics of the tumor, etc. [30].
As any other ML approach, radiomics studies require ample and diverse data for learning the underlying patterns of the disease, and to overcome the so-called “curse of dimensionality” problem, i.e. a remarkably larger number of features compared to the number of samples. Clinical data collected at a single institution is usually limited in the number and diversity, thereby, hampering generalization of the ML methods [9]. These challenges motivated the formation of ReSPOND (Radiomics Signatures for PrecisiON Diagnostics) consortium, as an international initiative for machine learning in GBM imaging [32]. ReSPOND is a collaborative effort of over 10 international institutions that aims to gather brain MRI scans from over 3000 de novo GBM patients and develop radiomics biomarkers for personalized prognostication. The main four areas of focus for ReSPOND include prediction of OS and PFS, early prediction of tumor recurrence to help in adopting aggressive treatments of GBMs through an extended resection and escalation of the dose within the peritumoral regions that are suspected of recurrence, differentiation of true tumor progression from pseudo-progression, and prediction of molecular characteristics of the GBM tumors [32, 33].
6 Conclusion
Artificial intelligence, in the forms of radiomics and radiogenomics, has introduced appealing solutions to the current clinical problems for management of GBM tumors and has raised the hopes for accomplishing the purpose of tailoring diagnosis and treatments for the patients at an individual level. Risk stratification of the GBM patients by upfront projection of their OS and PFS, early prediction of tumor recurrence, distinguishing TP from PsP, and prediction of the molecular properties of the tumor and the spatial heterogeneity are among the key applications of radiomics and radiogenomics. Nonetheless, these promising tools face the challenges of reproducibility and generalizability that need to be carefully addressed by the community.
References
Stupp, R., Taillibert, S., Kanner, A.A., et al.: Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma a randomized clinical trial. JAMA – J. Am. Med. Assoc. 314, 2535–2543 (2015). https://doi.org/10.1001/jama.2015.16669
Stupp, R., Taillibert, S., Kanner, A., et al.: Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma a randomized clinical trial. JAMA – J. Am. Med. Assoc. 318, 2306–2316 (2017). https://doi.org/10.1001/jama.2017.18718
Davatzikos, C., Sotiras, A., Fan, Y., et al.: Precision diagnostics based on machine learning-derived imaging. Magn. Reson. Imaging 64, 49–61 (2019). https://doi.org/10.1016/j.mri.2019.04.012
Kazerooni, A.F., Nabil, M., Zadeh, M.Z., et al.: Characterization of active and infiltrative tumorous subregions from normal tissue in brain gliomas using multiparametric MRI. J. Magn. Reson. Imaging 48, 938–950 (2018). https://doi.org/10.1002/jmri.25963
Fathi Kazerooni, A., Mohseni, M., Rezaei, S., Bakhshandehpour, G., Saligheh Rad, H.: Multi-parametric (ADC/PWI/T2-w) image fusion approach for accurate semi-automatic segmentation of tumorous regions in glioblastoma multiforme. Magn. Reson. Mater. Phys., Biol. Med. 28(1), 13–22 (2014). https://doi.org/10.1007/s10334-014-0442-7
Gillies, R.J., Kinahan, P.E., Hricak, H.: Radiomics: images are more than pictures, they are data. Radiology 278, 563–577 (2016). https://doi.org/10.1148/radiol.2015151169
Kumar, V., Gu, Y., Basu, S., et al.: Radiomics: the process and the challenges. Magn. Reson. Imaging 30, 1234–1248 (2012). https://doi.org/10.1016/j.mri.2012.06.010
Gatenby, R.A., Grove, O., Gillies, R.J.: Quantitative imaging in cancer evolution and ecology. Radiology 269, 8–15 (2013). https://doi.org/10.1148/radiol.13122697
Fathi Kazerooni, A., Bakas, S., Saligheh Rad, H., Davatzikos, C.: Imaging signatures of glioblastoma molecular characteristics: a radiogenomics review. J. Magn. Reson. Imaging 52, 54–69 (2019). https://doi.org/10.1002/jmri.26907
Han, K., Ren, M., Wick, W., et al.: Progression-free survival as a surrogate endpoint for overall survival in glioblastoma: a literature-based meta-analysis from 91 trials. Neuro Oncol. 16, 696–706 (2014). https://doi.org/10.1093/neuonc/not236
Macyszyn, L., Akbari, H., Pisapia, J.M., et al.: Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol. 18, 417–425 (2016). https://doi.org/10.1093/neuonc/nov127
Bakas, S., Shukla, G., Akbari, H., Erus, G.: Overall survival prediction in glioblastoma patients using structural magnetic resonance imaging (MRI): advanced radiomic features may compensate for lack of advanced MRI modalities. J. Med. Imaging 7, 1–18 (2020). https://doi.org/10.1117/1.JMI.7.3.031505
Bakas, S., Akbari, H., Shukla, G., et al.: Deriving stable multi-parametric MRI radiomic signatures in the presence of inter-scanner variations: survival prediction of glioblastoma via imaging pattern analysis and machine learning techniques, p. 1057509:8 (2018). https://doi.org/10.1117/12.2293661
Fathi Kazerooni, A., Akbari, H., Shukla, G., et al.: Cancer imaging phenomics via CaPTk: multi-institutional prediction of progression-free survival and pattern of recurrence in glioblastoma. JCO Clin. Cancer Inform. 234–244 (2020). https://doi.org/10.1200/cci.19.00121
Fathi Kazerooni, A., et al.: NIMG-35. Quantitative estimation of progression-free survival based on radiomics analysis of preoperative multi-parametric MRI in patients with glioblastoma. Neuro Oncol. 21(Suppl_6), vi168–vi169 (2019). https://doi.org/10.1093/neuonc/noz175.705
Kickingereder, P., Burth, S., Wick, A., et al.: Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 280, 880–889 (2016). https://doi.org/10.1148/radiol.2016160845
Kickingereder, P., Neuberger, U., Bonekamp, D., et al.: Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol 20, 848–857 (2018). https://doi.org/10.1093/neuonc/nox188
Bae, S., Choi, Y.S., Ahn, S.S., et al.: Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology 289, 797–806 (2018). https://doi.org/10.1148/radiol.2018180200
Akbari, H., Macyszyn, L., Da, X., et al.: Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 78, 572–580 (2016). https://doi.org/10.1227/NEU.0000000000001202
Akbari, H., Rathore, S., Bakas, S., et al.: Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer 126, 2625–2636 (2020). https://doi.org/10.1002/cncr.32790
Li, M., Tang, H., Chan, M.D., et al.: DC-AL GAN: pseudoprogression and true tumor progression of glioblastoma multiform image classification based on DCGAN and AlexNet. Med. Phys. 47, 1139–1150 (2020). https://doi.org/10.1002/mp.14003
Pinker, K., Shitano, F., Sala, E., et al.: Background, current role, and potential applications of radiogenomics. J. Magn. Reson. Imaging 47, 604–620 (2018)
Akbari, H., Bakas, S., Pisapia, J.M., et al.: In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro Oncol. 20, 1068–1079 (2018). https://doi.org/10.1093/neuonc/noy033
Bakas, S., Akbari, H., Pisapia, J., et al.: In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index. Clin. Cancer Res. 23, 4724–4734 (2017)
Rathore, S., Akbari, H., Rozycki, M., et al.: Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1. Sci. Rep. 8, 1–2 (2018)
Zinn, P.O., Mahmood, Z., Elbanan, M.G., Colen, R.R.: Imaging genomics in gliomas. Cancer J. 21, 225–234 (2015)
Smits, M., van den Bent, M.J.: Imaging correlates of adult glioma genotypes. Radiology 284, 316–331 (2017)
Ellingson, B.M.: Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr. Neurol. Neurosci. Rep. 15, 506 (2015)
Park, J.E., Kickingereder, P., Kim, H.S.: Radiomics and deep learning from research to clinical workflow: neuro-oncologic imaging. Korean J. Radiol. 21, 1126–1137 (2020). https://doi.org/10.3348/kjr.2019.0847
Davatzikos, C., et al.: Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 5(01), 1 (2018). https://doi.org/10.1117/1.JMI.5.1.011018
Zwanenburg, A., Vallières, M., Abdalah, M.A., et al.: The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295, 328–338 (2020)
Davatzikos, C., Barnholtz-Sloan, J.S., Bakas, S., et al.: AI-based prognostic imaging biomarkers for precision neuro-oncology: the ReSPOND consortium. Neuro Oncol. 22, 886–888 (2020). https://doi.org/10.1093/neuonc/noaa045
Davatzikos, C., et al.: NIMG-66. AI-based prognostic imaging biomarkers for precision neurooncology and the respond consortium. Neuro Oncol. 22(Suppl_2), ii162–ii163 (2020). https://doi.org/10.1093/neuonc/noaa215.679
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Kazerooni, A.F., Davatzikos, C. (2021). Computational Diagnostics of GBM Tumors in the Era of Radiomics and Radiogenomics. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12658. Springer, Cham. https://doi.org/10.1007/978-3-030-72084-1_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-72084-1_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72083-4
Online ISBN: 978-3-030-72084-1
eBook Packages: Computer ScienceComputer Science (R0)