Abstract
The invention of CRISPR/Cas technologies and its rapid advancement enables us to modify plant genomes like never before. The Conventional CRISPR/Cas tool has an unparalleled ability to generate targeted mutations in the genome. In contrast, advanced tools like the base editor can perform single DNA letter swap, and prime editing can generate precise insertion or deletion or DNA letter swapping. This chapter intends to provide the readers with the basics of CRISPR/Cas9 genome editing (GE) technologies, a brief introduction to various CRISPR-derived advanced tools, and how they are implemented to generate site-specific DNA modifications for plant biotechnological applications. In addition, we highlight how genome-edited crops are different from genetically modified organisms (GMOs).
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- CRISPR/Cas tools
- Targeted mutations
- CRISPR-mediated plant biotechnology innovation
- Crop improvement
- Stress tolerant crops
- CRISPR in Agriculture
1 Introduction
The world has witnessed ample growth in agricultural productivity from the last 50 years. The inception of technologies for crop genetic improvement has especially led to a drastic increase in yield for major staple crops, for instance, wheat and rice. This achievement evolved in the form of the Green Revolution (1966–1985). Later, recombinant DNA-based biotechnology developed in the 1970s gave rise to genetically modified (GM) crops, appreciations to innovators like Marc Van Montagu, Jozef Schell, and Mary-Dell Chilton, who co-developed Agrobacterium-mediated plant transformation technology. While transgenic technology has heralded a new era in crop improvement, GM crops’ development is expensive, and they also face societal acceptance issues in many countries. In the meantime, conventional breeding approaches cannot keep pace with biggest challenges such as global population growth and climate change, e.g., the current percentage of annual increase in yield for four major crops (wheat, rice, maize, and soybean) must be doubled to meet the future demand in 2050. All these concerns demand the evolution of new breeding techniques (NBTs) that can likely transform agriculture. GE is one such technology that enables rewriting the code of life, which in most cases depends on the ability to induce DNA double-strand breaks (DSBs) in a sequence-specific manner (Jiang and Doudna 2017). For more details on DSB, see chapter, ‘New technologies for precision plant breeding’, in this book by S. Filler Hayut et al. Sequence-specific nucleases (SSNs) are molecular clippers that are engineered to create targeted DSBs in DNA. A functional unit of DNA is called a gene. SSNs such as zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and CRISPR (clustered regularly interspaced short palindromic repeats)-Cas systems have been successfully used in many plant species to enable efficient genome engineering. Developed in 2012 (Jinek et al. 2012) and applied to eukaryotic cells (mammalian cells) in 2013 (Cong et al. 2013), CRISPR-Cas GE technology has since been transforming plant biology. It improves reverse genetics research in both model and non-model plants and constitutes an efficient breeding tool for crop improvement. In recent years, the number of peer-reviewed papers exploiting CRISPR in plants has skyrocketed. However, it can be challenging and puzzling for new users to select a CRISPR system to achieve a specific GE outcome in a plant of interest. This chapter discusses the development of CRISPR/Cas GE tools with a historical perspective, how these tools precisely alter DNA, and the potential fields of applications in plant biotechnology and crop improvement.
2 Genome Editing Technologies: Historical Perspective and the Rise of Genome Editing Tools
Eukaryotic genomes are composed of billions of DNA bases. The ability to change these DNA bases with precision holds tremendous implications for molecular biology, medicine, and agriculture. Introducing desired genomic changes, i.e., “genome editing”, has been a long-sought-after goal in molecular biology. An earlier article by Urnov (2018) can be consulted to get an exhaustive historical overview of GE technology. To this end, the breakthrough in the form of restriction enzymes that defend bacteria against invading viruses (bacteriophages) in the late 1970s marked the beginning of an era of recombinant DNA technology. For the first time in history, scientists were successful in manipulating DNA molecules in test tubes. Although such accomplishment drove several discoveries in molecular biology and genetics, the ability to precisely alter DNA in eukaryotic organisms came a few decades later. Recent progress in GE tools brings a new revolution in biological research. The GE toolbox was developed between 1994 and 2010 in a mutual attempt between academia and industry, using meganucleases as a prototype and zinc finger nucleases (ZFNs) to edit native loci. While meganucleases provided valuable information on the efficiency and mechanism of DSB repair, but have not been widely adopted as a gene-editing platform owing to the lack of a clear correspondence between meganuclease protein residues and their target DNA sequence specificity. Even though several techniques have been developed to account for these limitations, assembly of functional zinc finger proteins with the preferred DNA binding specificity remains a major problem that involves an extensive screening process. Zinc finger nucleases (ZFNs) were the first of the genome-editing nucleases to hit the scene. Zinc fingers (ZF) are the most common DNA binding domain in eukaryotes. They usually are comprised of ~30 amino acid units that interact with nucleotide triplets. ZFs have been designed to recognize all of the 64 possible trinucleotide combinations. By stringing different zinc finger moieties, one can create ZFs that specifically recognize any specific DNA triplets’ sequence. Each ZF typically recognizes 3–6 nucleotide triplets. ZFN monomer consists of two distinct functional domains: an artificial zinc finger (ZF) domain at the N-terminal portion to bind a target DNA and a FokI DNA cleavage domain (Fok1) at the C-terminal region to create Double-strand break in the DNA. ZFNs have been widely adopted and proved to be the most versatile for GE for more than a decade in living organisms, including animal and plant systems. In 2010–2012, the GE toolkit was rapidly added with a third nuclease class known as transcription activator-like effector nucleases (TALENs). DNA binding characteristic of TALE (transcription activator-like effector) protein is used in constructing TALEN. TALENs are related to ZFNs in that they use DNA binding motifs to direct the same non-specific nuclease (Fok1) to cleave the genome at a specific site. Instead of recognizing DNA triplets as in ZF, each TALE domain recognizes a single nucleotide. In 1987–1989, an “unusual arrangement with repeated sequences” was noticed at a specific locus in the Escherichia coli genome—an array now known as a “clustered regularly interspaced short palindromic repeat,” or CRISPR. This effort on investigating CRISPR-based bacterial immunity against phages finally led to the discovery, in 2012, that a key enzyme of a specific CRISPR-based system, Cas9, is an RNA-guided endonuclease. CRISPR immunity is RNA-based defensive machinery in bacteria designed to recognize and degrade foreign DNA elements from invading bacteriophage and plasmids. The bacterial genome codes for both Cas endonuclease and the guide RNA by a “CRISPR/array.” This system can be co-opted to cut any targeted DNA sequence of choice by modifying it. Thus, in 2012, Martin Jinek, Jennifer Doudna, Emmanuelle Charpentier, and colleagues wrote: “Zinc-finger nucleases and transcription-activator–like effector nucleases have attracted significant attention as artificial enzymes engineered to manipulate genomes. We propose an alternative methodology based on RNA-programmed Cas9 that could offer considerable potential for gene-targeting and genome-editing applications”. It was the beginning of a GE revolution that has taken biologists around the world by storm.
3 CRISPR/Cas-Based Genome Engineering
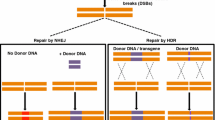
GE technologies refer to those enabling gene knockout, chromosomal recombination, and site-directed insertion/substitution at precise gene locations and chromosomal regions. DNA double-strand breaks (DSBs) are one of the powerful forces that figure plant genomes. Briefly, GE technologies create DNA double-strand breaks (DSBs) via sequence-specific nucleases (SSNs), and the DSBs are then repaired either by the error-prone ‘nonhomologous end joining’ (NHEJ) pathway or by the error-free ‘homology-directed repair’ (HDR) pathway (Molla and Yang 2020). Error in the coding region during DNA repair may cause codon mutations or frameshift mutations of the gene. Therefore, the gene becomes non-functional. DNA DSB repair systems have been extensively studied in many organisms, including plants. Investigations in plants have characterized the genes associated with DSB repair through Non-Homologous-End-Joining (NHEJ) or Homologous Recombination (HR) and tested the result of DSB repair in both somatic and meiotic tissues. NHEJ has been characterized in a wide variety of species and tissues (mostly somatic), using multiple DSB inducing agents including site specific meganucleases, transposon excision and custom-designed nucleases, such as zinc-finger nucleases, transcription activator-like effector nucleases (TALENs) and Clustered Regulatory Interspaced Short Palindromic Repeat associated protein Cas9 (CRISPR-Cas). In contrast to random mutagenesis by chemical (e.g., EMS, EES, Bisulfan) or physical agents (e.g., gamma-ray, x-ray, UV ray), GE tools can install mutations at specific chromosomal sites; therefore, have loads of advantages in functional genomics and molecular breeding. In fact, the genome fixed-point editing technique not only has a greater chance to cause mutations but also is more specific and more potent than random mutagenesis.
The recently developed CRISPR/Cas9 system has rapidly replaced the earlier ZFNs and TALENs. Owing to its simplicity, high efficiency, low cost, and possibility to target multiple genes at once, the CRISPR/Cas9-based GE platform has become an unprecedented tool in plant science research. The use of the CRISPR/Cas9 system for gene-editing tool was first established in human cells and then utilized in plants. Hitherto, CRISPR/Cas9 vector systems have been applied to generate gene knockouts, deletions, disruption of cis-regulatory elements, as well as gene replacements (knock-ins).
Fundamentally, the CRISPR/Cas9 system is relatively simple and is composed of only two major components viz. the Cas9 protein and a single guide RNA (sgRNA), forming a Cas9/sgRNA complex. The 20-nucleotide sequence at the 5′ end of a sgRNA can precisely hybridize with a homologous DNA sequence. This RNA–DNA hybrid activates Cas9 to generate a DSB in the target sequence. The target sequence must be present immediately upstream of an adjacent protospacer motif (PAM; NGG for SpCas9 from Streptococcus pyogenes and TTTV for Cas12a). During DSB repair, NHEJ frequently generates small insertion or deletion (Indel) of nucleobases. Notably, multiple sgRNAs with different target sequences can be designed for the simultaneous editing of more than one gene or DNA region. Once a gene sequence is disrupted due to indel formation, the resultant change in ‘the appearance of a plant’ (phenotype) is correlated. If the correlation is established, it is assumed that the phenotype is controlled by the gene in the study.
Guide RNAs are artificially designed to explicitly direct Cas9 to the target sequence to be edited. Available bioinformatic programs are used to design candidate guide RNAs while considering the likelihood of off-targets. Plant cell transformation to express guide RNAs and Cas9 follows a procedure analogous to the well-known methods for developing transgenic plants. The expression cassettes contain constitutive or inducible promoters, transcription terminators, and antibiotics and/or herbicide resistance markers used for selection purposes.
A vector harboring the DNA sequences for Cas9 protein, and the guide RNA is then incorporated into Agrobacterium bacterial cells. Then, plants are genetically transformed through the Agrobacterium-mediated method, and the identification of first-generation transformed plants is made by using either antibiotic or herbicide selection. Cells or calluses carrying sgRNA-Cas9 cassettes can also be distinguished using green fluorescent protein (GFP). In all instances, target gene/DNA region sequencing is necessary to identify Cas9-induced mutations. The plants generated via sgRNA/Cas9 mediated GE is called transgenic as they carry the sgRNA-Cas9 cassette.
On the other hand, the sgRNA-Cas9 transgene cassette can be eliminated through sexual segregation after identifying CRISPR-edited sexually propagated plants. Selection is made for plants that are edited but do not carry the sgRNA/Cas9 cassette. This would lead to transgene removal in the second or subsequent generations, resulting in transgene-free genome-edited plants’ production. Hence, they bear a resemblance to those with mutations created by natural means. In some countries, the introduction of the sgRNA-Cas9 DNA cassette as a transgene may be treated as GMOs under existing biosafety regulations.
Protocols have been developed to edit plant genomes using guide RNA-Cas9 ribonucleoprotein (RNA plus protein) complexes or transient expression resulting in DNA-free plants. Preassembled Cas9-guide RNA ribonucleoproteins complexes can be introduced into protoplasts via polyethylene glycol-calcium-mediated transfection.
Notable genetic alterations have been achieved through CRISPR-Cas9 to improve metabolic pathways, biotic (fungal, bacterial, and viral pathogens) and abiotic stress (cold, drought, salt) tolerance, nutritional content, yield, and grain quality, obtain haploid seeds, herbicide resistance, and others. Here, we highlight the technical features of the CRISPR/Cas-based GE systems and their crop improvement applications.
4 Various CRISPR/Cas-Based Tools and Their Utilities
Besides the conventional CRISPR/Cas-mediated gene disruption, creative designing and ingenious protein engineering have generated versatile genome disruptors, transcriptional regulators, epigenetic modifiers, base editors, and prime editors (Molla et al. 2020a).
While many Cas proteins have been discovered, Cas9 and Cas12a (also known as Cpf1), are widely used for genome engineering and transcriptional regulation. Cas9 and Cas12a are remarkably easy to program and can be directed to target DNA through Watson–Crick base pairing between the target sequence and gRNA. We refer the readers to an earlier book chapter for more information on orthologous and engineered Cas protein and their utilities in genome targeting (Molla et al. 2020a). In CRISPR systems, gRNAs are composed of a crRNA:tracrRNA complex, that can be fused to form a sgRNA (single gRNA), whereas those for Cas12a consist exclusively of a crRNA. After forming an RNA–protein complex with a gRNA, Cas9 and Cas12a carry out a double-strand break (DSB) adjacent to a protospacer adjacent motif (PAM). Subsequently, DSB repair results in the edited genome.
4.1 Multiplex Editing
The Cas9 and gRNA expression cassettes are the two primary components for CRISPR/Cas9-mediated genome editing. In general, Cas9 expression is driven by an RNA polymerase II (Pol II) promoter. The gRNAs are normally expressed by an RNA polymerase III (Pol III) promoter. For editing multiple genomic loci simultaneously, multiple gRNAs need to be expressed. Different gRNA expression systems are used to achieve multiplex genome editing. The Tandem array of single gRNA expression cassettes, where each guide is controlled by a pol-III promoter, is often used. However, a number of techniques have been developed for single promoter driven expression of multiple gRNAs by engineering different RNA processing machineries, including Csy4 RNase from a bacterium, self-cleavable ribozyme from a virus, and the endogenous tRNA processing enzymes (Vicki and Yang 2020). These RNA processing reagents are employed to generate many gRNAs from single primary polycistronic transcript driven by a Pol II or Pol III promoter. Recently, the polycistronic tRNA-gRNA (PTG) system and ribozyme-mediated assembly of multiple gRNAs under the control of a single promoter gained popularity. In the PTG system, post-transcriptional cleavage of tRNAs by cellular RNAse-P and RNAseZ releases individual gRNA. Ribozymes are RNA molecules with nuclease activity that catalyze its own cleavage. Scientists took advantages of ribozyme’s self-cleaving activity and designed an array of ‘ribozyme-gRNA-ribozyme-gRNA-ribozyme’ to produce multiple gRNAs. When transcribed, the primary transcripts contained the designed gRNA flanked with a ribozyme at each end. After self-cleaving of ribozyme, each gRNA becomes free and they guide Cas9 to their respective coded locus. Csy4 based excision system was also developed for multiplexing. On the other hand, the Cas12a system could be used directly with an array of crRNAs without intervening sequences due to its self-processing ability. We recommend the readers to follow a nice review on different strategies for expressing multiple gRNAs (Vicki and Yang 2020).
4.2 Transcriptional Activation and Repression
Cas9 and Cas12a are made catalytically inactive by changing critical amino acid residues. They are designated as deactivated Cas (dCas9 and dCas12a) proteins. The fusion of dCas enzymes and effector proteins harnesses efficient transcriptional regulation. Two techniques, CRISPR interference (CRISPRi) and CRISPR-activator (CRISPRa) are widely used for regulating gene expression.
Mechanistically, dCas9 enzymes limit transcription by impeding the binding of RNA polymerase or, if targeted to a coding sequence, by interfering with transcription elongation. In eukaryotes, dCas9 is usually fused to an effector protein to augment repression by recruiting chromatin remodeling proteins. Likewise, effector proteins for CRISPRa work by recruiting endogenous transcriptional activators.
4.3 Epigenome Editing
Different epigenetic factors determine distinct phenotypes despite sharing the same DNA. DNA base methylation and histone residue modification are the two well-known epigenetic modifications. CRISPR/dCas system enables transporting a modifier protein to a target epigenetic locus. This could be achieved through the fusion of the modifier protein with the dCas9. The modifier proteins include but not limited to, histone methyltransferase, demethylase, DNA methyltransferase, TET enzyme, etc. The dCas9 tethered with modifier protein, called epigenome editor, can modify epigenetic factors’ status and thus alter the phenotypes.
4.4 Base Editing
Homology-directed repair (HDR) is utilized to precisely change one nucleotide to another in a target DNA; it needs an adequate supply of donor DNA template harboring the change. However, HDR-mediated editing is extremely inefficient in the plant system. Base editing enables precise alteration of single nucleotides with high efficiency and does not require the donor DNA template. For a comprehensive review of base editing, Molla and Yang (2019) may be consulted. The base editor comprises of two essential components—a deaminase tethered with Cas9 nickase (nCas9). nCas9, a catalytically impaired version of Cas9, generates single-strand nick at the target DNA. Cytosine base editor (CBE) causes C-to-T conversion. Adenine base editor (ABE) performs A-to-G alteration. Three recent studies reported the development of a C-to-G base editor (CGBE) for C-to-G editing in the DNA (Molla et al. 2020b). These CBE, ABE, and CGBE enable us to alter a plethora of functional single nucleotide polymorphism (SNP) to improve desired traits in crops.
4.5 Prime Editing
Indeed, the development of base editors reduces our dependency on inefficient HDR-mediated editing. For generating precise indels (as opposed to the random indels caused by Cas9-DSB repair via NHEJ), scientists rely on HDR. A recently developed system, known as prime editing, was demonstrated to efficiently perform all types of base substitutions, 1-44 bp insertions, and 1-80 bp deletions in human cells with far better efficiency than HDR (Anzalone et al. 2019). Several studies also reported the success of prime editing in plant systems, although with lesser efficiency. The prime editing system does not require a supply of donor template. Prime editors contain nCas9 fused with reverse transcriptase (RT). Unlike all other CRISPR-derived techniques that require the same gRNA, prime editing needs a special type of prime editing-guide RNA or pegRNA. Besides specifying the target, PegRNA encodes the reverse transcriptase template (RT), which harbors the desired edit. ‘Two extra elements, 10–13 nt primer binding site (PBS) and 10–16 nt long RT template with the desired changes, are added with the traditional gRNA to construct pegRNA’ (Molla et al. 2020a). Sequence complementary to the nicked genomic DNA strand acts as a primer binding site (PBS). Hybridization of PBS sequence to the target site serves as the point of initiation for reverse transcription. RT copies the desired changes (coded in the RT template) directly in one strand of DNA. Subsequent nick and flap resolution by cellular repair machinery incorporate and fix the editing into the genome.
5 Better Crops with CRISPR/Cas Techniques
The biological world is being modernized through the field of GE technology. CRISPR/Cas9 has been proved to be the best choice for GE with high efficiency, accuracy, and ease of use. World agriculture is witnessing alarming issues, including increasing population growth rate, unpredictable weather, increasing biotic and abiotic stresses, and decreasing arable land availability. To overcome such threatening issues, GE technologies have great potential to be profitable heed to global food security. We recommend readers to consult a recent review article on GE in agriculture (Chen et al. 2019). CRISPR/Cas9 edited non-browning mushroom (Agaricus bisporus) by Dr. Yinong Yang from Pennsylvania State University, received a green signal from the United States Department of Agriculture (USDA). The department also approved CRISPR-edited corn, soybeans, tomatoes, pennycress, and Camelina and emphasized that the transgene free-genome edited crops would not be considered transgenic crops. Recently, a CRISPR-breed Petunia plant with pale pinky-purple colored flower has been approved by USDA. CRISPR/Cas9 system of gene editing has been adopted in many crop species such as rice, maize, wheat, soybean, citrus, tomato, potato, cotton, alfalfa, watermelon, grapes, cassava, ipomoea, barley, lettuce, cacao, carrot, banana, flax, rapeseed, Camelina, cucumber and many other crops for various traits including yield and nutritional quality improvement, biotic and abiotic stress management (Fig. 5.1).
Schematic showing different facets of application of CRISPR-Cas tools for crop improvement. This figure was created using BioRender (https://biorender.com/)
5.1 Rapid Domestication of Crops
With the increasing number of crop genomes sequenced, GE offers one the efficient approaches to plant domestication by opening up the vast genetic diversity from wild or semi-domesticated species, thus producing crops with desired traits.
Modern tomato cultivars produced from the long domestication process resulted in the loss of genetic diversity for stress tolerance. Wild tomato plants naturally exhibit a high degree of tolerance to different stresses; thus, they can serve as ideal materials for de novo domestication via targeting of so-called domestication genes using CRISPR. Zsogon et al. (2018) demonstrated accelerated de-novo domestication of wild tomato with CRISPR/Cas9-mediated multiplex editing, simultaneously targeting genes involved in flowering time, plant architecture, and fruit size that resulted in loss-of-function mutations. The CRISPR edited plants showed earlier and synchronized flowering, increased fruit size, and determinate plant architecture without losing the stress tolerance of the wild germplasm.
Similarly, rapid de-novo domestication of ground cherry (Physalis pruinosa) using the CRISPR/Cas approach was undertaken. Inactivating three genes resulted in determinate growth habit, increased flower petal number, and the number of fruits per plant. Another eye-catching candidate for researchers is a winter annual pennycress (Thlaspi arvense L.) weed for rapid domestication. Illinois state university researchers have been able to make the pennycress oil edible. Thus, CRISPR mediated de-novo domestication events in wild plants offer novel exhilarating possibilities for plant breeding. On one side, exploiting wild crop relatives as an important source of allele mining could expand the germplasm pool for the genetic impoverishment of various crops and resistance against a wide range of biotic/abiotic stresses. On the other side, de-novo domestication facilitates catapulting neglected, semi-domesticated, and wild relatives of crops into the spotlight of mainstream agriculture in a rapid time frame.
5.2 Improving Disease Resistance
The goal of providing adequate food supply to feed the growing global population is made even more challenging owing to crop loss due to various diseases. The development of host plant resistance to pathogens is one of the most sustainable ways to reduce the impact of the diseases. CRISPR/Cas9 is being extensively used to enhance the resistance power of host plants. Readers are suggested to consult the review article (Mushtaq et al. 2019). Few notable case studies are discussed below.
5.2.1 Bacterial Resistance
Phytopathogenic bacteria are highly diverse, with a high multiplication rate and are difficult to control. For example, in rice, Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight disease resulting in 10–20% yield loss. The virulence of Xoo pathogen strongly depends on secretion of its TALE proteins inside the susceptible host. The secreted TALE proteins bind to the promoter of the SWEET genes of rice and trigger their expression.
Higher expression of SWEET genes promotes a favorable environment for disease development. To interrupt the binding of TALE proteins with SWEET gene promoter, the sequence of the binding region was altered by CRISPR/Cas9. The resultant mutant lines were resistant to bacterial blight. A similar strategy was used to develop resistance against citrus canker bacteria.
5.2.2 Fungal Resistance
CRISPR/Cas9 system has shown great potential in mitigating the impacts of fungal diseases as well. Many fungi use host plant genes (known as susceptibility genes) for facilitating their establishment in the host. For example, mildew resistance locus O (Mlo) gene, a susceptibility gene, has been mutated through CRISPR/Cas9 system, and the developed mutants were more resistant to powdery disease-causing fungus Blumeria graminis f. sp. tritici and Oidiumneo lycopersici of wheat and tomato, respectively. Targeted mutagenesis of negative regulator OsERF922 showed disease resistance against rice blast. These examples validate the potential use of the CRISPR/Cas9 system for fungal disease resistance.
5.2.3 Virus Resistance
Plant viruses pose a significant threat to modern agriculture. Resistance in Nicotiana benthamiana against beet severe curly top virus (BSCTV) was demonstrated by the precise targeting of replication genes of viruses via CRISPR/Cas9. Similarly, rice plants resistant to Tungro virus disease was developed through CRISPR/Cas9 mediated disruption of a gene that is used by the virus for their multiplication. A recent review article may be consulted to get more insight on the application of GE for virus resistance (Mushtaq et al. 2020).
5.2.4 Nematode and Parasitic Weed Resistance
Various GE technologies have been adopted to improve crop resistance against nematodes. CRISPR/Cas9-mediated GE technology has been used for targeting GmSHMT08 to study soybean resistance to soybean cyst nematode (Kang 2016). CRISPR/Cas9 enabled mutagenesis of the tomato’s CCD8 gene has been used to provide resistance against the weed Phelipanche aegyptiaca.
5.3 Developing Climate-Smart Crops
Abiotic stresses, including salinity, drought, temperature, and heavy metals, pose a major threat to global food security. Developing cultivars tolerant against these various environmental stresses is the most sustainable and environmentally friendly approach to cope with this challenge. However, little work has been done regarding abiotic stress tolerance development employing CRISPR/Cas tools. Recently, tomato fruit setting under heat stress has been improved by targeting SlAGAMOUS-LIKE 6 gene using CRISPR-Cas9 mediated genome editing. Researchers have recently developed and tested drought-tolerant maize through precise gene-editing of AGROS8 (Shi et al. 2017). Nonetheless, the technique is also hugely utilized by researchers to decipher the functions of genes potentially involved in abiotic stress tolerance. Readers may consult a recent review to get more insight (Mushtaq et al. 2018).
5.4 Quality Enhancement
5.4.1 Color and Texture
The color and texture of fresh tomatoes many times determine consumer preferences. Consumers from diverse areas have distinct color preferences. For example, Americans and Europeans prefer red tomatoes, while Asians like pink-colored tomatoes. Researchers have effectively produced yellow, pink, and purple tomatoes by targeting different pigment biosynthetic genes.
5.4.2 Increasing Nutritional Composition and Removing Anti-Nutrient (Allergens) Factors
Consumption of gluten proteins from wheat, rye, and barley causes coeliac disease in genetically predisposed individuals. Patients with a strict gluten-free diet can recover from this disease. As there are a large number of gluten genes and wheat genome being more complex, wheat that is coeliac-safe but retains baking quality cannot be produced by conventional breeding alone. Recently, CRISPR/Cas tool was used to develop low gluten wheat with the baking quality intact (Chen et al. 2019).
Maize (Zea mays) is a major cereal crop, and phytic acid-P form more than 70% of the total phosphorus in maize seed. It is supposed to be anti-nutritional because it is not digested by monogastric animals and is also an environmental pollutant. Scientists have used the CRISPR technique for targeted knock out of genes governing phytic acid synthesis in maize.
Anthocyanin, malate, lycopene, and γ-aminobutyric acid (GABA) are the tomato's well-known bioactive compounds. CRISPR-Cas9 technology has been employed to generate tomato fruits with an enhanced level of those bioactive compounds by regulating the key genes responsible for their metabolism.
Potato starch quality is an important trait and a central area of research. Scientists have targeted the granule-bound starch synthase (GBSS) gene using CRISPR-mediated gene-editing, which resulted in waxy genotype in potato. Similarly, DuPont/Corteva agriscience developed waxy corn with more than 97% amylopectin by targeted disruption of the Wx1 gene (https://synbiobeta.com/dupont-pioneer-unveils-first-product-developed-crispr-cas/). These waxy crops have high values in processed food, adhesive, and high gloss paper making industry.
Accumulation of cesium and cadmium in rice plants grown in contaminated soil is a severe health concern. Recently, researchers from two distinct groups used the CRISPR-Cas system to inactivate transporter proteins that carry them to allow their inflow from soil to the plant. Mutant plants exhibited a remarkably lower accumulation of cesium and cadmium.
Recently, Yield10 Bioscience, MA, USA, announced the successful development of CRISPR-edited canola plant lines with increased oil content. They received a non-GMO regulatory response from USDA (https://geneticliteracyproject.org/2020/08/19/crispr-edited-canola-slated-for-2021-field-trials-moving-crop-closer-to-commercialization/).
Cassava is the primary source of nutrition for ~40% of Africans. It contains toxic cyanogen—excessive consumption of under processed cassava results in an epidemic paralytic disease, Konzo. Researchers from Innovative genomic institutes, California, are working on the removal of cassava cyanogen by disrupting its biosynthetic genes (https://innovativegenomics.org/news/crispr-cyanide-free-cassava/). Similarly, Tropic bioscience in the UK is working on producing a CRISPR-edited naturally decaffeinated coffee.
5.4.3 Enhancing Self-Life
Postharvest browning of mushroom causes decreased consumer acceptance and market value. Yinong Yang and his team at the Pennsylvania state university developed a non-browning mushroom by CRISPR/Cas-induced inactivation of genes responsible for mushroom-browning.
The food industry highly desires prolonged shelf life for fleshy fruits. Tomato lines with extended shelf life were generated by CRISPR/Cas mediated manipulation of ripening pathway genes.
5.5 Yield Improvement
Yield is one of the most important traits for crop plants. Traditional breeding has been used for decades to improve yield and develop plants suitable for particular growth environments, which is a time-consuming process. Researchers used CRISPR/Cas9 system to knockout four negative regulators of yield (the genes Gn1a, DEP1, and GS3) in rice and obtained mutants with improved grain number, dense, erect panicles, and larger grain size, respectively. Likewise, another group of scientists used CRISPR/Cas9-mediated multiplex genome-editing system to simultaneously knock out three major rice negative regulators of grain weight (GW2, GW5, and TGW6), resulting in a significant increase in thousand-grain weight. Researchers targeted three homoalleles of GASR7, a negative regulator of kernel width and weight in bread wheat, by employing CRISPR/Cas9 that increased the thousand-kernel weight. Researchers at the Cold Spring Harbor Laboratory employed CRISPR-Cas9 gene-editing tool to produce larger tomato fruits by destructing the classical CLAVATA-WUSCHEL (CLV-WUS) stem cell circuit.
Hybrid varieties provide yield advantages over traditional varieties. However, farmers cannot save hybrid seeds for the next generation because yield advantages are lost in subsequent generations due to genetic segregation. Scientists have long sought for a technology to propagate hybrid seeds clonally. Remarkably, two recent studies demonstrated the clonal seed production of hybrid rice employing CRISPR/Cas9 technology (Chen et al. 2019). For more details on yield improvement using CRISPR/Cas9, see Chen et al. (2019).
5.6 Early Disease Detection in Plant
The discovery of orthologous Cas proteins (Cas12a and Cas13) with collateral nucleic acid cleavage activities enabled the development of nucleic acid diagnostic tools. Studies showed they could be used to develop robust, highly sensitive, low-cost, a practical diagnostic tool for disease and pathogen detection. Application of DETECTR (based on Cas12a) and SHERLOCK (based on Cas13) system would enable trait detection, pest surveillance, GMO detection, and pathogen identification (Kocak and Gersbach 2018).
5.7 Controlling Invasive Species in Agri Field
Invasive species continue to be one of the greatest challenges to global biodiversity. CRISPR/Cas9-based gene drive is a powerful technology that allows biased inheritance of a gene and spread rapidly through a population. Gene drive is used to insert and spread a desired modification faster than the usual rate of Mendelian inheritance (50%). Gene drive technology is competent to control pest species to increase agricultural production. In contrast to other pest management strategies, it is cheaper, more precise than pesticide use. Gene drive‐mediated pest control can thus be eye-catching for agribusiness, owing to its direct manipulation of pest species. It could also potentially be used against invasive weeds. For instance, pigweed (Amaranthus) could be engineered by CRISPR/Cas-based gene drives to become susceptible to the widely used herbicide glyphosate. The principle for CRISPR–Cas9‐based suppression of weed species is based on the assumption that gene drives could be used to bring in and spread a fitness load that can limit the establishment, abundance, dispersal, persistence and/or impact of weed populations.
5.8 Weed Management
Besides using gene drives to eradicate a target weed population, CRISPR/Cas tools can also be used to generate herbicide-tolerant crop species; so that herbicide could be used to control weeds in the agricultural field effectively. Herbicide usually kills plants by inhibiting one or more crucial plant metabolic enzymes. Herbicide resistance is developed by a single or few point mutations in the enzyme’s herbicide binding site. Precise installation of those mutations was achieved either by Base editors or HDR-mediated precise editing. Rice plants resistant to Imazamox, haloxyfop, and bispyribac sodium have been generated by employing those CRISPR tools (Chen et al. 2019).
6 Genome Edited Versus Transgenic Crops
Both transgenic and gene-edited crops are developed by forms of genetic engineering that have possible applications in agriculture and plant biotechnology. Still, they both vary in one way or the other. In the process of developing both kind of crops, an initial transformation of foreign DNA constructs is done. For transgenic crops, the construct needs to be integrated and remained present in the genome for expressing the desired trait. On the other hand, in most of the genome-edited crops, the construct is no longer needed after it successfully induces editing. GE is more rapid than conventional breeding, is less controversial than techniques such as transgenesis which are considered as ‘GMOs’ from a regulatory point of view in many jurisdictions. GMOs involve introducing genes from the same or other species into DNA. Gene editing, in contrast, allows scientists to alter the organism's DNA without inserting genes from a different organism. GE enables modification of an existing gene. Gene-editing technology is advantageous over genetic modification on various grounds. As we discussed earlier in this chapter, editing is done by the influence of Cas9. Once the Cas9 makes a DSB at a target DNA and repaired erroneously, the gRNA/Cas9 transgene is no longer needed in the cell. By sexual inheritance, the transgene cassette can be segregated out. This technology is more specific (precise) than GMO processes, and continue to become more reliable. It is also relatively inexpensive in contrast to other methods, suggesting more scientists could gain access to it. Such advantages represent more potential innovation. How booming gene editing is, however, will also depend in large on how it's perceived. Crops developed by GE could face similar kinds of opposition as perceived by GMOs. Since they don’t insert foreign genes into the crop, consumers might find them more natural and consequently more appealing which remains the advantage of GE crops over GMOs. On March 28, 2018, U.S. Secretary of Agriculture declared that the USDA wouldn't regulate crop varieties generated employing GE that would yield plants indistinguishable from those developed through traditional breeding methods.
7 Concluding Remarks
Although GE tools existed before CRISPR, it has democratized the field by its efficiency, ease of use, and accessibility. Although it has tremendous potential in human therapeutics, fruits of GE would be visualized quicker in agriculture. It will have many similar applications like GMOs but optimistically with broader public acceptance. While CRISPR could be a major boon for increasing agricultural production, a lack of public acceptance might choke further improvement of CRISPR crops before commercialization can become a certainty. Plant genome editing’s societal concerns stem in part due to the unawareness of its principles and applications. Spreading the knowledge to the general public on the GE principles might correct and stop the spread of fallacy. We need to keep in mind that biotechnology familiarity and perceptions of safety, although not sufficient, is a crucial parameter for public acceptance.
References
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576(7785):149–157
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annual Rev Plant Biol 70:667–697
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Jiang F, Doudna JA (2017) CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Kang J (2016) Application of CRISPR/Cas9-mediated genome editing for studying soybean resistance to soybean cyst Nematode. University of Missouri-Columbia: Columbia, MO, USA
Kocak DD, Gersbach CA (2018) Scissors become sensors. Nature 557(7704):168–169
Molla KA, Yang Y (2019) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37:1121–1142
Molla KA, Yang Y (2020) Predicting CRISPR/Cas9—induced mutations for precise genome editing. Trends Biotechnol. 38:136–141
Molla KA, Qi Y, Karmakar S, Baig MJ (2020a) Base Editing Landscape Extends to Perform Transversion Mutation. Trends Genet. https://doi.org/10.1016/j.tig.2020.09.001
Molla KA, Karmakar S, Islam MT (2020b) Wide horizons of CRISPR-Cas-derived technologies for basic biology, agriculture, and medicine. In: CRISPR-Cas Methods. Humana, New York, NY, pp 1–23
Mushtaq M, Bhat JA, Mir ZA, Sakina A, Ali S, Singh AK, Tyagi A, Salgotra RK, Dar AA, Bhat R (2018) CRISPR/Cas approach: a new way of looking at plant-abiotic interactions. J Plant Physiol 224–225:156–162
Mushtaq M, Sakina A, Wani SH, Shikari AB, Tripathi P, Zaid A, Galla A, Abdelrahman M, Sharma M, Singh AK, Salgotra RK (2019) Harnessing genome editing techniques to engineer disease resistance in plants. Front Plant Sci 10:550
Mushtaq M, Mukhtar S, Sakina A, Dar AA, Bhat R, Deshmukh R, Molla K, Kundoo AA, Dar MS (2020) Tweaking genome-editing approaches for virus interference in crop plants. Plant Physiol Biochem 147:242–250
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE (2017) ARGOS8 variants generated by CRISPR—Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15(2):207–216
Urnov FD (2018) GEBC (before CRISPR): lasting lessons from the “old testament.” CRISPR J 1(1):34–46
Vicki HF, Yang Y (2020) Efficient expression of multiple guide RNAs for CRISPR/Cas genome editing. aBIOTECH 1:123–134
Zsogon A, Cermak T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36:1211–2121
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Mushtaq, M., Molla, K.A. (2021). CRISPR Technologies for Plant Biotechnology Innovation. In: Ricroch, A., Chopra, S., Kuntz, M. (eds) Plant Biotechnology. Springer, Cham. https://doi.org/10.1007/978-3-030-68345-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-68345-0_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-68344-3
Online ISBN: 978-3-030-68345-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)