Abstract
Semantic segmentation plays an essential role in brain tumor diagnosis and treatment planning. Yet, manual segmentation is a time-consuming task. That fact leads to hire the Deep Neural Networks to segment brain tumor. In this work, we proposed a variety of 3D U-Net, which can achieve comparable segmentation accuracy with less graphic memory cost. To be more specific, our model employs a modified attention block to refine the feature map representation along the skip-connection bridge, which consists of parallelly connected spatial and channel attention blocks. Dice coefficients for enhancing tumor, whole tumor, and tumor core reached 0.752, 0.879 and 0.779 respectively on the BRATS- 2020 valid dataset.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Comprising about 30% of all intracranial tumors, gliomas are one of the most common type of intracranial tumor with a highly variable clinical prognosis, and only one-fifth of gliomas are benign. Gliomas could lead to various symptoms, such as headaches, vomiting, seizures, and cranial nerve disorders. According to WHO’s classification, the gliomas have four grades. Grade I and II are the Low-Grade Gliomas, namely LGG, which bring lower threatens to patients. Likewise, Grade III and IV are the High-Grade Gliomas, namely HGG, which bring higher threat. Gliomas could be divided into several components: the enhancing tumor (ET), tumor core (TC), and the whole tumor (WT). The TC subregion describes the bulk of the tumor and is usually removed. The TC subregion entails the ET, along with the fluid-filled and the solid parts of the tumor. The WT subregion describes the whole extent of gliomas.
Magnetic Resonance Imaging (MRI) is widely used in clinical diagnosis and it is an effective method to portray the inner heterogeneity of gliomas using different radiographic phenotypes. Based on their features, distinct images and different appearance of certain subjects could be obtained easily by changing the sequence of MRI scanning, and this makes it possible to depict valuable images of subregions of gliomas with different modalities. To employ MRI scans to segment gliomas, it is a critical procedure for its therapy. With gliomas’ high heterogeneity in different tumor appearances and shapes, there are many challenging tasks in the diagnosis.
Brain Tumor Segmentation Challenge (BRATS) is organized for years, focusing on finding state-of-the-art methods handling brain tumors in multi-parametric MR scans with computer technologies [1,2,3,4,5]. BRATS-2020 provides training datasets consist of both LGG and HGG MR scans, containing 60 subjects and 309 subjects respectively, and a valid set consists of 125 subjects. Each subject has four 3D brain sequences data in Nifty (nii.gz) format with segmentation masks for all subregions as well. The four modalities are structural (T1) images, T1-weighted contrast-enhanced (T1ce) images, T2-weighted images and fluid-attenuated inversion recovery (Flair). In the BRATS challenge, MR scans originate from 19 institutions, and are all annotated manually. One of the main tasks in the BRATS challenge is to segment brain tumors of different subjects into sub-components using MR scans, as it plays an essential role in diagnosis treatment planning.
Popular methods of brain tumor segmentation can be classified into either generative or discriminative models [4]. With Deep Neural Network (DNN) technology developing, it seems the most popular method during the last few BraTS challenges. Based on the Convolution Neural Networks (CNN), different models are proposed and various theories are extended. Many of the CNN give the state-of-the-art performance in the semantic segmentation domain, such as VGG [5], FCN [6], DeepMedic [7], U-net [8], etc. Among all these methods, U-net is the choice of the majority to handle medical image analysis issues, due to its fitness towards medical images. Provided with sufficient data, U-net will learn to generalize to the unseen type of gliomas. Thus, there came out of lots of variety of U-net.
Section 2 involves the details about our model, including the backbone architecture, the dual pathway attention gate, the res block and the loss function we choose. Section 3 shows the result on train, valid and test set, along with the training strategy and the metrics evaluating the proposed model. Comparison between different performances of different models is also given in Sect. 3. Section 4 mainly involves the analysis and discussion of result on the proposed model.
In this work, we combine the 3D U-net with a Dual Pathway Attention (DPA) inspired by 3D Convolutional Block Attention Module (CBAM) [9] and use the residual module [10], for brain tumor segmentation from patches of MR images, as an extension of basic 3D U-net. Contribution of this work is presented as follow:
-
Based on the U-shaped structure of the U-net, an additional dual residual pathway is added to its encoder layers to enhancing transmission of the high-level features during down-sampling.
-
Meanwhile, a variety of attentions block is employed to skip-connection to weight the feature map conveying from down-sample block to up-sample. The attention block is based on the idea of CBAM, which is composed of spatial attention block and channel attention block, we further refined it by replacing the spatial attention block with the original attention block to conserve precious graphic memory instead of 7 × 7 big kernel.
2 Method
2.1 Backbone Architecture for Segmentation
Due to the high performance of the U-net in medical image segmentation, we use the 3D variant of it as the backbone of our proposed network, which takes four modalities of brain tumor MR scans along with the ground truth segmentation as input, and train them to segment subregions like WT, TC and ET simultaneously with a single network.
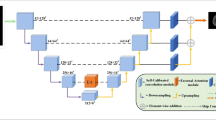
The proposed architecture in this work is shown in Fig. 1. As mentioned above, proposed network is a variant of 3D U-net. Based on its encoder-decoder structure with skip-connection, we improved its performance mainly by two methods: ① modified residual pathway along the down-sample layers, called dual-path residual module; ② a modified attention module. As it is shown in Fig. 1 (a), our net basically encompasses five down sample layers and four up-sample layers, namely Dual Pathway Attention (DPA) block.
2.2 Dual Residual Block
Inspired by the MFnet [11] and residual net, the residual module is modified into two pathways, each transmits a different part of the feature map along down-sampling layers. The structure of the dual-path residual module is shown in Fig. 1 (b), an extra pathway is added after the first convolution to reserve low-level features which contain more spatial information to locate the Region of Interest. Besides, this structure help solve the degradation problem that would occur to deep neural networks as well. Exerting this module to up sampling module would cause out of memory error, so the experiment goes only on down-sample layers for now. We plan to add it to up sample layers after the network is optimized and consumption of memory is reduced.
Each down-sample layer contains a max-pooling layer, a Rectified Linear Unit (ReLU) module and three basic 3D convolution layers followed by group normalization [12], additionally dual-path residual module is added to its encoder in order to ease the degradation problem for deep networks.
Each up-sample layer contains a basic trilinear up sample, two convolution layers to compute feature map from last down sample layer and its combination with skip-connection, followed by a ReLU layer.
2.3 Dual-Path Attention (DPA) Block for Tumor Segmentation
Embedding attention block into U-net is not a new idea, and it works fine according to former works [13,14,15]. Realizing that attention will help recalibration of feature maps, a modified attention block is introduced to skip connection. Though skip-connection bridges between high-level feature and low-level feature by concatenation, low-level information could work less effectively. To deal with that, the Dual-Path attention module is employed to help skip connections focus on the relevant regions and features instead of just sum them up.
Figure 2 shows that, there are mainly two parts of the dual pathway attention block: the spatial attention module and the channel attention module. Two modules connect parallelly, with an additional residual pathway to transmit the original feature map as the base bone of the channel-wise and spatial weights.
Channel Attention Module.
Inspired by the idea of squeeze-and-Excitation (SE) network [16], a small channel-wise attention module is employed to extract the hidden features along the channel. The SE module is a bottleneck attention gate variety, comprised of three main steps as could be told from its name: squeeze, excitation, and scale. Channel attention pathway in this work is implemented as a bottleneck attention gate, using 3D max-pooling and average-pooling together to shrink the size of input feature map finally into 1, and the outputs of each pooling layer are followed by a bottleneck consist of two convolution layers to learn the relation between different channels and find out which feature maps are relevant. Different from the original SE module, two kinds of pool strategies are used at the same time instead of using a single one. In the CBAM module, the two pool layers shared the same bottleneck to take the average result of two different pool strategies, but in this work separated bottlenecks to deal with the outputs of two pool layers instead. We believe that this improvement makes results more robust than the original implementation. The channel attention outputs the computed weight of each input channel, and the channel weights are used to measure the feature information each channel contains. After getting the channel weight, the model is supposed to focus on the channels containing more features and improve the final segment result.
Spatial Attention Module.
As a classic module, the attention gate is popular among all computer vision domains. Employing attention module to the network could provide automatically focus mechanism towards target regions. U-net uses skip connection to fuse high-level features and low-level features. Generally, low-level features contain more spatial information that helps network locate the target area. Simply conjoin low-level features and high-level features together would not put them into fully utilization. The spatial attention module in front of concatenation will provide improvement. The spatial attention module we employ is a variety of original attention gate, it takes both low-level features from skip-connection which has been reweighted by channel attention module, and high-level features from up sample layers as input, outputs computed weight of conducted feature maps to reweight the spatial dimension to achieve the goal of focus.
The Channel Attention Module and the Spatial Attention module are basically parallelly connected in our model. First, the input of whole attention module is accepted by the channel attention gate and a residual pathway to reserve its original feature maps. As mentioned above, channel attention module computes the weights of each channel by a bottleneck structure and connects the weights with the original feature maps to reweight them. And the spatial attention gate outputs the computed weight of a single MRI slice or feature map to make sure model focus on the most informed regions of each feature map. Contacting the outputs of two attention gates, the model is supposed to focus on the most informed channels and regions of the current layer that are well trained.
2.4 Loss Function
A negative patch is a patch that centers on the non-tumor region, which tends to raise the possibility of false positive. Likewise, having too much positive patch do the opposite. Thus, both negative and positive patches would be needed to balance the model. As randomly cropped original data to 128 × 128 × 128 patches as our input, a large amount of input patches is found containing no area of interest, this leads training to an imbalance issue. To moderating the imbalance issue between the negative and positive patches, we employ combined multi-class dice loss as the loss function.
Here in the first equation, \({L}_{dice}\) represents multi-class dice loss, \({L}_{ce}\) represents cross- entropy loss, \({\lambda }_{dice}\) and \({\lambda }_{ce}\) their weight respectively.
3 Experiments
3.1 Datasets Detail
The datasets used in this experiment are provided by the BraTS 2020 Challenge. The training dataset contains 369 subjects in total, and each subject consists of four different modalities of MR scans, respectively T1, T2, T1-enhanced (T1C), T2 with Fluid-Attenuated Inversion Recovery (FLAIR), and the segmentation mask segmented manually. The size of each MR image is 240 × 240 × 150. And the valid data include 125 subjects and share the same format with training data despite it does not include ground truth. In the experiment, training datasets is used to train and optimize our model to its best performance, and evaluate it using valid data on the BraTS 2020 official portal.
3.2 Preprocessing Methods
The model takes all four modalities as input, each modality is treated as a single channel of the model, that makes the model input channel as 4. Due to the distribution imbalance of values of different modalities, normalization such as z-score should be exerted on the original data before we feed it into networks. Also, we pack the data in sequence into the middle format in order to save data-reading time and resources.
Comparing to the 2D version, 3D U-Net consumes much more memory. In order to run 3D U-net on single 1080Ti with 11GB memory, we randomly crop the original MR images in sizes of 240 × 240 × 155 into 128 × 128 × 128 patches and feed it into our model.
3.3 Evaluating Metrics
Three metrics are used to evaluate the submissions of different models in the segmentation task:
Dice Coefficient Score.
The dice coefficient score is employed to evaluate the similarity on the area of intersections between two graphics, here it refers to the similarity between the ground truth segment mask and the prediction segment mask. It receives popularity in medical image segmentation domains.
Hausdorff Distance.
Differ with the Dice coefficient, Hausdorff distance measures the similarity by computing the distance between two different subsets. Due to its nature, Hausdorff distance is highly sensitive to the error of the graphic border. BraTS use 95% Hausdorff distance as the criteria, which is a variety of Hausdorff distance based on 95% of the distances between subsets to eliminate the influence of a small subset of outliers.
Sensitivity and Specificity.
Sensitivity measures the proportion of true positives, and specificity measures the proportion of true negatives.
3.4 Experimental Results
Proposed network is build using PyTorch, trained on 1080Ti GPU with 11G RAM with BraTS2020 train set. As putting voxel into GPU could consume a lot of memory, batch size is set only to 2. The initial learning rate is set to 0.01, using SGD optimizer with momentum set to 0.80, batch normalization is the Group Batch Norm.
Table 1 shows the average results of different models on BraTS 2020 valid data, including mean dice accuracies and mean hausdorff_95. Our model achieves to outperformance original 3D U-net or 3D U-net with single Attention or with Residual module. The results show that our model raises the dice coefficient score of ET and TC by 1% and 3% respectively, thus we think our model achieves a relatively good improvement on BraTS segmentation task. Trained on the same dataset with same hyper-parameters, our model still outperforms the best of its baseline.
Table 2 show the detailed statistics result of our model, evaluated on BraTS 2020 valid dataset on BraTS official portal. The standard deviation is 0.282, 0.112 and 0.199 for dice coefficient score of ET, WT and TC respectively. This means that our method could provide a relatively stable and reliable results on unseen brain tumors.
Meantime, the Hausdorff distance on ET subregion is high, which is considered having outliers on our segmentation results caused by inappropriate sample strategy or training methods. We plan to refine that using cascade by dividing the model into two steps: locate and segment.
Not very competitive, though, we hope our method to provide improvement to others’ methods.
Trained the proposed model is used to segment the training data as well, and Table 3 shows the result. Among all 369 subjects, the results contain several outliers that dice coefficient score are zero or near zero. This leads to the fact that the Mean Hausdorff_95 is relatively high but the Median Hausdorff_95 remains a reasonable range and far lower than the mean value.
Also, the proposed model is evaluated on testing data provided by BraTS2020. Testing data contains 166 subjects in total. Likewise, each testing data contains four modalities without ground truth. The detailed result is shown in Table 4.
4 Discussion
Inspired by the works of predecessors, we present a variety of 3D U-net with CBAM based attention gate and dual-path residual module, to handle the segmentation tasks of brain tumor with multi modalities with a single model and limited resources.
The attention module used here is modified to two parallel pathways contacted with extra residual. Adding a channel-wise attention would help us reweight the feature map in up sample layers, and focus on the feature maps contains more information. In order to reduce the random error, a dual pool pathway, max pool and average pool is proposed. Meanwhile, the spatial attention module is implemented by the original attention gate. Yet, we found that this modification brings some troubles to segmenting small objects. Thus, in the future, we plan to develop it to a better performance using a split attention mechanism.
According to the results of three datasets, the proposed model provides a significant improvement comparing to the base model. Also, we find that there is a common issue with the segment method. The hausdorrf_95 distance is relatively high comparing to others’ methods. We find that among all subjects used to train, there are several subjects is wrongly handled, makes ET subregions of these subjects remains zero or near zero and that is the main reason makes the mean ET dice score of each data set lower and makes the mean hausdorff_95 relatively high. The fact that the mean hausdorff_95 is much higher than the median also reveals the potential cause, namely the performance on ET subregion is influenced by the outliers. The most possible reason is the inappropriate sampling strategy. Input images are cropped into 128 × 128 pieces randomly to save memory, that might lead to potential information lost and result in incorrect or insufficient feature. We mean to refine that issue later by use a randomly crop with center to prevent feature lost.
Meanwhile, training a 3D model with limited resources is a tough way to go. 1080Ti with 11GB RAM could work setting the batch size of 2, but it’s still touching the edge of out of memory. We will improve the proposed model to prevent this problem by introducing dilated convolution to our model. After all, 3D models usually outperform 2D ones at accuracies, which makes all consumption worthy. In the future, we will perform some refinement to this model like introducing GAN [17], split attention mechanism or CRF to it to improve the final score. Cascade network is also a good choice to try.
5 Conclusion
In this work, a refined attention mechanism with dual pathway and a double pathway residual block are introduced to improve the performance on brain tumor segmentation. Dual pathway attention gate could help the network focus not only on spatial feature area but also on target-related channels. Double pathway residual block embedded in the down sample layers of the U-net to prompt the feature transmission. These two blocks are introduced to a 3D U-net. As for the training strategy, we use random crop to avoid feeding whole subjects’ voxels to graphic card and to reduce the proportion of false positive samples. The proposed model evaluated on both train, valid and test datasets. The dice score of ET, WT and TC on train set are 0.823, 0.912 and 0.878 respectively. The dice score of ET, WT and TC on valid set are 0.752, 0.878 and 0.779 respectively. The dice score of ET, WT and TC on test set are 0.773, 0.861 and 0.790 respectively.
References
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Arch. 286 (2017)
Bakas, S., Akbari, H., Sotiras, A., et al.: Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. CoRR abs/1811.02629 (2018). (1811)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. Nat. Sci. Data 4, 170117 (2017)
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Agravat, R.R., Raval, M.S.: Deep learning for automated brain tumor segmentation in MRI images. In: Soft Computing Based Medical Image Analysis, pp. 183–201. Elsevier (2018)
Kamnitsas, K., et al.: DeepMedic for brain tumor segmentation. In: Crimi, A., Menze, B., Maier, O., Reyes, M., Winzeck, S., Handels, H. (eds.) BrainLes 2016. LNCS, vol. 10154, pp 138–149. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-55524-9_14
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Woo, S., Park, J., Lee, J.-Y., Kweon, I.S.: CBAM: convolutional block attention module. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11211, pp. 3–19. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01234-2_1
He, K., et al.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2016)
Ha, Q., et al.: MFNet: towards real-time semantic segmentation for autonomous vehicles with multi-spectral scenes. In: 2017 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS). IEEE (2017)
Wu, Y., He, K.: Group normalization. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11217, pp. 3–19. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01261-8_1
Oktay, O., et al.: Attention U-Net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
Abraham, N., Khan, N.M.: A novel focal Tversky loss function with improved attention u-net for lesion segmentation. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE (2019)
Li, S., et al.: Attention dense-u-net for automatic breast mass segmentation in digital mammogram. IEEE Access 7, 59037–59047 (2019)
Cheng, D., et al.: SeNet: structured edge network for sea–land segmentation. IEEE Geosci. Remote Sens. Lett. 14(2), 247–251 (2016)
Mondal, A.K., Dolz, J., Desrosiers, C.: Few-shot 3D multi-modal medical image segmentation using generative adversarial learning. CoRR abs/1810.12241 (2018). https://arxiv.org/abs/1810.12241
Acknowledge
The project is supported by Sichuan Science and Technology Program. It is partially funded by Grant SCITLAB-0013 of Intelligent Terminal Key Laboratory of SiChuan Province.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Jun, W., Haoxiang, X., Wang, Z. (2021). Brain Tumor Segmentation Using Dual-Path Attention U-Net in 3D MRI Images. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12658. Springer, Cham. https://doi.org/10.1007/978-3-030-72084-1_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-72084-1_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72083-4
Online ISBN: 978-3-030-72084-1
eBook Packages: Computer ScienceComputer Science (R0)