Abstract
The release of extracellular vesicles (EVs) is a process conserved across the three domains of life. Amongst prokaryotes, EVs produced by Gram-negative bacteria, termed outer membrane vesicles (OMVs), were identified more than 50 years ago and a wealth of literature exists regarding their biogenesis, composition and functions. OMVs have been implicated in benefiting numerous metabolic functions of their parent bacterium. Additionally, OMVs produced by pathogenic bacteria have been reported to contribute to pathology within the disease setting. By contrast, the release of EVs from Gram-positive bacteria, known as membrane vesicles (MVs), has only been widely accepted within the last decade. As such, there is a significant disproportion in knowledge regarding MVs compared to OMVs. Here we provide an overview of the literature regarding bacterial membrane vesicles (BMVs) produced by pathogenic and commensal bacteria. We highlight the mechanisms of BMV biogenesis and their roles in assisting bacterial survival, in addition to discussing their functions in promoting disease pathologies and their potential use as novel therapeutic strategies.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Bacterial membrane vesicles (BMVs)
- Outer membrane vesicles (OMVs)
- Membrane vesicles (MVs)
- Bacterial pathogenesis
- Therapeutic applications of BMVs
Introduction
What are Bacterial Membrane Vesicles?
The release of extracellular vesicles (EVs) is a process conserved across the three domains of life (reviewed in (Deatherage and Cookson 2012)). Microbiologists first identified the ability of Gram-negative bacteria to release EVs from their cell membrane more than 50 years ago (Knox et al. 1966), and since then their production by both Gram-negative and Gram-positive bacteria has become widely accepted. Due to bacterial EVs originating from the cell membrane of bacteria, EVs produced by both Gram-negative and Gram-positive bacteria are collectively referred to as bacterial membrane vesicles (BMVs). BMVs can be further classified on the basis of the cellular architecture of the bacterium from which they are derived, whereby Gram-negative and Gram-positive bacteria produce vesicles known as outer membrane vesicles (OMVs) and membrane vesicles (MVs), respectively. Specifically, the release of OMVs by Gram-negative bacteria occurs at the cells outermost membrane, whilst MVs released by Gram-positive bacteria are derived from the single cytoplasmic cell membrane, which is surrounded by a peptidoglycan-rich cell wall (reviewed in (Kaparakis-Liaskos and Ferrero 2015; Bitto and Kaparakis-Liaskos 2017)).
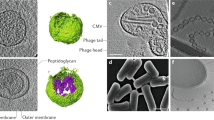
OMVs are nanoparticles released by Gram-negative bacteria ranging between 30–300 nm in size, therefore having a far narrower size distribution compared to eukaryotic EVs (Fig. 6.1a) (reviewed in (Zavan et al. 2020; Raposo and Stoorvogel 2013)). OMVs are capable of packaging cargo such as proteins, nucleic acids and toxins within a lipid membrane (Fig. 6.1b) (reviewed in (Kaparakis-Liaskos and Ferrero 2015)). First discovered in 1966, the production of OMVs by the Gram-negative bacterium Escherichia coli was initially considered an artefact of bacterial growth, without substantive physiological relevance (Knox et al. 1966). In 1975, interest in OMVs was sparked by the identification of OMVs within the cerebrospinal fluid of patients with meningococcal disease (DeVoe and Gilchrist 1975). Subsequent studies of patients infected with Gram-negative pathogens, including Helicobacter pylori, Haemophilus influenzae and Neisseria meningitidis identified the presence of OMVs within patient tissue or biological fluids (Fiocca et al. 1999; Ren et al. 2012; Stephens et al. 1982; Namork and Brandtzaeg 2002). However, the functions of OMVs from these pathogens in a disease setting remained unclear.
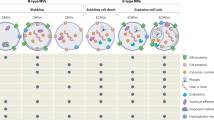
Size and composition of Bacterial membrane vesicles. (a) Schematic overview of the size and characteristics of EVs secreted by eukaryotic and prokaryotic cells. Eukaryotic EV release is defined by populations of vesicles emanating from intracellular compartments, termed exosomes (30–150 nm), or from the cell membrane, termed microvesicles (100–1000 nm) or apoptotic bodies (<1–5μm) which are produced during apoptotic cell death. By contrast, prokaryotic bacteria produce bacterial membrane vesicles (BMVs) via their cell membrane (30–300 nm). For comparison, the size of bacteriophages and soluble proteins are depicted at right (<30 nm). (b) Gram-negative bacteria produce OMVs which package cargo within a lipid bilayer. OMVs package contents derived from their parent bacterium which include membrane-bound proteins, cytoplasmic or periplasmic proteins, toxins, nucleic acids and peptidoglycan. (c) Gram-positive bacteria produce MVs which package cargo including nucleic acids, toxins, cytoplasmic or membrane-bound proteins and lipoproteins. MVs may also contain cell wall components such as peptidoglycan and lipoteichoic acids
More recent studies have confirmed that OMVs significantly contribute to fundamental biological processes of their parent bacterium. For example, it has been shown that OMVs can function to facilitate horizontal gene transfer (HGT) , the formation of bacterial biofilms and the sequestration of antibiotics (Renelli et al. 2004; Schooling and Beveridge 2006; Manning and Kuehn 2011; Kulkarni et al. 2015). Furthermore, OMVs may promote bacterial pathogenesis by acting as vehicles for the delivery of toxins and immunomodulatory products (reviewed in (Ellis and Kuehn 2010)), or by packaging conserved microbe-associated molecular patterns (MAMPs) from their parent bacterium, that can be detected by host pathogen recognition receptors (PRRs) and mediate an innate immune response (reviewed in (Kaparakis-Liaskos and Ferrero 2015; Johnston et al. 2020)). Non-pathogenic bacteria , such as those constituting the human microbiota, also produce OMVs containing MAMPs and research in this field has increased significantly within the last 5 years. It was first identified that OMVs from the commensal bacterium Bacteroides fragilis could modulate host immunity (Shen et al. 2012) and numerous studies have since investigated the effects of OMVs produced by the human microbiota in shaping immunological processes within the gut and in other organs (reviewed in (Stentz et al. 2018)). Due to their intrinsic immunogenic properties, OMV-based vaccines have been developed and licenced for human use (Oster et al. 2005), with considerable interest in furthering their development to protect against antibiotic-resistant bacterial infections.
MVs are nanoparticles produced by Gram-positive bacteria that range between 30–300 nm in size (Fig. 6.1c) (reviewed in (Brown et al. 2015)). The release of MVs from Gram-positive bacteria was first reported as early as 1990 (Dorward and Garon 1990). However, in an era where prokaryotic extracellular vesicles were viewed as cellular artefacts, the suggestion of vesicle release from the thick and rigid cell wall of Gram-positive bacteria was unaccepted by the scientific community. While interest in OMVs gained momentum, MV research stalled, with limited studies reporting the characterisation and functions of MVs (Klieve et al. 2005; Mayer and Gottschalk 2003). It was not until 2009, nearly 20 years after their initial identification, that this dogma was challenged by electron microscopy images demonstrating the release of MVs by Staphylococcus aureus (Lee et al. 2009). Following this finding, subsequent studies identified MV production by a range of Gram-positive bacteria , including the pathogens Bacillus anthracis (Rivera et al. 2010), Streptococcus pyogenes (Resch et al. 2016), Listeria monocytogenes (Vdovikova et al. 2017) and Clostridium perfringens (Jiang et al. 2014) and even the commensal strains Lactobacillus reuteri (Grande et al. 2017) and Enterococcus faecium (Wagner et al. 2018). Akin to OMVs, MVs have been implicated in a range of biological functions including inter-bacterial communication and host-pathogen interactions (Liao et al. 2014; Obana et al. 2017; Wang et al. 2020). Yet there still remains a significant gap in knowledge surrounding the biogenesis, composition and biological functions of MVs, with significant efforts focused on expanding knowledge in this area.
The Study of BMVs
Despite more than 50 years of BMV research, difficulties remain within the field in terms of uniformity and standardisation of techniques. By comparison to the field of eukaryotic EVs , the field of bacterial membrane vesicles has not seen the development of guidelines regarding appropriate methods and minimal standards that need be applied for their isolation and characterisation. This is compounded by the isolation and examination of BMVs from a vast number of Gram-negative and Gram-positive bacterial species of interest with varying growth and culturing conditions. Therefore, the techniques used to isolate and characterise vesicles produced by one bacterial species may not be inherently identical to those produced by another. As a result, variables in bacterial strains, culture conditions and vesicle purification methods prevent effective comparisons to be made between BMV studies. With the rapid expansion of studies examining the characteristics and functions of OMVs and MVs, it is becoming increasingly necessary to ensure standardisation and uniformity across the field. Below we outline general techniques used to isolate, purify and analyse OMVs and MVs.
Isolation and Purification
The majority of the first reported OMV preparations were achieved by ultracentrifugation of bacterial supernatants from broth cultures. Whilst succeeding in harvesting OMVs from these species, these seminal studies were unaware of the contaminating debris, including soluble proteins and bacterial structures such as flagella and pili within their vesicle preparations (reviewed in (Kulp and Kuehn 2010)). Gradually, researchers have improved the purity and yield of their vesicle preparations by adopting techniques including ultrafiltration and low-speed differential centrifugation prior to ultracentrifugation (reviewed in (Klimentová and Stulík 2015)). Density-gradient separation of crude BMV preparations, using iodixanol or sucrose mediums, has allowed for greater purification of vesicles for proteomic and immunological analyses (reviewed in (Klimentová and Stulík 2015)), in addition to the separation of different sized OMVs (Kaparakis et al. 2010; Turner et al. 2018). Other groups have also implemented size exclusion chromatography (SEC) techniques to isolate vesicles (Post et al. 2005; Hong et al. 2019; Schulz et al. 2018). A recent study comparing BMV purification methods reported that vesicle purification by density-gradient separation or SEC were both equally suitable for the isolation of highly purified BMV preparations (Dauros Singorenko et al. 2017). However, irrespective of the technique used to isolate and purify BMVs, scientific rigor and careful reporting of the techniques used to isolate and purify these vesicles are necessary to publish meaningful and reproducible findings that can enable comparisons to be made between independent studies.
Characterisation & Analysis
As researchers begin to investigate the functions of BMVs more broadly, it has become increasingly important to thoroughly detail their methods of production and characterisation before reporting BMV functions. For example, the protein content of OMVs and MVs can vary between bacterial species, strains and even by the same strain cultured using different growth media and to varying growth stages (Yara et al. 2019; Taheri et al. 2019; Klimentova et al. 2019; Zavan et al. 2019; Jeon et al. 2016; Jiang et al. 2014). Furthermore, genetic engineering of bacteria enables the generation of hypervesiculating bacterial mutants , which has greatly increased the yield of OMV preparations (Bernadac et al. 1998; Turner et al. 2015; Moon et al. 2012; Ojima et al. 2020; Mitra et al. 2016). These differences in BMV production highlight that it is imperative that researchers clearly define the conditions used to generate, isolate and purify their BMVs, as this may impair the reproducibility of results and limit comparisons between studies if they are not clearly reported.
There are numerous techniques employed to characterise OMVs and MVs which include electron microscopy (EM) to visually assesses vesicle morphology, and nanoparticle tracking analysis (NTA) instruments such as NanoSight™ and ZetaView™ that assess the yield and size of BMVs. Additionally, techniques to characterise vesicular cargo include Western-immunoblot or mass-spectrometry (proteins) (Kaparakis et al. 2010; Zavan et al. 2019; Lee et al. 2009; Choi et al. 2011), lipidomics (lipids) (Jasim et al. 2018; Roier et al. 2015; Jeon et al. 2018) and next-generation DNA and RNA sequencing methods (Koeppen et al. 2016; Bitto et al. 2017). Collectively, the tools and techniques used to isolate and characterise BMVs vary, and this highlights the need for detailed methodology and uniformity in the purification, characterisation and functional analyses of BMVs to enable better comparisons between studies.
BMV Biogenesis
Mechanisms of OMV Biogenesis
The production of OMVs is common to all Gram-negative bacteria, and although many key determinants of OMV biogenesis have been suggested, the molecular pathways underlying vesiculation are only recently being uncovered. Unlike eukaryotic EVs, BMVs do not originate from an intracellular organelle. Two mechanisms of OMV biogenesis have been identified to date. First, OMVs secreted by Gram-negative bacteria may be released via the outer membrane (OM) of their parent bacterium by OM bulging, eventuating in the formation and budding of a new vesicle. OMV budding from the bacterial OM has been described for numerous species and is thought to be a conserved mechanism of OMV biogenesis amongst Gram-negative bacteria (reviewed in (Schwechheimer and Kuehn 2015)).
Blebbing of the bacterial OM can be regulated via several different pathways and OM molecules such as peptidoglycan (PG), proteins and lipids. The bacterial OM is anchored to the periplasmic peptidoglycan layer via strong covalent bonding. Thus, disruptions to the linkages between the OM and PG layer can affect OMV biogenesis, as demonstrated by investigations of Acinetobacter baumannii , E. coli and Porphyromonas gingivalis, amongst other bacterial species (Moon et al. 2012; Schwechheimer et al. 2014; Iwami et al. 2007; Suzuki et al. 1978). Furthermore, degradation of the PG layer itself, or accumulation of PG fragments within the periplasm, were shown to augment the production of E. coli and P. gingivalis OMVs (Schwechheimer et al. 2014; Hayashi et al. 2002). OMV biogenesis may also be upregulated in response to other envelope stresses, such as the accumulation of misfolded proteins or OM curvature-inducing molecules (McBroom and Kuehn 2007; Mashburn-Warren et al. 2008; Mashburn and Whiteley 2005). Additionally, the transmembrane Tol-Pal protein system, an important regulator of bacterial membrane integrity, has been identified to play a role in the regulation of OMV biogenesis of both non-pathogenic E. coli and pathogenic bacteria including H. pylori and Shigella boydii (Bernadac et al. 1998; Turner et al. 2015; Mitra et al. 2016).
Another regulator of OMV production is the composition of LPS at the site of vesicle budding, as demonstrated by lipid remodelling in the OM of the pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) being necessary for OMV release (Elhenawy et al. 2016). Interestingly, another recent study identified a universal regulator of OMV blebbing common to most Gram-negative bacteria via a retrograde lipid transporter system (Roier et al. 2016). Deletion or repression of the VacJ/Yrb ABC phospholipid transporter system in H. influenzae and Vibrio cholerae increased OMV production due to phospholipid accumulation in the bacterial OM of both species (Roier et al. 2016). The authors suggested that this system may serve as a universal regulator of OMV production by Gram-negative bacteria.
Finally, OMV production can also be modulated by environmental stresses including exposure to antibiotics or unfavourable bacterial growth conditions such as aberrant temperature or pH (McBroom and Kuehn 2007; Kadurugamuwa and Beveridge 1995; Klimentova et al. 2019). Overall, these studies highlight the role of alterations to the cell membrane in regulating OMV biogenesis via blebbing from the OM.
An alternative mechanism of OMV biogenesis was recently identified via stress-induced explosive cell lysis of bacterial cells (Turnbull et al. 2016). The production of Pseudomonas aeruginosa OMVs via explosive cell lysis is mediated by a prophage-encoded endolysin whereby membrane fragments of exploded bacteria re-fused to form vesicles, thus capturing cytoplasmic contents (Turnbull et al. 2016). However, it remains to be determined if this mechanism of OMV biogenesis is common to all Gram-negative bacteria or specific to P. aeruginosa.
Mechanisms of MV Biogenesis
In contrast to the vast knowledge surrounding OMV biogenesis, few studies defining the mechanisms of MV biogenesis exist to date. The structural differences between the Gram-positive and Gram-negative cell wall imply that there are mechanistic differences between MV and OMV biogenesis (reviewed in (Brown et al. 2015)). While OMVs can be released from the outer membrane of the cell surface, MVs are derived from the cytoplasmic membrane and therefore must traverse the thick layer of peptidoglycan to be released (Brown et al. 2015). However, recent evidence has shown that accumulation of phenol-soluble modulin (PSMs) in the cytoplasmic membrane leads to MV formation by increasing membrane fluidity (Wang et al. 2018; Schlatterer et al. 2018). The first electron microscopy images depicting the release of MVs by S. aureus suggest that disruption of the peptidoglycan layer allows for the protrusion of the cytoplasmic membrane through the cell wall, and therefore MV release (Lee et al. 2009). This was followed by similar findings in other species of Gram-positive bacteria and is thought to occur via protrusions through the cell wall mediated by autolysins that hydrolyse peptidoglycan crosslinks (Wang et al. 2018; Toyofuku et al. 2017; Resch et al. 2016; Lee et al. 2009).
An alternative model of MV biogenesis has been observed in Bacillus subtilis that involves the disruption of the peptidoglycan cell wall via a prophage-encoded endolysin (Toyofuku et al. 2017). Similar to the formation of OMVs via prophage-mediated explosive cell lysis reported in P. aeruginosa (Turnbull et al. 2016), B. subtilis MV production by this mechanism is dependent on activation of a defective prophage triggered by genotoxic stress. However, unlike the process in P. aeruginosa, explosive cell lysis was not observed in B. subtilis, but rather cell death eventuated from the loss of membrane integrity (Toyofuku et al. 2017). This alternative mechanism of MV biogenesis is believed to occur in addition to other mechanisms that do not result in cell death (Toyofuku et al. 2017; Andreoni et al. 2019). Furthermore, a putative phage-encoded endolysin is enriched in S. pyogenes MVs (Resch et al. 2016), suggesting a similar mechanism may occur in this species. More studies are required to determine whether the prophage-mediated liberation of MVs via cell death is common to other Gram-positive species.
Bacterial extracellular vesicles are becoming increasingly recognised as a bacterial secretion system for the long-distance delivery of effector molecules. While there have been significant breakthroughs in our understanding of MV biogenesis over the last 10 years, there is still much to uncover. Elucidating the mechanisms by which MV biogenesis occurs can provide a better understanding of the role of MVs in bacterial survival and pathogenesis.
Cargo Packaging into BMVs
BMVs are often enriched in cargo that facilitate specific biological functions. Certain molecules may act as chaperones to facilitate packaging of necessary structural components, while others may directly enact functions such as toxins packaged by pathogens.
Since their discovery, research in the field of BMVs has predominantly focused on defining their contribution to pathogenesis. Consequently, numerous studies have identified virulence effectors associated with OMVs and MVs from various pathogens. Indeed, proteomic profiling of BMVs produced by pathogenic bacteria including E. coli, S. aureus, Streptococcus sp., Neisseria gonorrhoeae and even members of the non-pathogenic Bacteroidetes genus has revealed selective enrichment of proteins in BMVs compared to the outer membrane of their parent bacteria (Lee et al. 2007, 2009; Thay et al. 2013; Olaya-Abril et al. 2014; Biagini et al. 2015; Zielke et al. 2014; Elhenawy et al. 2014). However, the mechanisms underpinning the packaging of these virulence effector proteins into OMVs and MVs remain largely unclear.
One proposed mechanism for cargo selection into OMVs is via outer membrane lipid chaperones which guide molecules to sites of OMV biogenesis. For example, Klebsiella pneumoniae is a nosocomial pathogen whose OMVs differentially package protein cargo in response to LPS modifications (Cahill et al. 2015). Specifically, with the use of a K. pneumoniae mutant strain deficient in the O antigen of LPS (ΔwbbO strain), it was shown that the LPS composition of the bacterial outer membrane impacted the protein composition of OMVs produced (Cahill et al. 2015). Similar results have been reported for OMV and MV production by P. gingivalis and Group A streptococcus, whereby the lipid composition of the bacterial membrane was responsible for the selective enrichment of specific proteins or RNA species (Haurat et al. 2011; Resch et al. 2016).
It has long been reported that BMVs can associate with bacterial genetic material including DNA, and more recently RNA (Dorward and Garon 1990; Sjöström et al. 2015; Liao et al. 2014; Choi et al. 2018; Dorward and Garon 1989; Kolling and Matthews 1999). In particular, bacterial DNA can be packaged into OMVs, delivered to eukaryotic host cells and thereafter trafficked to the host cell nucleus (Bitto et al. 2017). Similarly, the delivery of bacterial regulatory RNAs to eukaryotic host cells via OMVs was recently demonstrated, resulting in the modulation of the host immune response (Koeppen et al. 2016; Choi et al. 2017; Zhang et al. 2020). Packaging of nucleic acids into MVs has also been reported to occur by a limited number of Gram-positive species, including C. perfringens, Streptococcus mutans and L. reuteri (Jiang et al. 2014; Liao et al. 2014; Grande et al. 2017). Although there is limited understanding of its biological functions, evidence suggests that MV-bound DNA may serve in biofilm formation (Liao et al. 2014). In addition, RNA was recently detected in MVs from Streptococcus sp., however its role is yet to be elucidated (Choi et al. 2018; Resch et al. 2016). Overall, it remains unclear how nucleic acids are packaged into BMVs, however their functions in bacterial communications are becoming increasingly clear.
Collectively, it is clear that cargo packaging into BMVs serves to facilitate bacterial pathogenesis and contribute to disease pathology. In the following sections, we will further discuss the varied mechanisms by which the contents associated with OMVs and MVs drive pathology in the human host .
Physiological Roles of BMVs
Physiological Roles of OMVs Produced by Gram-Negative Bacteria
One of the earliest reviews about OMVs speculated that they possess bacteriolytic activity, binding capability, proteolytic activity and a possible role in genetic exchange (Mayrand and Grenier 1989). Thirty years later, researchers have unravelled many of their multi-faceted roles that facilitate pathogenesis, including inter-bacterial communication, biofilm formation, nutrient acquisition and horizontal gene transfer.
Biofilm Formation
Biofilm formation enables bacteria to thrive in a protected microenvironment and is therefore strongly linked to bacterial pathogenesis, resistance and survival (Whitchurch 2002). The role of OMVs in biofilm formation was first observed in 1987, whereby OMVs from the periodontal pathogen P. gingivalis were shown to facilitate adherence between bacterial cells (Grenier and Mayrand 1987). Almost 10 years later, this observation was confirmed by transmission electron microscopy imaging that revealed OMVs were common constituents of the extracellular matrix of P. aeruginosa biofilms (Schooling and Beveridge 2006). A clear link between biofilm formation and OMV production has since been demonstrated in a number of bacterial species (Yonezawa et al. 2017; Nakao et al. 2018; Esoda and Kuehn 2019). Furthermore, the contribution of OMVs to biofilm formation in vivo was demonstrated in a study that showed P. aeruginosa OMVs could fuse with mouse corneas to prime the ocular surface for bacterial adhesion and biofilm formation (Metruccio et al. 2016).
Recent studies have identified components of OMVs that influence biofilm formation, such as the major biofilm matrix proteins identified in V. cholerae OMVs (Altindis et al. 2014), leucine aminopeptidase PaAP in P. aeruginosa OMVs and outer membrane proteins of H. pylori OMVs (Altindis et al. 2014; Esoda and Kuehn 2019; Yonezawa et al. 2017). Furthermore, OMV-associated DNA forms part of the biofilm matrix and its negative charge confers adhesive properties to OMVs (Turnbull et al. 2016; Grande et al. 2015; Schooling et al. 2009). Advances in super-resolution microscopy have led to insights into the release of OMV-bound DNA within biofilms by explosive cell lysis (Turnbull et al. 2016). While these studies shed light on novel roles that OMVs serve in host colonization, more research is needed to understand how OMVs may regulate biofilm establishment.
Nutrient Acquisition
Bacteria are often faced with nutrient-limited conditions in their host, a hurdle which must be overcome to effectively establish colonisation. It is therefore not surprising that one of the earliest putative roles for OMVs was in nutrient acquisition (Forsberg et al. 1981; Thompson et al. 1985). Upregulation of OMV production is observed in nutrient-limited conditions, whilst bacterial enzymes required for nutrient acquisition including proteases, glycosidases and ion-chelators are common components of OMVs, suggesting that nutrient acquisition is a key function (Gerritzen et al. 2019; Kadurugamuwa and Beveridge 1995; Mashburn et al. 2005; Schaar et al. 2011; Elhenawy et al. 2014). However, there have been few studies investigating this aspect of OMV function. Studies have identified OMVs that possess enzymatic activity to liberate energy sources from the environment, such as lignin-degrading ability to liberate carbon and redox-reactivity to sequester metal ions (Salvachúa et al. 2020; Gorby et al. 2008; Subramanian et al. 2018). A recent study showed that the human opportunistic pathogen P. aeruginosa uses OMVs to sequester iron and supplement bacterial growth in iron-deficient media, suggesting that OMVs may play a role in iron acquisition in the host environment (Lin et al. 2017). In this study, the delivery of iron to bacteria occurred via receptor-mediated entry, a finding that sheds light on the poorly understood process of OMV entry into bacterial cells and suggests selectivity in nutrient delivery (Lin et al. 2017). Whilst nutrient acquisition by OMVs is a poorly understood process , it presents a novel therapeutic target to limit the growth of bacteria in host systems and therefore warrants further investigation.
Competition and Predation
In their natural environment, bacteria must compete with other species for nutrients and space. This is particularly important in the mixed microbial communities within the host, where bacteria can use BMVs to establish a niche in the host. OMVs can be trafficked out of biofilms (Schooling and Beveridge 2006) and therefore have the potential to serve as long distance antimicrobials against competing species. In addition to their role in inter-species competition, OMVs are also utilised by predatory bacteria to lyse surrounding bacterial species (Berleman et al. 2014; Evans et al. 2012; Vasilyeva et al. 2008; Schulz et al. 2018). The lytic function of OMVs is conferred by peptidoglycan hydolases that are delivered to target cells by membrane fusion and are capable of destroying the cell wall of Gram-positive and Gram-negative species (Li et al. 1998; Kadurugamuwa and Beveridge 1996; Kadurugamuwa et al. 1998). However, rather than indiscriminately lysing all bacteria, peptidoglycan hydrolases contained within OMVs possess specificity in their mode of action by targeting the peptidoglycan of particular bacterial species (Li et al. 1998). The antimicrobial specificity of OMVs towards certain species is of relevance for the development of novel, targeted OMV-based antibiotics; a potential application of OMVs that remains largely unexplored (Schulz et al. 2018). Overall, there is much to uncover about the antimicrobial properties of OMVs in maintaining homeostasis within mixed microbial communities, particularly in the context of the microbiome, and their possible application as novel antibiotics.
Bacterial Defence
Bacteria have developed sophisticated ways to use OMVs to evade recognition and attack by the host immune system. OMV production increases in response to the host environment (Martinez et al. 2019) and early studies revealed that they confer protection against bacteriolytic factors in human serum (Grenier and Bélanger 1991; Pettit and Judd 1992b). Proteolytic enzymes, LPS and outer membrane proteins carried by OMVs are involved in their ability to confer serum resistance (Grenier and Bélanger 1991; Pettit and Judd 1992a; Lekmeechai et al. 2018). OMV-mediated serum resistance has been implicated in facilitating co-infections of Moraxella catarrhalis and H. influenzae (Tan et al. 2007), as well as facilitating the spread of the etiological agent of infective endocarditis, Aggregatibacter actinomycetemcomitans, through the bloodstream (Lindholm et al. 2020).
OMVs can also protect bacteria against antimicrobial treatment, by harbouring enzymes that neutralise antibiotics or by serving as decoys to sequester antimicrobials away from bacteria (Roszkowiak et al. 2019; Stentz et al. 2015). There are many reports identifying the presence of antibiotic-degrading β-lactamase enzymes within OMVs , a feature which is also linked to bacterial virulence (Yun et al. 2018; Stentz et al. 2015; Ciofu et al. 2000; Giwercman et al. 1992). Antibiotic treatment has also been demonstrated to increase OMV production (Devos et al. 2017; Manning and Kuehn 2011), while blocking OMV release can increase bacterial susceptibility to antibiotics (Kosgodage et al. 2019), indicating that OMVs contribute to antibiotic resistance. P. aeruginosa OMVs harbouring enzymatically active β-lactamase have been found to form a layer around P. aeruginosa biofilms in the lungs of cystic fibrosis patients, thereby protecting the underlying bacteria from antibiotics (Giwercman et al. 1992). OMVs have also been shown to confer inter-species protection against antibiotics in microbial communities, such as in the gastrointestinal tract (Stentz et al. 2015; Roszkowiak et al. 2019). These findings highlight the clinical importance of characterising the antimicrobial neutralising properties of OMVs and determining the bacterial strains that contain antimicrobial enzymes in order to understand their role in antibiotic resistance.
Additionally, OMVs can protect bacteria against infection by bacteriophages, which are ubiquitous in all natural and host environments (Barr et al. 2013). Specifically, OMVs can sequester phages to defend bacteria from infection, although there are limited studies demonstrating this interaction. Proteomic profiling has revealed OMVs contain phage-targeting receptors (Lee et al. 2007), and the rapid and irreversible binding of phages to OMVs have been observed by electron microscopy (Manning and Kuehn 2011; Biller et al. 2014). V. cholerae OMVs contain phage receptors that bind and inactivate phages naturally found in human stools, giving evidence of phage-OMV interaction in host environments (Reyes-Robles et al. 2018). Collectively, these studies suggest that OMVs play a protective role against bacteriophages , particularly in phage-rich environments such as the gastrointestinal tract. However, the interplay between OMVs and phages in host environments remains poorly understood and requires further research.
Horizontal Gene Transfer
OMVs are known to carry DNA that is protected from nuclease digestion and trafficked out of biofilms, suggesting the prospect of their far-reaching dissemination of virulence-related genes and their role in HGT (Schooling et al. 2009; Dorward and Garon 1989). OMV-mediated transfer of virulence genes was demonstrated in studies showing E. coli OMVs could transfer plasmid DNA to S. enterica serovar Enteritidis, thereby conferring cytotoxic activity and antibiotic resistance to recipient bacteria (Yaron et al. 2000). A similar study demonstrated OMV-mediated transfer of plasmid DNA containing antibiotic resistance genes from Acinetobacter baylyi to E. coli (Fulsundar et al. 2014). The potential for OMVs to facilitate wide-spread HGT is highlighted in a study that demonstrated A. baumannii OMVs transferred plasmids carrying virulence and antibiotic resistance genes to stably transform recipient strains. These transformed recipient strains were subsequently able to act as donors, producing OMVs laden with the acquired plasmid and transforming other bacteria (Rumbo et al. 2011). The rate and efficiency of OMV-mediated transformation has been suggested to be similar to that of bacterial transduction, adding further credence to its biological significance (Tran and Boedicker 2017). However, there appears to be differences in the genetic transformation potential of OMVs from different bacteria (Renelli et al. 2004; Klieve et al. 2005; Tran and Boedicker 2017), which may be due to factors such as restriction modifications conferred by the donor strain, OMV charge and natural competency of the recipient strain (Klieve et al. 2005; Tran and Boedicker 2017; Fulsundar et al. 2014; Tashiro et al. 2017). Collectively, these studies highlight the potential biological relevance of OMVs in HGT, however further studies are required to define these processes in vivo.
Physiological Roles of MVs Produced by Gram-Positive Bacteria
There is little known about the non-pathogenic functions of MVs produced by Gram-positive bacteria. Limited reports suggest they serve similar roles to OMVs in biofilm formation, nutrient acquisition, inter-bacterial communication and bacteria defence. However, more studies are required to build an understanding of the functions of MVs in microbial populations.
Membrane vesicles produced by S. aureus and S. mutans have been identified in biofilms (He et al. 2017; Liao et al. 2014). S. aureus MVs have also been shown to enhance bacterial adhesion and mediate cell aggregation to promote biofilm formation (He et al. 2017). Moreover, MV release by S. aureus increased upon treatment with vancomycin, which corresponded with an increase in biofilm formation (He et al. 2017). Biofilm formation by S. mutants can also be enhanced by the release of extracellular DNA associated with MVs, which aids in facilitating adhesion to surfaces (Liao et al. 2014). Similarly, E. faecium MVs contain biofilm-promoting proteins and extracellular matrix-binding proteins that point toward their role in supporting biofilm formation (Wagner et al. 2018).
The proteomic profiles of MVs from a variety of Gram-positive species suggest they function in nutrient acquisition. S. aureus MVs contain iron-binding factors that indicate a role in iron sequestration (Lee et al. 2009) and Streptomyces coelicolor MVs are enriched in proteins involved in the acquisition of phosphates, iron and carbon sources (Schrempf et al. 2011). Similarly, Mycobacterium tuberculosis MVs package the iron-binding compound mycobactin and have been shown to directly participate in the delivery of iron to recipient cells to support their growth in iron-limited conditions (Prados-Rosales et al. 2014).
Although MVs have not been directly implicated in inter-species competition, their contents points to a possible role in lysis of competing species. B. subtilis MVs contain endolysins that are known to degrade the cell wall of bacteria (Toyofuku et al. 2017), and E. faecium MVs contain glycopeptides that hydrolyse bacterial peptidoglycan (Wagner et al. 2018). Similarly, proteomic analysis of S. aureus, Streptococcus pneumoniae and Streptococcus suis MVs show they contain autolysins that also degrade bacterial peptidoglycan (Haas and Grenier 2015; Lee et al. 2009; Olaya-Abril et al. 2014; Wang et al. 2018). Clearly further studies are needed to further explore the antimicrobial role of MVs.
MVs can also contain enzymes that hydrolyse antibiotics and there is evidence that MVs play a role in bacterial defence against antibiotic treatment and host immune attack (Andreoni et al. 2019; Kosgodage et al. 2019; Lee et al. 2013; Wagner et al. 2018). S. aureus MVs reduce bacterial lysis in whole-blood, suggesting that they can protect bacteria against host innate immune molecules (Andreoni et al. 2019). E. faecium MVs contain eleven antimicrobial resistance-related proteins, including proteins that confer vancomycin resistance (Wagner et al. 2018), while β-lactamase carried by S. aureus MVs enables protection of bacteria, such as E. coli, S. enterica serovar Enteritidis and other S. aureus strains against ampicillin (Lee et al. 2013). S. aureus MVs can also confer resistance against the last-resort antibiotic daptomycin, highlighting the clinical relevance of MVs in promoting antibiotic resistance (Andreoni et al. 2019). Inhibition of bacterial vesiculation may be a potential target to increase the efficacy of antibiotics, as a recent study demonstrated that inhibition of S. aureus MV production corresponded with an increased susceptibility to a wide range of antibiotics (Kosgodage et al. 2019).
Gram-positive MVs are reported to contain DNA, however little is known about the forms of DNA they carry and their role in HGT (Klieve et al. 2005; Liao et al. 2014; Sisquella et al. 2017). A study examining MVs from the bovine rumen-derived bacteria Ruminococcus albus was the first to demonstrate HGT by MVs (Klieve et al. 2005). DNA associated with R. albus MVs was genomic in origin and contained genes encoding for cellulose-digesting enzymes (Klieve et al. 2005). MV-mediated HGT conferred cellulase activity to strains that previously lacked cellulase activity, and this trait was stable and inheritable in subsequent bacterial generations (Klieve et al. 2005). However, R. albus MVs were not able to transform the rumen-derived Butyrivibrio fibrisolvens, suggesting a possible mechanism for selective transformation of species (Klieve et al. 2005). This study is the first to indicate a role for MV-mediated HGT in microbial communities (Klieve et al. 2005). However, further studies are required to substantiate these findings in other Gram-positive species.
We have merely touched the surface of understanding the multi-faceted functions of MVs in interbacterial communication, biofilm formation and bacterial defence that facilitate pathogenesis. Future research is needed to provide a greater understanding of the ways in which Gram-positive bacteria utilise MVs to their advantage, and to shed light on their possible antimicrobial targets and therapeutic applications.
Pathogen-Derived BMVs Can Modulate the Host Immune Response
Since the discovery of BMVs more than 50 years ago, research investigating their functions has largely centred upon their role in promoting pathogenesis and their detrimental effects on human health. Several clinically relevant Gram-negative and Gram-positive pathogens have been shown to utilise OMVs and MVs as vehicles to deliver virulence effectors to host cells (Lindmark et al. 2009; Wai et al. 2003; Elluri et al. 2014; Thay et al. 2013; Jin et al. 2011; Fiocca et al. 1999; Vdovikova et al. 2017; Rivera et al. 2010). Packaging of bacterial toxins into OMVs and MVs allows pathogenic bacteria to access distal sites within the body without directly translocating away from their environmental niche.
Pathogenic Functions of BMVs at the Mucosal Surface
Pathogenic BMVs Can Disrupt the Epithelial Barrier
The human body is protected from the external environment by skin and the mucosal surfaces of the respiratory, gastrointestinal and urogenital tracts. These mucosal surfaces are important interfaces supporting host-microbe interactions; however, in many instances they are also the site of first contact with pathogens. Whilst some pathogenic bacteria are invasive intracellular pathogens, many bacteria mediate their virulence extracellularly whilst remaining within the mucosa (reviewed in (Acheson and Luccioli 2004)). Such bacteria have developed means to elicit pathogenic effects, via the secretion of soluble proteins or using complex secretion systems that act as a molecular syringe to ‘inject’ virulence factors inside host epithelial cells, or via the secretion of OMVs or MVs to deliver cargo to the host epithelium without the need for direct cell to cell contact (reviewed in (Thanassi and Hultgren 2000; Kulp and Kuehn 2010)).
Pathogen-derived OMVs and MVs may contain cargo including toxins and immunostimulatory molecules that induce cellular reprogramming and disrupt the integrity of epithelial barriers. For example, H. pylori is a Gram-negative pathogen that infects approximately 50% of the world’s population, causing chronic inflammation and peptic ulceration (reviewed in (Correa and Piazuelo 2008)). OMVs from H. pylori have been studied extensively as they perform numerous functions in facilitating the pathogenesis of their parent bacterium. H. pylori OMVs can damage the gastric epithelium via the disruption of tight junction (TJ) proteins, such as zonula occludens (ZO)-1, that act to maintain paracellular junctions (Turkina et al. 2015). Furthermore, H. pylori OMVs containing the oncogenic virulence factor ‘cytotoxin-associated gene A’ (cagA) induce the redistribution of ZO-1 from paracellular junctions to the cytoplasm, resulting in increased epithelial permeability in vitro (Turkina et al. 2015).
Campylobacter jejuni is another Gram-negative intestinal pathogen whose OMVs were demonstrated to disrupt epithelial integrity. For example, treating the intestinal epithelial cell lines T84 and Caco-2 with C. jejuni OMVs was sufficient to induce cleavage of adherens protein E-cadherin and TJ protein occludin (Elmi et al. 2016). Notably, the disruption of intestinal epithelial cells by C. jejuni OMVs allowed for increased invasiveness of their parent bacteria, indicating that the production of OMVs was directly facilitating bacterial pathogenesis. Periodontal pathogens Treponema denticola and P. gingivalis also produce OMVs that have been shown to disrupt and dislodge epithelial cell layers, allowing their parent bacteria to invade into the underlying tissue and cause further damage to host tissues (Chi et al. 2003; Nakao et al. 2014).
Another mechanism used by pathogenic bacteria to disrupt the epithelial cell barrier is to cause morphological changes to the cytoskeleton of host epithelial cells. Many pathogens are known to elicit this effect via Type I-VI secretion systems or by the secretion of soluble toxins, however little is known about the roles of OMVs in cytoskeletal rearrangements at the epithelial cell barrier. One prominent pathogen whose OMVs have been investigated is Bordetella pertussis, the causative agent of whooping cough. Interestingly, B. pertussis OMVs have been isolated post-mortem from a patient with lethal B. pertussis infection (Donato et al. 2012). B. pertussis OMVs were found to contain adenylate cyclase toxin-hemolysin (CyaA), which is responsible for decreasing epithelial barrier integrity, disrupting TJ proteins and upregulating host cell cyclic AMP (cAMP) signalling, resulting in rearrangement of the actin cytoskeleton (Donato et al. 2012; Hasan et al. 2018). The effects mediated by OMV-associated CyaA toxin facilitate bacterial translocation across the airway epithelium, demonstrating a role for B. pertussis OMVs in contributing to the pulmonary inflammation observed during whooping cough disease.
BMVs Can Enter Host Epithelial Cells
As well as mediating pathogenic effects by disrupting epithelial layers, BMVs can also enter epithelial cells, thereby releasing their cargo intracellularly and modulating host cell metabolism (reviewed in (O’Donoghue and Krachler 2016; Kaparakis-Liaskos and Ferrero 2015)). Several routes of cell entry have been suggested for BMVs from different bacterial species. OMV entry into host cells can occur via cholesterol-rich lipid rafts in the host cell membrane. This was first demonstrated using P. aeruginosa OMVs, whereby OMVs entered lung epithelial cells in a cholesterol-dependent manner (Bauman and Kuehn 2009). Lipid raft-independent mechanisms of BMV endocytosis have also been described, including macropinocytosis whereby the host cell cytoskeleton engulfs extracellular BMVs, or by receptor-mediated formation of clathrin or caveolin pits (Bomberger et al. 2009; Furuta et al. 2009; Pollak et al. 2012; Bielaszewska et al. 2013; Kunsmann et al. 2015; Vanaja et al. 2016; Kesty et al. 2004; Chatterjee and Chaudhuri 2011; Schaar et al. 2011; Wang et al. 2020; Turner et al. 2018). Direct fusion of the OMV membrane with the host cell membrane has also been observed for membrane vesicles from multiple species (Bomberger et al. 2009; Rompikuntal et al. 2012; Jäger et al. 2015). The specific endocytic mechanisms utilised by BMVs to enter host cells have been discussed extensively elsewhere and therefore will not be explored in great detail here (reviewed in (O’Donoghue and Krachler 2016)).
Interestingly, OMVs or MVs produced by the same species can enter host cells using various endocytic pathways. A study examining the mechanisms used by H. pylori OMVs to enter host cells revealed that small (20–100 nm) and large (90–450 nm) OMVs enter gastric epithelial cells via distinct pathways (Turner et al. 2018). Furthermore, it has been reported that the composition of OMVs can also alter their mechanism of uptake by host cells. Notably, H. pylori OMVs harbouring the vacuolating cytotoxin (VacA) virulence factor entered gastric epithelial cells via a different mechanism of endocytosis compared to OMVs that did not package VacA toxin (Parker et al. 2010). Furthermore, the lipid composition of OMVs produced by different E. coli serotypes has been reported to affect the mechanism and kinetics of OMV-mediated entry into HeLa cells (O’Donoghue et al. 2017). In contrast to knowledge identifying that OMV entry into host cells can be regulated by their cargo or size, very few studies have investigated whether size and cargo composition of MVs produced by Gram positive bacteria can also determine their mechanisms of cellular entry. However, it is known that MVs produced by the Gram-positive pathogens B. anthracis and S. aureus are endocytosed by host cells via different pathways, entering via receptor-mediated and dynamin-dependent pathways, respectively (Rivera et al. 2010; Wang et al. 2020). These studies suggest that MVs also have the potential to enter host cells via a range of endocytic mechanisms.
BMVs Mediate Innate Immune Signalling in Epithelial Cells
OMVs and MVs produced by pathogens are loaded with immunostimulatory cargo from their parent bacteria, known as MAMPs, including molecules such as LPS, PG and nucleic acids. BMV-associated MAMPs are recognised by host innate immune pathogen recognition receptors (PRRs), which detect foreign molecules and elicit a cytokine-mediated immune response, recruiting immune cells to the site of infection. Some of the PRRs at the epithelial barrier include the transmembrane Toll-like receptors (TLRs) and cytoplasmic nucleotide-binding oligomerisation domain (NOD) receptors. Innate immune receptors of both the TLR and NOD-like receptor (NLR) families have been identified as key responders to BMVs and subsequently have important roles in OMV and MV-mediated pathologies which we discuss below.
TLRs at the Apical and Endosomal Membranes Detect BMVs
Epithelial cells expressing Toll-like receptors are the innate immune system’s first responders to pathogenic microbes. The TLR family constitutes 10 transmembrane receptors, which can be located on the apical membrane of epithelial cells, detecting MAMPs such as LPS, flagella and lipoproteins associated with bacterial cells or BMVs (reviewed in (Janeway Jr and Medzhitov 2002)). Ligand-mediated TLR activation leads to the recruitment of adaptor proteins such as Myeloid Differentiation primary response protein (MyD)-88, inducing a signal cascade that eventuates in the phosphorylation and nuclear translocation of pro-inflammatory transcription factors including nuclear factor kappa B (NF-κB) (reviewed in (Akira et al. 2006)). These transcription factors subsequently mediate the secretion of inflammatory cytokines from epithelial cells, which is critical for the recruitment of immune cells to the site of infection.
As BMVs produced by pathogenic bacteria are loaded with MAMPs from their parent bacteria, they are capable of stimulating a pro-inflammatory immune response in the host (Fig. 6.2). This was first reported by Ismail et al. who identified a fundamental role for H. pylori OMVs in stimulating an interleukin (IL)-8-mediated inflammatory response by gastric epithelial cells in vitro (Ismail et al. 2003). Many other pathogen derived BMVs have since been discovered to elicit inflammatory immune responses by epithelial cells . One clinically relevant example is the opportunistic pathogen P. aeruginosa, whose OMVs have a well characterised role in promoting inflammatory responses in the host. OMVs produced by P. aeruginosa isolates obtained from patients with cystic fibrosis induced IL-8 secretion by primary human bronchial epithelial cells (Bauman and Kuehn 2006). P. aeruginosa OMVs were also shown to engage human epithelial cells in a TLR4-dependent manner, resulting in the increased expression of TLR4 in a positive-feedback mechanism to drive further inflammation, while also increasing host expression of MyD88, NF-κB and IL-1β (Zhao et al. 2013). Thus, it is proposed that the aberrant inflammation driven by OMVs during P. aeruginosa infection may contribute to the pathology mediated by this opportunistic pathogen.
Bacterial membrane vesicles (BMVs) facilitate bacterial pathogenesis via many mechanisms. BMVs can interact with TLRs at the apical membrane of host epithelial cells or enter host cells and interact with TLRs in the endosomal lumen. Intracellular BMVs can also release their cargo resulting in the activation of cytoplasmic NOD receptors. These processes lead to the phosphorylation and nuclear translocation of the transcription factor NF-κB, resulting in the production and release of pro-inflammatory cytokines including IL-6, IL-8 and TNF-α. Additionally, BMVs can disrupt TJ proteins, resulting in a loss of epithelial barrier integrity and ultimately allowing BMVs and bacteria to cross the epithelial barrier. In this way, BMVs can indirectly and directly modulate innate and adaptive immune cell functions. However, BMVs may also deliver bacterial sRNA to host epithelial cells, which upon binding to host mRNA, may dampen host inflammatory responses
TLRs are also located at the endosomal membrane and are capable of detecting microbial products that have entered intracellularly. Once endocytosed by epithelial or immune cells, microbial products are trafficked to endosomes where they are processed for degradation (Nakamura et al. 2014). Similarly, upon entry into host epithelial cells, OMVs produced by Gram-negative bacteria can be degraded via the host cellular degradation pathway of autophagy (Fig. 6.2) (Irving et al. 2014). OMV-mediated autophagy was found to be essential for the initiation of an inflammatory immune response to OMVs (Irving et al. 2014). In addition, OMVs may also be detected by intracellular TLRs -7, −8 and − 9, which respond to microbial nucleic acids (Fig. 6.2) (Cecil et al. 2016; Akira et al. 2006). Recent studies investigating the PRRs activated by OMVs produced by the three prominent periodontal pathogens P. gingivalis, T. denticola and Tannerella forsythia revealed that they could all induce inflammatory immune responses by host immune cells, characterised by the release of IL-1β, as well as tumour necrosis factor (TNF)-α and IL-8 (Cecil et al. 2017). Moreover, the activation of TLR7, TLR8 and TLR9 by P. gingivalis and T. forsythia OMVs were found to play a direct role in pathogen detection and the subsequent generation of an inflammatory immune response by the host (Cecil et al. 2016).
Bacterial sRNA Delivery by OMVs and MVs
The delivery of bacterial RNA to host cells via the release of membrane vesicles is an emerging and exciting field. RNA was first identified within OMVs only 5 years ago, though the importance of RNA delivery by BMVs has quickly become apparent (Sjöström et al. 2015). Bacterial small RNA (sRNA), which is similar in size to eukaryotic micro (mi)RNA, can regulate gene expression by binding to host cell messenger RNA (mRNA) (reviewed in (Repoila and Darfeuille 2009)). Furthermore, recent evidence suggests bacterial pathogens differentially package sRNAs into OMVs and MVs to modulate the host immune response. Notably, P. aeruginosa OMVs differentially package regulatory sRNAs that can be stably transferred into human airway epithelial cells (Koeppen et al. 2016). It was reported that P. aeruginosa OMVs containing sRNA52320 were able to downregulate both LPS-mediated and OMV-mediated inflammatory cytokine production by airway epithelial cells. Furthermore, OMVs carrying sRNA52320 attenuated OMV-induced cytokine secretion and neutrophil infiltration in vivo (Koeppen et al. 2016). The immunomodulatory roles of OMV-associated sRNAs may also extend to adaptive immune responses, whereby sRNAs associated with OMVs produced by several periodontal pathogens were isolated, characterised and their immunological effects on T cells were investigated (Choi et al. 2017). Isolation of these sRNAs and their transfection into Jurkat T cells resulted in reduced basal expression of IL-5, IL-13 and IL-15 compared to cells that were transfected with a non-coding sRNA as a control (Choi et al. 2017). The authors proposed that OMV-mediated delivery of sRNAs facilitate the pathogenic functions of their parent bacterium (Choi et al. 2017).
More recently, a role for small non-coding RNAs (sncRNA) in H. pylori pathogenesis and evasion of the host immune system has been identified (Zhang et al. 2020). The authors demonstrated that two sncRNA species, sR-2509025 and sR-989262 were enriched in H. pylori OMVs compared to their parent bacterium (Zhang et al. 2020). With the use of deletion mutant strains, it was shown that OMVs produced by H. pylori strains lacking these sncRNA species induced a heightened immune response when introduced to gastric epithelial cells, compared to OMVs produced by the wild-type strain that were enriched with these immunomodulatory sncRNA species (Zhang et al. 2020). Therefore, it was suggested that differential packaging of particular sncRNA into OMVs is a strategy employed by H. pylori to dampen host immunity and promote long-term persistence in the host.
Finally, the delivery of sRNA to host cells by BMVs is not unique to Gram-negative bacteria. The Gram-positive species group A streptococcus differentially regulates RNA packaging into MVs (Resch et al. 2016). However, very few studies have investigated the biological significance of RNA packaging into MVs produced by Gram-positive bacteria and this remains an interesting avenue for further research.
Intracellular Activation of NLRs
The cytoplasmic innate immune receptors NOD1 and NOD2 are responsible for the detection of bacterial PG, a complex molecule constituting the cell walls of both Gram-negative and Gram-positive bacteria (Girardin et al. 2003a, b). Examination of OMVs produced by the mucosal pathogens H. pylori, P. aeruginosa and N. gonorrhoeae revealed that OMVs could package PG that could be detected by the host innate immune receptor NOD1 (Kaparakis et al. 2010). Specifically, it was identified that once OMVs entered human epithelial cells, PG contained within OMVs was detected by NOD1, resulting in the induction of a NOD1-dependent pro-inflammatory immune response (Fig. 6.2) (Kaparakis et al. 2010). Furthermore, NOD1 was also found to be essential for the generation of OMV-specific adaptive immune responses in vivo, as oral administration of H. pylori OMVs to mice lacking the NOD1 receptor did not result in the induction of innate and OMV-specific adaptive immune responses (Kaparakis et al. 2010). Collectively, these findings revealed that OMVs are a conserved mechanism used by Gram-negative pathogens to mediate NOD1-driven inflammation and pathology in vivo (Kaparakis et al. 2010). Subsequently, studies also identified the ability of NOD1 and NOD2 to mediate immune responses to both OMVs produced by A. actinomycetemcomitans, V. cholerae and MVs from Gram-positive S. aureus, amongst other bacteria (Thay et al. 2014; Bielig et al. 2011; Jun et al. 2017).
BMVs can also interact with another member of the NLR family known as NLR-pyrin domain containing 3 (NLRP3) (Wang et al. 2020; Cecil et al. 2017; Fleetwood et al. 2017; Bitto et al. 2018). Activation of NLRP3 by OMVs or MVs leads to the formation of an intracellular complex termed the inflammasome, a protein complex responsible for the maturation of caspase-1 and the subsequent secretion of mature IL-1 family cytokines including IL-1β and IL-18 (reviewed in (Schroder et al. 2010)). For example, S. aureus MVs were internalised by human macrophages which resulted in activation of the NLRP3 inflammasome via K+ efflux (Wang et al. 2020). Additionally, P. gingivalis OMVs were found to induce inflammasome activation both in vitro and in vivo, leading to recruitment of inflammasome adaptor proteins and the secretion of mature IL-1β from murine peritoneal macrophages (Cecil et al. 2017). Collectively, these studies highlight that there are a broad range of PRRs and multiple mechanisms by which BMVs mediate the induction of pro-inflammatory responses in the host.
BMVs can cross the epithelial cell barrier
At the mucosal interface, BMVs produced by pathogenic bacteria may face various fates. As described above, OMVs and MVs can interact with epithelial cell surface receptors, or they may be internalised by host cells and release their cargo into the cell cytoplasm. However, BMVs can also directly cross the epithelial layer and access immune cells within the submucosa (Fig. 6.2) (Kaparakis-Liaskos and Ferrero 2015). It has been hypothesised that BMVs can cross epithelial barriers by different methods including transcytosis or by diffusion through paracellular junctions (reviewed in (Stentz et al. 2018)). Once the epithelial barrier has been crossed, bacterial OMVs and MVs can modulate innate immune cells, which can subsequently affect T and B cell functions and ultimately manipulate host immunity.
BMVs and Macrophages
Macrophages are innate immune cells which survey host tissues and rapidly detect and respond to invading pathogens and their secreted products. Macrophages can phagocytose extracellular bacterial products including BMVs, resulting in the production of inflammatory cytokines to ultimately facilitate the induction of a pathogen-specific adaptive immune response (reviewed in (Arango Duque and Descoteaux 2014)). However, some pathogens can specifically target macrophages to facilitate the onset of pathogenesis in the host. One such bacterium that mediates pathogenesis by macrophages is P. gingivalis which produces OMVs that can interact with macrophages in the subgingival cavity (Imayoshi et al. 2011). It was found that P. gingivalis OMVs induced the production of inflammatory mediators such as nitric oxide (NO) and inducible nitric oxide synthase (iNOS) by murine macrophages in vitro (Imayoshi et al. 2011). Furthermore, S. Typhimurium OMVs were also found to interact with murine macrophages via their ability to induce the production of TNF and NO production, whilst OMVs from a range of periodontal pathogens induced the secretion of TNF-α, IL-8 and IL-1β from THP-1 monocytes (Alaniz et al. 2007; Cecil et al. 2017). Additionally, BMVs may deliver their cytotoxic cargo to macrophages, as is the case with OMVs produced by the pathogen A. baumannii (Jin et al. 2011). Specifically, OMVs produced by wild-type A. baumannii were found to package the virulence effector protein AbOmpA, resulting in a significant increase in cytotoxic cell death of U937 macrophage-like cells compared to OMVs isolated from ΔOmpA mutant A. baumannii strains lacking AbOmpA (Jin et al. 2011).
Whilst BMVs facilitate pathogenesis by driving inflammation in the host to promote tissue damage, they can also downregulate the immune response generated by macrophages to prolong bacterial survival within the host. For example, OMVs produced by H. pylori induce anti-inflammatory IL-10 secretion by human peripheral blood mononuclear cells, whilst P. gingivalis OMVs can degrade the CD14 receptor, rendering macrophages incapable of responding to bacterial LPS and thus avoiding hyperinflammatory immune responses (Winter et al. 2014; Duncan et al. 2004).
In contrast to BMVs produced by gastrointestinal or periodontal pathogens , BMVs produced by respiratory pathogens can interact with alveolar macrophages. For example, Legionella pneumophila, a causative agent of severe pneumonia, produces OMVs that induce TLR2- and NF-κB-dependent increases in inflammatory cytokine production by alveolar macrophages (Jung et al. 2016). However, by contrast, prolonged exposure to L. pneumophila OMVs may facilitate bacterial replication within alveolar macrophages by downregulating host immune responses (Jung et al. 2016). Similarly, M. tuberculosis is a chronic intracellular pathogen that infects alveolar macrophages (reviewed in (Pieters 2008)). EVs secreted by M. tuberculosis-infected macrophages are immunostimulatory to the human host, however it was unclear until recently whether this immunogenicity was attributed to host-derived exosomes containing bacterial cargo or directly attributed to BMVs emanating from the intracellular bacterium (Athman et al. 2015). Using density-gradient separation to resolve prokaryotic BMVs from eukaryotic EVs, it was elucidated that M. tuberculosis BMVs are responsible for this immunogenicity (Athman et al. 2015). Collectively, these studies highlight the roles of BMVs in promoting bacterial pathogenesis via their interactions with macrophages (Fig. 6.2).
BMVs and Neutrophils
The interactions between BMVs and neutrophils can be categorised into two groups: immunomodulatory effects and Neutrophil Extracellular Trap (NET)-related. Foremost, neutrophils are involved in inflammatory immune responses as they migrate to the site of infection in response to chemoattractant cytokines and chemokines release by epithelial cells. However, BMVs can also directly mediate inflammatory cytokine production by neutrophils, as demonstrated using N. meningitidis OMVs and S. aureus MVs (Lapinet et al. 2000; Askarian et al. 2018). Second, NETs are released by neutrophils upon cell death, whereby fibrillar networks composed of bacterial DNA and antimicrobial peptides (AMPs) extend from the cell membrane to trap and kill surrounding pathogens, preventing their spread throughout the body (reviewed in (Papayannopoulos 2018)). The pathogens S. pneumoniae and P. aeruginosa are captured by NETs to prevent their dissemination through the host, however they have evolved mechanisms to evade NET entrapment via the secretion of BMVs (Jhelum et al. 2018; Shan et al. 2014). Specifically, S. pneumoniae packages the extracellular DNase TatD into its MVs to degrade the structure of neutrophil NETs, whereas OMVs produced by highly cytotoxic strains of P. aeruginosa bind NETs to reduce the available amount of NET that can bind to the parent bacteria (Jhelum et al. 2018; Shan et al. 2014). Overall, BMVs can interact with neutrophils to potentiate disease pathologies by eliciting inflammatory immune signalling, or they may subvert normal immune responses by disrupting NET formation and thereby modulating the host’s response to infection.
BMVs and DCs
Dendritic cells (DCs) are professional antigen presenting cells and are considered to bridge the innate and adaptive immune systems. One of the earliest reports of DC activation by BMVs was demonstrated using the pathogen S. Typhimurium (Alaniz et al. 2007). The authors showed that S. Typhimurium OMVs induced the maturation of murine bone marrow-derived DCs (BMDCs) and stimulated pro-inflammatory cytokine signalling (Alaniz et al. 2007). Interestingly, it has also been reported that the commensal bacterium B. fragilis produces OMVs that can interact with DCs, inducing anti-inflammatory immune signalling which may represent an important regulator of host immunity by the microbiota (Shen et al. 2012; Chu et al. 2016). These studies suggest that the outcome of the interactions between OMVs and DCs may be dependent on the pathogen from which the OMVs are derived. By contrast, there is still very limited knowledge regarding the interactions between DCs and MVs released by Gram-positive bacteria and how these may modulate immunity in the host.
BMVs and Adaptive Immune Cells
The innate immune system functions as an early response to mitigate infection, whereas adaptive immunity functions to provide the host with long term protection from pathogens. The following section will briefly describe the ability of BMVs to modulate T cell and B cells responses of the humoral immune system.
While direct interactions between humoral B cells and BMVs are not yet well elucidated, BMVs from multiple pathogens have been implicated in modulating B cell responses. OMVs from pathogens including N. meningitidis and M. catarrhalis have been shown to promote B cell activation via OMV-associated DNA delivery to intracellular TLR9, and to induce the production of long-term memory B cells, respectively (Vidakovics et al. 2010; Romeu et al. 2014). More recent studies examining the efficacy of BMV-based vaccines have demonstrated significant antibody production by B cells in response to OMV or MV administration (Wang et al. 2017; Stevenson et al. 2018).
By contrast, the roles of T cells in BMV-mediated immune responses are better understood. T cells are critical in the response to pathogenic infection and several studies have indicated that BMVs from various pathogens can induce T cell activation (Alaniz et al. 2007; Kim et al. 2013; Schetters et al. 2019). However, in contrast, chronic pathogens including H. pylori and M. tuberculosis release BMVs which can inhibit T cell proliferation and function. For example, H. pylori OMVs can induce apoptosis of Jurkat T cells or supress T cell function in an IL-10 and cyclo-oxygenase-2 (COX-2)-dependent manner (Winter et al. 2014; Hock et al. 2017). Similarly, M. tuberculosis vesicles inhibit CD4+ T cell responses to facilitate bacterial survival in vivo (Athman et al. 2017). Collectively, these studies and others have demonstrated that BMVs modulate cellular immune responses . The therapeutic potential of BMVs for use in stimulating protective immunity against bacterial infections will be covered in greater detail later in this chapter.
Microbiota-Derived BMVs Modulate Immune Responses
The human microbiota is a community of organisms that inhabit various sites throughout the body. This complex community is comprised of bacteria, viruses, fungi and bacteriophages whose metabolic functions are closely linked to the environment they inhabit. In particular, both Gram-negative and Gram-positive bacteria occupy niches within the gastrointestinal tract, lungs, skin, urinary tract and other such mucosal surfaces. The mutualistic relationships between host and microbiota are broad, whereby the microbiota facilitates the human host in the acquisition of nutrients and the maintenance of immunological homeostasis. Interestingly, recent evidence suggests that microbial secreted factors, including OMVs and MVs, are key determinants in many of the health benefits conferred by the microbiota.
BMVs in Host-Microbiota Interactions at Mucosal Surfaces
Membrane vesicles produced by the microbiota are able to gain direct access to the adjacent epithelium, whereas their parent bacteria may be physically inhibited by mucus barriers. Akin to pathogen-derived OMVs, epithelial cells can endocytose commensal OMVs resulting in the release of their content into the host cell cytoplasm (Cañas et al. 2016; Cañas et al. 2018; Jones et al. 2020). The cargo associated with commensal OMVs and MVs is constituted of molecules derived from their parent bacterium, including membrane components such as LPS and PG which can be immunogenic in the host. However, despite ongoing production of BMVs by commensal bacteria, they are not regarded to elicit uncontrolled hyper-inflammatory immune responses within the host during normal conditions. Rather, commensal-derived BMVs can induce both pro- and anti-inflammatory immune responses by the host, contributing to immunological homeostasis.
OMVs produced by commensal E. coli strain ‘C25’ were identified to elicit moderate IL-8 secretion from multiple intestinal epithelial cell lines in vitro (Patten et al. 2017). However, pre-treatment of the intestinal cell line Caco-2 with E. coli C25 OMVs inhibited subsequent internalisation of live bacteria by Caco-2 cells, indicating that OMV-mediated priming of the intestinal epithelium may confer protection against challenge by invasive bacteria (Patten et al. 2017). The authors suggested that the mild inflammatory response induced by these commensal OMVs mimicked the homeostatic conditions of the healthy intestine, implicating a role for OMVs in immunological maintenance in the gut.
Membrane vesicles from other commensal and probiotic E. coli strains have also been studied extensively to elucidate their functions in promoting intestinal homeostasis. For example, OMVs produced by the probiotic E. coli strain Nissle 1917 and commensal E. coli strain ECOR12 are internalised by intestinal epithelial cells via clathrin-mediated endocytosis (Cañas et al. 2016). Subsequent studies by the same group identified that OMVs from both E. coli Nissle 1917 and ECOR12 modulated innate immune responses in multiple epithelial cell models toward an anti-inflammatory profile by regulating the expression of cytokines such as IL-10 and antimicrobial peptides including human beta-defensin-2 (Fábrega et al. 2016). Furthermore, OMVs produced by the probiotic E. coli Nissle 1917 improved intestinal barrier integrity by upregulating the expression of TJ proteins ZO-1 and claudin-14 and downregulating the expression of pro-inflammatory transcription factors iNOS and COX-2, as well as various pro-inflammatory cytokines (Alvarez et al. 2016; Fábrega et al. 2017). Moreover, probiotic E. coli Nissle 1917 OMVs conferred protection against chemically induced colitis in mice (Fábrega et al. 2017). Similar to pathogen-derived OMVs, it was reported that OMVs produced by both E. coli strains Nissle 1917 and ECOR12 induced NOD1-dependent innate immune responses characterised by the activation of NF-κB and the secretion of IL-6 and IL-8 (Cañas et al. 2018). These findings indicate that the interkingdom communications mediated by BMVs are complex, whereby both pro-inflammatory and anti-inflammatory signalling pathways may be induced by the commensal microbiota to maintain immunological homeostasis. Further studies are required to better elucidate the signalling pathways induced by BMVs produced by the microbiota to broaden our understanding of their fundamental roles in maintaining homeostasis in the dynamic environment of the gut.
Members of the Gram-positive commensal Lactobacillus genus have long been considered to be probiotic due to the health benefits they confer to the host. Recently, MVs produced by several Lactobacillus species have been shown to elicit similar probiotic effects. Lactobacillus plantarum MVs were first reported to upregulate antimicrobial defence genes in Caco-2 cells in vitro (Li et al. 2017). Interestingly, in a Caenorhabditis elegans model of infection, L. plantarum MVs upregulated antimicrobial defence genes, conferring moderate protection against vancomycin-resistant E. faecium infection (Li et al. 2017). This study demonstrates the potential for the incorporation of L. plantarum MVs into novel therapeutics against Enterococcal infection. Amongst other Lactobacillus species, Lactobacillus sakei-derived MVs promote the production of IgA antibodies from Peyer’s Patch cells of the murine intestine, which function to neutralise extracellular toxins and protect epithelial cells from bacterial invasion (Yamasaki-Yashiki et al. 2019). Furthermore, in an ex vivo model of HIV-1 infection of human cervico-vaginal and tonsillar tissues, incubation with Lactobacillus crispatus or Lactobacillus gasseri MVs for 1 hour was sufficient to protect against viral replication (Ñahui Palomino et al. 2019). These studies indicate the protective properties of MVs produced by the Lactobacillus genus and provide insights into the mechanisms by which these probiotic bacteria confer health benefits to the host.
The Gram-negative anaerobe B. fragilis is a member of the intestinal microbiota. B. fragilis bacteria are encapsulated by a polysaccharide A (PSA) capsule which mediates anti-inflammatory effects via a TLR2-dependent interaction with dendritic cells, leading to the activation of regulatory T cells and subsequent secretion of the immunosuppressive cytokine IL-10 (Mazmanian et al. 2008; Ochoa-Repáraz et al. 2010; Round and Mazmanian 2010). However, B. fragilis is a non-motile bacterium and early studies could not identify the method of PSA delivery to the host immune system (Shen et al. 2012; Cerdeño-Tárraga et al. 2005). Ultimately, OMVs produced by B. fragilis were found to associate with PSA and elicit immunosuppressive effects via PSA-TLR2 dependent interactions (Shen et al. 2012). Oral administration of B. fragilis OMVs to mice conferred protection against colitis via the activation of regulatory T cells and the production of IL-10 (Shen et al. 2012). This study was the first to identify a specific bacterial species and mechanism by which microbiota-derived OMVs conferred health benefits to the host (Shen et al. 2012).
With the advent of more powerful DNA sequencing technologies, alleles for Crohn’s disease susceptibility have been implicated in the immunomodulatory pathway subverted by B. fragilis and their OMVs (Chu et al. 2016). Specifically, the genes Atg16l1 and Nod2 involved in the non-canonical autophagy pathway are essential to suppress mucosal inflammation via dendritic cell-dependent induction of regulatory T cells (Chu et al. 2016). Immune cells from human subjects with a major Crohn’s disease risk variant in the Atg16l1 allele were defective in regulatory T cell responses to B. fragilis OMVs in an ex vivo model, highlighting that gene-microbiota interactions mediated by BMVs are critical for the induction of mucosal tolerance and may be relevant to the aetiology of inflammatory bowel diseases (Chu et al. 2016).
Finally, B. fragilis has been implicated in mediating protection from multiple sclerosis, a demyelinating disease of the central nervous system (CNS). Using a murine model of multiple sclerosis, administration of B. fragilis bacteria was sufficient to confer protection against neuronal demyelination, however this protection was abrogated in IL-10 deficient mice or upon the administration of PSA-deficient B. fragilis strains (Ochoa-Repáraz et al. 2010). The author’s suggested that the protection conferred by B. fragilis was dependent upon PSA. Notably, the authors did not investigate the potential for OMV-associated PSA in protection against neuronal demyelination. The potential for B. fragilis OMV-based therapies against demyelinating conditions such as multiple sclerosis remains an avenue for further research.
Commensal BMVs Mediate Long-Distance Anti-Inflammatory Effects
There is mounting interest in the potential for extracellular factors secreted by the gut microbiota to travel to distal sites within the body to mediate their effects (reviewed in (Ahmadi Badi et al. 2017)). BMVs may represent one mechanism responsible for long-distance transport of such factors, protecting them from degradation by the host. However, whilst many studies have speculated on the effects of microbiota-derived BMVs outside of the intestinal lumen, very few studies have investigated these interactions experimentally. Nonetheless, microbiota-derived BMVs have been collected from both urine and blood samples, highlighting that they can cross the epithelial boundary and enter the bloodstream to be distributed throughout the body (Yoo et al. 2016; Lee et al. 2017; Park et al. 2017). Using DNA extraction and sequencing methods, these studies provide insights into the composition of the microbiota during states of both health and disease. A more recent study provided a protocol to isolate BMVs from biological materials and eukaryotic EVs found within bodily fluids (Tulkens et al. 2020). The authors reported that orthogonal implementation of ultrafiltration, size-exclusion chromatography and density-gradient ultracentrifugation resulted in the isolation of BMVs from bodily fluids within 72 hours, commenting that this method may prove valuable in the characterisation of BMVs within bodily fluids and drive the development of novel clinical diagnostic kits (Tulkens et al. 2020).
Depressive and neurological disorders have been linked to dysregulated metabolic functions of the microbiota, termed ‘dysbiosis’, the symptoms of which may improve with restoration of a more normal microbiota community. Thus, many commensal bacteria and their secreted BMVs are under investigation as novel strategies for the treatment of extra-intestinal diseases. For example, administration of L. plantarum MVs to mice was found to reduce stress-induced depressive behaviours and restored the normal expression of brain-derived neurotrophic factor (BDNF), a molecule depleted during depression (Choi et al. 2019). Additionally, Akkermansia muciniphila is a Gram-negative commensal bacterium with anti-inflammatory properties that are becoming increasingly well recognised. As well as eliciting immunomodulatory effects directly in the gut (Kang et al. 2013), A. muciniphila OMVs administered to C57BL/6 mice were shown to alleviate symptoms of cardiovascular disease such as weight gain, adipose tissue inflammation and increased blood glucose and cholesterol (Ashrafian et al. 2019). Interestingly, the beneficial effects mediated by A. muciniphila OMVs were stronger than those mediated by their parent bacteria, however it should be noted that it remains difficult to normalise between administration of OMVs and live bacteria.
Recent technological advances have revealed that areas of the body that were previously perceived to be sterile, such as the brain, have been found to harbour microbial products (Branton et al. 2013; Emery et al. 2017). Notably, studies investigating Alzheimer’s disease have discovered that brains of both healthy and diseased patients showed evidence of microbial products including E. coli LPS and K99 pili protein (Zhan et al. 2016). However, it remains contentious whether the brain may harbour its own resident microbiota or if the microbial molecules discovered in the brain were secreted from bacteria at distal sites, potentially via BMVs.
Evidently, the potential health benefits conferred by microbiota derived OMVs and MVs are broad and our understanding of their implications is still in its infancy. With the advent of more powerful resolving technologies being employed within the EV research field, we may soon be able to precisely identify the specific origins of BMVs within the bloodstream and characterise their functions after crossing the epithelial boundary. Limited studies have thus far demonstrated tissue tropism for microbiota derived BMVs in the bloodstream and this remains an avenue of important future research. Furthermore, few studies have investigated whether OMVs or MVs within the blood can cross the endothelial blood-brain barrier (BBB). Answering these emerging questions raises exciting new research avenues aiming to better understand the contribution of BMVs to systemic and neurological disorders.
Applications of BMVs
BMVs as Vaccine Candidates
A global rise in the incidence of antibiotic resistance necessitates that new therapies be developed to combat drug-resistant microbes. Such therapies may involve the use of BMVs either as novel vaccine candidates or as therapeutics to facilitate drug delivery.
OMVs and MVs have become interesting vaccine candidates in the last 20 years due to many of their innate immunostimulatory properties discussed throughout this chapter. First, they are relatively simple and cost-effective to produce, and more recent studies have demonstrated that upscaling their production is within the means of modern technologies (Baart et al. 2007; van de Waterbeemd et al. 2013). Second, BMVs package antigenic contents from their parent bacterium in a non-replicative form and are able to protect their cargo from degradative enzymes and potentially deliver them to specific sites within the body. Accordingly, BMV-associated proteins can be delivered to host cells in their native form which avoids the need for attenuation or inactivation of live bacterial cells. Finally, OMVs in vaccine formulations have proven to be stable for long periods of time and more importantly, safe for administration to humans (Alves et al. 2016; Arigita et al. 2004; Oster et al. 2005).
As discussed previously in this chapter, OMVs and MVs induce innate and adaptive immune responses in the host as they contain MAMPs which can interact with host innate immune PRRs to mediate an immune response. Indeed, vaccination of mice with E. coli OMVs was shown to not only mediate the production of cytokines that are indicative of Th1 and Th17 responses, but could also protect mice from lethal challenge via the generation of T cell-dependent immunity (Kim et al. 2013). Similar observations have also been observed using murine models whereby OMVs and MVs produced by S. Typhimurium, V. cholerae and S. aureus have been used to protect against subsequent bacterial infection (Alaniz et al. 2007; Schild et al. 2008; Sinha et al. 2015; Pritsch et al. 2016; Choi et al. 2015). A more recent study identified a role for A. baumannii OMVs as a potential therapeutic to combat multi-drug resistant bacterial infection (Huang et al. 2019). Mice were immunised with A. baumannii OMVs, resulting in the production of high titre IgG antibodies which, when purified from vaccinated animals and re-introduced in combination with quinolone antibiotics, significantly improved bacterial susceptibility to antibiotics in multiple murine models of infection (Huang et al. 2019). The authors noted that anti-A. baumannii OMV antibodies were likely exhibiting their function by blockade of outer membrane porins, thus preventing antibiotic efflux and improving antibiotic susceptibility (Huang et al. 2019). This study provides an interesting example of the use of BMVs as a novel therapeutic agent to combat antibiotic resistant pathogens (Huang et al. 2019).
The ability of OMVs and MVs to deliver antigenic cargo to host cells makes them dually useful in vaccine development as both an antigen and an adjuvant. To date, the only OMV-based vaccine licensed for human use confers protection against meningococcal serotype B disease, caused by infection with the Gram-negative pathogen N. meningitidis. The MeNZB OMV-based vaccine was first implemented in New Zealand whereby reports indicated that the vaccine was protective against infection in both adults and children (Oster et al. 2005; Oster et al. 2007). OMV-based vaccines against meningococcal serogroup B disease are now available globally (reviewed in (Holst et al. 2013)). Interestingly, a retrospective study investigating the efficacy of MeNZB in New Zealand identified that protection conferred by vaccine administration was also 31% effective against the closely related N. gonorrhoeae (Petousis-Harris et al. 2017). This was the first report of a vaccine showing effectiveness against gonorrhoea, albeit limited.
BMVs as Novel Therapeutics
Another advantage of OMV and MV-based vaccines is the ease with which bacteria can be genetically manipulated to express certain antigenic epitopes. These antigenic epitopes can be anchored to native OM proteins, including the ClyA protein of E. coli or fHbp protein of N. meningitidis, resulting in their association with OMVs at a high concentration (Kim et al. 2008; Chen et al. 2010; Huang et al. 2016; Rappazzo et al. 2016; Salverda et al. 2016). Using this method, it is hoped that a more successful influenza vaccine may be developed using E. coli OMVs as a platform (Rappazzo et al. 2016). The current influenza vaccine is designed against a strain specific glycoprotein haemagglutinin, however these molecules are highly variable and thus the vaccine must be re-designed annually (reviewed in (Treanor 2015)). The conserved ectodomain matrix 2 (M2e) ion channel motif is a highly conserved membrane protein of the influenza A strain (Mezhenskaya et al. 2019). Engineering of the commensal E. coli Nissle 1917 to express a multimeric construct of the M2e receptor via fusion with ClyA, a transmembrane protein enriched in E. coli OMVs, conferred 100% protection against lethal challenge of a mouse-adapted H1N1 influenza strain PR8 (Rappazzo et al. 2016). Additionally, an E. coli OM autotransporter (Hbp) was also engineered to perform as a platform to display multiple antigenic epitopes. The Hbp protein associated with E. coli OMV surfaces at a high density, indicating that this model may benefit researchers wanting to display heterologous antigens in OMV-based vaccines in the future (Daleke-Schermerhorn et al. 2014).
An alternative model of engineering BMVs for their use as vaccines is via the generation of hypervesiculating mutants by disruption of bacterial outer membrane integrity (Turner et al. 2015; Watkins et al. 2017). However, as many outer membrane proteins and lipids are essential for cargo trafficking into vesicles, conflicting reports raise doubts as to whether the contents, structure and composition of BMVs produced by hypervesiculation, via the disruption of the bacterial outer membrane, remain identical to their counterpart wild-type BMVs (Schager et al. 2018; Pérez-Cruz et al. 2016). Collectively, these studies highlight the potential for the genetic engineering of antigenic epitopes into BMVs to provide benefit in future vaccine research. Moreover, consideration must to be given to how these vesicles are produced as this may ultimately define their composition and functions.
Drug Delivery
Whilst the biodistribution of BMVs in the human body has not been fully elucidated, multiple recent studies have identified tissue-specific tropisms (Jang et al. 2015; Jones et al. 2020). As such, the potential for OMVs and MVs to serve as drug delivery vehicles has attracted significant attention. Compared to more well-established methods of drug delivery using nanoparticles including liposomes or metal-based nanoparticles, OMVs and MVs have the advantage that they can target specific tissue types, and even cell types, before entering host cells and releasing their cargo (reviewed in (Yoo et al. 2011)). This concept was first described using E. coli engineered to express the enzyme phosphotriesterase (PTE), demonstrating that the activity of PTE was better preserved when associated with OMVs, compared to its soluble form (Alves et al. 2016). These results show that OMVs could be utilised in drug delivery to protect enzymatic function with applications spanning to environmental remediation and potential industrial applications.
Of particular interest to the field is the potential use of engineered OMV- and MV-based therapies as novel cancer treatments. The ability to selectively hone OMVs or MVs to specific tumour cells via the targeting of cancer cell receptors may allow heightened specificity compared to the indiscriminate chemotherapeutics currently in use. Recent studies have materialised this potential in the targeting of HER2 receptors expressed at high density on tumour cell surfaces, and EGF receptors overexpressed on epithelial tumours, by E. coli OMVs loaded with anti-tumour siRNA and anti-tumour drugs, respectively (Gujrati et al. 2014; Kim et al. 2017). In both cases, tumour-specific effects and a significant reduction in tumour size were observed, with no apparent side effects. Furthermore, MVs produced by commensal bacteria Lactobacillus rhamnosus GG were shown to display cytotoxic effects on hepatic cancer cell line HepG2 in vitro and may prove beneficial as drug delivery vehicles (Behzadi et al. 2017).
Novel Technologies Utilising BMVs
Researchers are beginning to realise the potential applications of BMVs using novel technologies . For example, a recent study highlighted a role for BMVs in optoacoustic imaging, whereby E. coli OMVs were bioengineered to encapsulate high levels of melanin suitable for imaging applications in vivo (Gujrati et al. 2019). The authors identified that BMVs containing melanin were sufficient to facilitate imaging of tumours in vivo (Gujrati et al. 2019). Another study identified that bioengineering of both the internal lumen and external surface of E. coli OMVs allowed for biosensing applications by functional capturing of antigen and simultaneous reporting via luminescence (Chen et al. 2017). Finally, a recent study revealed the potential use of BMVs in antibiotic delivery, as antibiotic-treated bacteria efflux antibiotics via OMVs, resulting in their potential use as bactericidal therapies (Huang et al. 2020). Indeed, oral administration of antibiotic-loaded vesicles provided protection against intestinal bacterial infections in vivo, demonstrating their efficiency in delivering antibiotics to distal sites (Huang et al. 2020).
These studies highlight the current and potential future applications of BMVs in designing novel therapeutics to presently untreatable infectious and non-communicable diseases. Whilst these seminal studies have provided the premise for future work, further research is needed to realise the broad therapeutic applications of BMVs.
Conclusions and Perspectives
In recent years, BMVs have emerged as key mediators of bacterial communication. OMVs and MVs have been demonstrated to contribute to several biological functions ranging from supporting nutrient acquisition from the external environment to the delivery of virulence effectors to host cells. In particular, a wealth of literature now supports the notion of pathogen-derived OMVs in driving disease pathologies either by the delivery of toxins or by the packaging of MAMPs to promote inflammation within the host. Furthermore, due to their native immunostimulatory properties, OMVs have been successfully utilised in vaccine development and remain promising candidates for drug delivery in cancer settings. In the last 15 years, MVs released by Gram-positive bacteria have become recognised as bona fide secretion systems. Since their discovery, studies have demonstrated roles for MVs in promoting antibiotic resistance, the formation of bacterial biofilms and the packaging of toxins to promote pathogenesis. However, whilst these functions of MVs are increasingly being appreciated, a significant knowledge gap still remains regarding their physiological roles in disease, as well as their mechanisms of biogenesis. Furthermore, OMVs and MVs produced by commensal bacteria have recently emerged as important messengers that function to promote host-microbiota communications, thereby preventing the onset and severity of pro-inflammatory diseases of bacterial or autoimmune aetiology. However, we have only begun to uncover the broad scope of host-microbiota interactions, including the roles for microbiota-derived BMVs in inter-kingdom communications. Whilst the field looks ahead to utilise BMVs in novel technologies such as vaccine development, drug delivery and rapid screening techniques for infectious and non-communicable diseases, it is important to continue to improve our fundamental understanding of the biogenesis and functions of both OMVs and MVs to ultimately limit the pathologies they mediate and to harness their therapeutic potential.
References
Acheson DW, Luccioli S (2004) Microbial-gut interactions in health and disease. Mucosal immune responses. Best Pract Res Clin Gastroenterol 18(2):387–404
Ahmadi Badi S, Moshiri A, Fateh A, Rahimi Jamnani F, Sarshar M, Vaziri F, Siadat SD (2017) Microbiota-Derived Extracellular Vesicles as New Systemic Regulators. Front Microbiol 8:1610. https://doi.org/10.3389/fmicb.2017.01610
Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124(4):783–801. https://doi.org/10.1016/j.cell.2006.02.015
Alaniz RC, Deatherage BL, Lara JC, Cookson BT (2007) Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. Journal of immunology (Baltimore, Md: 1950) 179. doi:https://doi.org/10.4049/jimmunol.179.11.7692
Altindis E, Fu Y, Mekalanos JJ (2014) Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc Natl Acad Sci 111(15):E1548–E1556. https://doi.org/10.1073/pnas.1403683111
Alvarez CS, Badia J, Bosch M, Giménez R, Baldomà L (2016) Outer Membrane Vesicles and Soluble Factors Released by Probiotic Escherichia coli Nissle 1917 and Commensal ECOR63 Enhance Barrier Function by Regulating Expression of Tight Junction Proteins in Intestinal Epithelial Cells. Front Microbiol 7:1981. https://doi.org/10.3389/fmicb.2016.01981
Alves NJ, Turner KB, Medintz IL, Walper SA (2016) Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci Rep 6:24866. https://doi.org/10.1038/srep24866
Andreoni F, Toyofuku M, Menzi C, Kalawong R, Mairpady Shambat S, François P, Zinkernagel AS, Eberl L (2019) Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob Agents Chemother 63(2):e01439–e01418. https://doi.org/10.1128/AAC.01439-18
Arango Duque G, Descoteaux A (2014) Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 5:491
Arigita C, Jiskoot W, Westdijk J, van Ingen C, Hennink WE, Crommelin DJA, Kersten GFA (2004) Stability of mono- and trivalent meningococcal outer membrane vesicle vaccines. Vaccine 22(5–6):629–642. https://doi.org/10.1016/j.vaccine.2003.08.027
Ashrafian F, Shahriary A, Behrouzi A, Moradi HR, Keshavarz Azizi Raftar S, Lari A, Hadifar S, Yaghoubfar R, Ahmadi Badi S, Khatami S, Vaziri F, Siadat SD (2019) Akkermansia muciniphila-Derived Extracellular Vesicles as a Mucosal Delivery Vector for Amelioration of Obesity in Mice. Front Microbiol 10:2155. https://doi.org/10.3389/fmicb.2019.02155
Askarian F, Lapek JD Jr, Dongre M, Tsai C-M, Kumaraswamy M, Kousha A, Valderrama JA, Ludviksen JA, Cavanagh JP, Uchiyama S, Mollnes TE, Gonzalez DJ, Wai SN, Nizet V, Johannessen M (2018) Staphylococcus aureus membrane-derived vesicles promote bacterial virulence and confer protective immunity in murine infection models. Front Microbiol 9:262–262. https://doi.org/10.3389/fmicb.2018.00262
Athman JJ, Sande OJ, Groft SG, Reba SM, Nagy N, Wearsch PA, Richardson ET, Rojas R, Boom WH, Shukla S, Harding CV (2017) Mycobacterium tuberculosis Membrane Vesicles Inhibit T Cell Activation. J Immunol (Baltimore, Md: 1950) 198(5):2028–2037. https://doi.org/10.4049/jimmunol.1601199
Athman JJ, Wang Y, McDonald DJ, Boom WH, Harding CV, Wearsch PA (2015) Bacterial Membrane Vesicles Mediate the Release of Mycobacterium tuberculosis Lipoglycans and Lipoproteins from Infected Macrophages. J Immunol (Baltimore, Md: 1950) 195(3):1044–1053. https://doi.org/10.4049/jimmunol.1402894
Baart GJE, de Jong G, Philippi M, Riet Kvt, van der Pol LA, Beuvery EC, Tramper J, Martens DE (2007) Scale-up for bulk production of vaccine against meningococcal disease. Vaccine 25 (34):6399–6408. doi:https://doi.org/10.1016/j.vaccine.2007.06.008
Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, Salamon P, Youle M, Rohwer F (2013) Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc Natl Acad Sci 110(26):10771. https://doi.org/10.1073/pnas.1305923110
Bauman SJ, Kuehn MJ (2006) Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect 8(9–10):2400–2408. https://doi.org/10.1016/j.micinf.2006.05.001
Bauman SJ, Kuehn MJ (2009) Pseudomonas aeruginosa vesicles associate with and are internalized by human lung epithelial cells. BMC Microbiol 9:26. https://doi.org/10.1186/1471-2180-9-26
Behzadi E, Mahmoodzadeh Hosseini H, Imani Fooladi AA (2017) The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb Pathog 110:1–6. https://doi.org/10.1016/j.micpath.2017.06.016
Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, Cunha J, Hadi MZ, Zusman DR, Northen TR, Witkowska HE, Auer M (2014) The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol 5:474. https://doi.org/10.3389/fmicb.2014.00474
Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubès R (1998) Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 180(18):4872–4878
Biagini M, Garibaldi M, Aprea S, Pezzicoli A, Doro F, Becherelli M, Taddei AR, Tani C, Tavarini S, Mora M (2015) The human pathogen Streptococcus pyogenes releases lipoproteins as lipoprotein-rich membrane vesicles. Mol Cell Proteomics 14(8):2138–2149
Bielaszewska M, Ruter C, Kunsmann L, Greune L, Bauwens A, Zhang W, Kuczius T, Kim KS, Mellmann A, Schmidt MA, Karch H (2013) Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLoS Pathog 9(12):e1003797. https://doi.org/10.1371/journal.ppat.1003797
Bielig H, Rompikuntal PK, Dongre M, Zurek B, Lindmark B, Ramstedt M, Wai SN, Kufer TA (2011) NOD-like receptor activation by outer membrane vesicles from Vibrio cholerae non-O1 non-O139 strains is modulated by the quorum-sensing regulator HapR. Infect Immun 79(4):1418–1427. https://doi.org/10.1128/iai.00754-10
Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW (2014) Bacterial vesicles in marine ecosystems. Science 343(6167):183–186. https://doi.org/10.1126/science.1243457
Bitto NJ, Baker PJ, Dowling JK, Wray-McCann G, De Paoli A, Tran LS, Leung PL, Stacey KJ, Mansell A, Masters SL, Ferrero RL (2018) Membrane vesicles from Pseudomonas aeruginosa activate the noncanonical inflammasome through caspase-5 in human monocytes. Immunol Cell Biol 96(10):1120–1130. https://doi.org/10.1111/imcb.12190
Bitto NJ, Chapman R, Pidot S, Costin A, Lo C, Choi J, D'Cruze T, Reynolds EC, Dashper SG, Turnbull L, Whitchurch CB, Stinear TP, Stacey KJ, Ferrero RL (2017) Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep 7(1):7072. https://doi.org/10.1038/s41598-017-07288-4
Bitto NJ, Kaparakis-Liaskos M (2017) The therapeutic benefit of bacterial membrane vesicles. Int J Mol Sci 18 (6):pii: E1287. doi:https://doi.org/10.3390/ijms18061287
Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA (2009) Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5(4):e1000382. https://doi.org/10.1371/journal.ppat.1000382
Branton WG, Ellestad KK, Maingat F, Wheatley BM, Rud E, Warren RL, Holt RA, Surette MG, Power C (2013) Brain microbial populations in HIV/AIDS: α-proteobacteria predominate independent of host immune status. PLoS One 8(1):e54673. https://doi.org/10.1371/journal.pone.0054673
Brown L, Wolf JM, Prados-Rosales R, Casadevall A (2015) Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Micro 13(10):620–630. https://doi.org/10.1038/nrmicro3480
Cahill BK, Seeley KW, Gutel D, Ellis TN (2015) Klebsiella pneumoniae O antigen loss alters the outer membrane protein composition and the selective packaging of proteins into secreted outer membrane vesicles. Microbiol Res 180:1–10. https://doi.org/10.1016/j.micres.2015.06.012
Cañas M-A, Fábrega M-J, Giménez R, Badia J, Baldomà L (2018) Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol 9(498). https://doi.org/10.3389/fmicb.2018.00498
Cañas M-A, Giménez R, Fábrega M-J, Toloza L, Baldomà L, Badia J (2016) Outer membrane vesicles from the probiotic Escherichia coli Nissle 1917 and the commensal ECOR12 enter intestinal epithelial cells via clathrin-dependent endocytosis and elicit differential effects on DNA damage. PLoS One 11(8):e0160374. https://doi.org/10.1371/journal.pone.0160374
Cecil JD, O’Brien-Simpson NM, Lenzo JC, Holden JA, Chen Y-Y, Singleton W, Gause KT, Yan Y, Caruso F, Reynolds EC (2016) Differential responses of pattern recognition rceptors to outer membrane vesicles of three periodontal pathogens. PLoS One 11(4):e0151967. https://doi.org/10.1371/journal.pone.0151967
Cecil JD, O’Brien-Simpson NM, Lenzo JC, Holden JA, Singleton W, Perez-Gonzalez A, Mansell A, Reynolds EC (2017) Outer Membrane Vesicles Prime and Activate Macrophage Inflammasomes and Cytokine Secretion In Vitro and In Vivo. Front Immunol 8:1017. https://doi.org/10.3389/fimmu.2017.01017
Cerdeño-Tárraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J (2005) Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307(5714):1463–1465. https://doi.org/10.1126/science.1107008
Chatterjee D, Chaudhuri K (2011) Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett 585(9):1357–1362. https://doi.org/10.1016/j.febslet.2011.04.017
Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, DeLisa MP, Putnam D (2010) Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc Natl Acad Sci U S A 107(7):3099–3104. https://doi.org/10.1073/pnas.0805532107
Chen Q, Rozovsky S, Chen W (2017) Engineering multi-functional bacterial outer membrane vesicles as modular nanodevices for biosensing and bioimaging. Chem Commun (Camb) 53(54):7569–7572. https://doi.org/10.1039/c7cc04246a
Chi B, Qi M, Kuramitsu HK (2003) Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol 154(9):637–643. https://doi.org/10.1016/j.resmic.2003.08.001
Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS (2011) Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11(16):3424–3429. https://doi.org/10.1002/pmic.201000212
Choi J, Kim YK, Han PL (2019) Extracellular Vesicles Derived from Lactobacillus plantarum Increase BDNF Expression in Cultured Hippocampal Neurons and Produce Antidepressant-like Effects in Mice. Exp Neurobiol 28(2):158–171. https://doi.org/10.5607/en.2019.28.2.158
Choi JW, Kim SC, Hong SH, Lee HJ (2017) Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J Dent Res 96(4):458–466. https://doi.org/10.1177/0022034516685071
Choi SJ, Kim MH, Jeon J, Kim OY, Choi Y, Seo J, Hong SW, Lee WH, Jeon SG, Gho YS, Jee YK, Kim YK (2015) Active immunization with extracellular vesicles derived from Staphylococcus aureus effectively protects against staphylococcal lung infections, mainly via Th1 cell-mediated immunity. PLoS One 10(9):e0136021. https://doi.org/10.1371/journal.pone.0136021
Choi J-W, Kwon T-Y, Hong S-H, Lee H-J (2018) Isolation and characterization of a microRNA-size secretable small RNA in Streptococcus sanguinis. Cell Biochem Biophys 76(1):293–301. https://doi.org/10.1007/s12013-016-0770-5
Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, Targan SR, Xavier RJ, Ernst PB, Green DR, McGovern DP, Virgin HW, Mazmanian SK (2016) Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352(6289):1116–1120. https://doi.org/10.1126/science.aad9948
Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N (2000) Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 45(1):9–13
Correa P, Piazuelo MB (2008) Natural history of Helicobacter pylori infection. Dig Liver Dis 40(7):490–496
Daleke-Schermerhorn MH, Felix T, Soprova Z, Ten Hagen-Jongman CM, Vikström D, Majlessi L, Beskers J, Follmann F, de Punder K, van der Wel NN, Baumgarten T, Pham TV, Piersma SR, Jiménez CR, van Ulsen P, de Gier JW, Leclerc C, Jong WS, Luirink J (2014) Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Appl Environ Microbiol 80(18):5854–5865. https://doi.org/10.1128/aem.01941-14
Dauros Singorenko P, Chang V, Whitcombe A, Simonov D, Hong J, Phillips A, Swift S, Blenkiron C (2017) Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. J Extracell Vesicles 6(1):1324731. https://doi.org/10.1080/20013078.2017.1324731
Deatherage BL, Cookson BT (2012) Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun 80:1948–1957. https://doi.org/10.1128/iai.06014-11
DeVoe IW, Gilchrist JE (1975) Pili on meningococci from primary cultures of nasopharyngeal carriers and cerebrospinal fluid of patients with acute disease. J Exp Med 141(2):297–305. https://doi.org/10.1084/jem.141.2.297
Devos S, Van Putte W, Vitse J, Van Driessche G, Stremersch S, Van Den Broek W, Raemdonck K, Braeckmans K, Stahlberg H, Kudryashev M, Savvides SN, Devreese B (2017) Membrane vesicle secretion and prophage induction in multidrug-resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environ Microbiol doi:https://doi.org/10.1111/1462-2920.13793
Donato GM, Goldsmith CS, Paddock CD, Eby JC, Gray MC, Hewlett EL (2012) Delivery of Bordetella pertussis adenylate cyclase toxin to target cells via outer membrane vesicles. FEBS Lett 586(4):459–465. https://doi.org/10.1016/j.febslet.2012.01.032
Dorward DW, Garon CF (1989) DNA-binding proteins in cells and membrane blebs of Neisseria gonorrhoeae. J Bacteriol 171(8):4196–4201
Dorward DW, Garon CF (1990) DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl Environ Microbiol 56(6):1960–1962
Duncan L, Yoshioka M, Chandad F, Grenier D (2004) Loss of lipopolysaccharide receptor CD14 from the surface of human macrophage-like cells mediated by Porphyromonas gingivalis outer membrane vesicles. Microb Pathog 36(6):319–325. https://doi.org/10.1016/j.micpath.2004.02.004
Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF (2016) LPS remodeling triggers formation of outer membrane vesicles in Salmonella. MBio 7(4). https://doi.org/10.1128/mBio.00940-16
Elhenawy W, Debelyy MO, Feldman MF (2014) Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. MBio 5(2):e00909–e00914. https://doi.org/10.1128/mBio.00909-14
Ellis TN, Kuehn MJ (2010) Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74(1):81–94. https://doi.org/10.1128/mmbr.00031-09
Elluri S, Enow C, Vdovikova S, Rompikuntal PK, Dongre M, Carlsson S, Pal A, Uhlin BE, Wai SN (2014) Outer membrane vesicles mediate transport of biologically active Vibrio cholerae cytolysin (VCC) from V. cholerae strains. PLoS One 9(9):e106731. https://doi.org/10.1371/journal.pone.0106731
Elmi A, Nasher F, Jagatia H, Gundogdu O, Bajaj-Elliott M, Wren B, Dorrell N (2016) Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occludin. Cell Microbiol 18(4):561–572
Emery DC, Shoemark DK, Batstone TE, Waterfall CM, Coghill JA, Cerajewska TL, Davies M, West NX, Allen SJ (2017) 16S rRNA Next Generation Sequencing Analysis Shows Bacteria in Alzheimer's Post-Mortem Brain. Front Aging Neurosci 9:195. https://doi.org/10.3389/fnagi.2017.00195
Esoda CN, Kuehn MJ (2019) Pseudomonas aeruginosa leucine aminopeptidase influences early biofilm composition and structure via vesicle-associated antibiofilm activity. MBio 10(6):e02548–e02519. https://doi.org/10.1128/mBio.02548-19
Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE (2012) Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158(11):2742–2752. https://doi.org/10.1099/mic.0.060343-0
Fábrega MJ, Aguilera L, Giménez R, Varela E, Alexandra Cañas M, Antolín M, Badía J, Baldomà L (2016) Activation of Immune and Defense Responses in the Intestinal Mucosa by Outer Membrane Vesicles of Commensal and Probiotic Escherichia coli Strains. Front Microbiol 7:705. https://doi.org/10.3389/fmicb.2016.00705
Fábrega MJ, Rodríguez-Nogales A, Garrido-Mesa J, Algieri F, Badía J, Giménez R, Gálvez J, Baldomà L (2017) Intestinal Anti-inflammatory Effects of Outer Membrane Vesicles from Escherichia coli Nissle 1917 in DSS-Experimental Colitis in Mice. Front Microbiol 8:1274. https://doi.org/10.3389/fmicb.2017.01274
Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover TL, Solcia E (1999) Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J Pathol 188(2):220–226. https://doi.org/10.1002/(sici)1096-9896(199906)188:2<220::Aid-path307>3.0.Co;2-c
Fleetwood AJ, Lee MKS, Singleton W, Achuthan A, Lee MC, O’Brien-Simpson NM, Cook AD, Murphy AJ, Dashper SG, Reynolds EC, Hamilton JA (2017) Metabolic remodeling, inflammasome activation, and pyroptosis in macrophages stimulated by Porphyromonas gingivalis and its outer membrane vesicles. Front Cell Infect Microbiol 7:351. https://doi.org/10.3389/fcimb.2017.00351
Forsberg CW, Beveridge TJ, Hellstrom A (1981) Cellulase and xylanase release from Bacteroides succinogenes and its importance in the rumen environment. Appl Environ Microbiol 42(5):886
Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, Nielsen KM (2014) Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl Environ Microbiol 80(11):3469. https://doi.org/10.1128/AEM.04248-13
Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A (2009) Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect Immun 77(10):4187–4196. https://doi.org/10.1128/iai.00009-09
Gerritzen MJH, Martens DE, Uittenbogaard JP, Wijffels RH, Stork M (2019) Sulfate depletion triggers overproduction of phospholipids and the release of outer membrane vesicles by Neisseria meningitidis. Sci Rep 9(1):4716. https://doi.org/10.1038/s41598-019-41233-x
Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ (2003a) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300(5625):1584–1587. https://doi.org/10.1126/science.1084677
Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ (2003b) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278(11):8869–8872. https://doi.org/10.1074/jbc.C200651200
Giwercman B, Meyer C, Lambert PA, Reinert C, Høiby N (1992) High-level beta-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob Agents Chemother 36(1):71–76. https://doi.org/10.1128/aac.36.1.71
Gorby Y, McLean J, Korenevsky A, Rosso K, El-Naggar MY, Beveridge TJ (2008) Redox-reactive membrane vesicles produced by Shewanella. Geobiology 6(3):232–241. https://doi.org/10.1111/j.1472-4669.2008.00158.x
Grande R, Celia C, Mincione G, Stringaro A, Di Marzio L, Colone M, Di Marcantonio MC, Savino L, Puca V, Santoliquido R, Locatelli M, Muraro R, Hall-Stoodley L, Stoodley P (2017) Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front Microbiol 8 (1040). doi:https://doi.org/10.3389/fmicb.2017.01040
Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L, Paolino D, Codagnone M, Muraro R, Stoodley P, Hall-Stoodley L, Mincione G (2015) Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front Microbiol 6:1369. doi:https://doi.org/10.3389/fmicb.2015.01369
Grenier D, Bélanger M (1991) Protective effect of Porphyromonas gingivalis outer membrane vesicles against bactericidal activity of human serum. Infect Immun 59(9):3004–3008
Grenier D, Mayrand D (1987) Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun 55(1):111–117
Gujrati V, Kim S, Kim S-H, Min JJ, Choy HE, Kim SC, Jon S (2014) Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 8(2):1525–1537. https://doi.org/10.1021/nn405724x
Gujrati V, Prakash J, Malekzadeh-Najafabadi J, Stiel A, Klemm U, Mettenleiter G, Aichler M, Walch A, Ntziachristos V (2019) Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat Commun 10(1):1114. https://doi.org/10.1038/s41467-019-09034-y
Haas B, Grenier D (2015) Isolation, characterization and biological properties of membrane vesicles produced by the swine pathogen Streptococcus suis. PLoS One 10(6):e0130528. https://doi.org/10.1371/journal.pone.0130528
Hasan S, Kulkarni NN, Asbjarnarson A, Linhartova I, Osicka R, Sebo P, Gudmundsson GH (2018) Bordetella pertussis adenylate cyclase toxin disrupts functional integrity of bronchial epithelial layers. Infect Immun 86(3):e00445–e00417
Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF (2011) Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem 286(2):1269–1276. https://doi.org/10.1074/jbc.M110.185744
Hayashi J, Hamada N, Kuramitsu HK (2002) The autolysin of Porphyromonas gingivalis is involved in outer membrane vesicle release. FEMS Microbiol Lett 216(2):217–222. https://doi.org/10.1111/j.1574-6968.2002.tb11438.x
He X, Yuan F, Lu F, Yin Y, Cao J (2017) Vancomycin-induced biofilm formation by methicillin-resistant Staphylococcus aureus is associated with the secretion of membrane vesicles. Microb Pathog 110:225–231. https://doi.org/10.1016/j.micpath.2017.07.004
Hock BD, McKenzie JL, Keenan JI (2017) Helicobacter pylori outer membrane vesicles inhibit human T cell responses via induction of monocyte COX-2 expression. Pathogens and Disease 75(4)
Holst J, Oster P, Arnold R, Tatley M, Næss L, Aaberge I, Galloway Y, McNicholas A, O'Hallahan J, Rosenqvist E (2013) Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother 9(6):1241–1253
Hong J, Dauros-Singorenko P, Whitcombe A, Payne L, Blenkiron C, Phillips A, Swift S (2019) Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions. J Extracell Vesicles 8(1):1632099. https://doi.org/10.1080/20013078.2019.1632099
Huang W, Wang S, Yao Y, Xia Y, Yang X, Li K, Sun P, Liu C, Sun W, Bai H, Chu X, Li Y, Ma Y (2016) Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci Rep 6:37242. https://doi.org/10.1038/srep37242
Huang W, Zhang Q, Li W, Chen Y, Shu C, Li Q, Zhou J, Ye C, Bai H, Sun W, Yang X, Ma Y (2019) Anti-outer Membrane Vesicle Antibodies Increase Antibiotic Sensitivity of Pan-Drug-Resistant Acinetobacter baumannii. Front Microbiol 10:1379. https://doi.org/10.3389/fmicb.2019.01379
Huang W, Zhang Q, Li W, Yuan M, Zhou J, Hua L, Chen Y, Ye C, Ma Y (2020) Development of novel nanoantibiotics using an outer membrane vesicle-based drug efflux mechanism. J Control Release 317:1–22. https://doi.org/10.1016/j.jconrel.2019.11.017
Imayoshi R, Cho T, Kaminishi H (2011) NO production in RAW264 cells stimulated with Porphyromonas gingivalis extracellular vesicles. Oral Dis 17(1):83–89. https://doi.org/10.1111/j.1601-0825.2010.01708.x
Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, Votta BJ, Bertin J, Boneca IG, Sasakawa C, Philpott DJ, Ferrero RL, Kaparakis-Liaskos M (2014) The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15(5):623–635. https://doi.org/10.1016/j.chom.2014.04.001
Ismail S, Hampton MB, Keenan JI (2003) Helicobacter pylori outer membrane vesicles modulate proliferation and interleukin-8 production by gastric epithelial cells. Infect Immun 71(10):5670–5675. https://doi.org/10.1128/IAI.71.10.5670-5675.2003
Iwami J, Murakami Y, Nagano K, Nakamura H, Yoshimura F (2007) Further evidence that major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis stabilize bacterial cells. Oral Microbiol Immunol 22(5):356–360. https://doi.org/10.1111/j.1399-302X.2007.00363.x
Jäger J, Keese S, Roessle M, Steinert M, Schromm AB (2015) Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell Microbiol 17(5):607–620. https://doi.org/10.1111/cmi.12392
Janeway CA Jr, Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol 20(1):197–216
Jang SC, Kim SR, Yoon YJ, Park KS, Kim JH, Lee J, Kim OY, Choi EJ, Kim DK, Choi DS, Kim YK, Park J, Di Vizio D, Gho YS (2015) In vivo kinetic biodistribution of nano-sized outer membrane vesicles derived from bacteria. Small (Weinheim an der Bergstrasse, Germany) 11(4):456–461. https://doi.org/10.1002/smll.201401803
Jasim R, Han M-L, Zhu Y, Hu X, Hussein MH, Lin Y-W, Zhou QT, Dong CYD, Li J, Velkov T (2018) Lipidomic analysis of the outer membrane vesicles from paired polymyxin-susceptible and-resistant Klebsiella pneumoniae clinical isolates. Int J Mol Sci 19(8):2356
Jeon H, Oh MH, Jun SH, Kim SI, Choi CW, Kwon HI, Na SH, Kim YJ, Nicholas A, Selasi GN, Lee JC (2016) Variation among Staphylococcus aureus membrane vesicle proteomes affects cytotoxicity of host cells. Microb Pathog 93:185–193. https://doi.org/10.1016/j.micpath.2016.02.014
Jeon J, Park SC, Her J, Lee JW, Han J-K, Kim Y-K, Kim KP, Ban C (2018) Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. RSC Adv 8(27):15241–15247
Jhelum H, Sori H, Sehgal D (2018) A novel extracellular vesicle-associated endodeoxyribonuclease helps Streptococcus pneumoniae evade neutrophil extracellular traps and is required for full virulence. Sci Rep 8(1):7985. https://doi.org/10.1038/s41598-018-25865-z
Jiang Y, Kong Q, Roland KL, Curtiss R (2014) Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 304(3–4):431–443. https://doi.org/10.1016/j.ijmm.2014.02.006
Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC (2011) Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6(2):e17027. https://doi.org/10.1371/journal.pone.0017027
Johnston EL, Kufer TA, Kaparakis-Liaskos M (2020) Immunodetection and Pathogenesis Mediated by Bacterial Membrane Vesicles. In: Kaparakis-Liaskos M, Kufer TA (eds) Bacterial Membrane Vesicles. Springer, Switzerland, pp 159–188
Jones EJ, Booth C, Fonseca S, Parker A, Cross K, Miquel-Clopés A, Hautefort I, Mayer U, Wileman T, Stentz R, Carding SR (2020) The Uptake, Trafficking, and Biodistribution of Bacteroides thetaiotaomicron Generated Outer Membrane Vesicles. Front Microbiol 11:57. https://doi.org/10.3389/fmicb.2020.00057
Jun SH, Lee JH, Kim SI, Choi CW, Park TI, Jung HR, Cho JW, Kim SH, Lee JC (2017) Staphylococcus aureus-derived membrane vesicles exacerbate skin inflammation in atopic dermatitis. Clinical and Experimental Allergy: journal of the British Society for Allergy and Clinical Immunology 47(1):85–96. https://doi.org/10.1111/cea.12851
Jung AL, Stoiber C, Herkt CE, Schulz C, Bertrams W, Schmeck B (2016) Legionella pneumophila-derived outer membrane vesicles promote bacterial replication in macrophages. PLoS Pathog 12(4):e1005592
Kadurugamuwa JL, Beveridge TJ (1995) Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 177(14):3998–4008
Kadurugamuwa JL, Beveridge TJ (1996) Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 178(10):2767–2774
Kadurugamuwa JL, Mayer A, Messner P, Sara M, Sleytr UB, Beveridge TJ (1998) S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J Bacteriol 180(9):2306–2311
Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, Gho YS, Kim JG, Kim YK (2013) Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One 8(10):e76520. https://doi.org/10.1371/journal.pone.0076520
Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J, Hutton ML, Davies JK, Crack PJ, Hertzog PJ, Philpott DJ, Girardin SE, Whitchurch CB, Ferrero RL (2010) Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol 12(3):372–385. https://doi.org/10.1111/j.1462-5822.2009.01404.x
Kaparakis-Liaskos M, Ferrero RL (2015) Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15(6):375–387. https://doi.org/10.1038/nri3837
Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ (2004) Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J 23(23):4538–4549. https://doi.org/10.1038/sj.emboj.7600471
Kim OY, Dinh NTH, Park HT, Choi SJ, Hong K, Gho YS (2017) Bacterial protoplast-derived nanovesicles for tumor targeted delivery of chemotherapeutics. Biomaterials 113:68–79. https://doi.org/10.1016/j.biomaterials.2016.10.037
Kim JY, Doody AM, Chen DJ, Cremona GH, Shuler ML, Putnam D, DeLisa MP (2008) Engineered bacterial outer membrane vesicles with enhanced functionality. J Mol Biol 380(1):51–66. https://doi.org/10.1016/j.jmb.2008.03.076
Kim OY, Hong BS, Park K-S, Yoon YJ, Choi SJ, Lee WH, Roh T-Y, Lötvall J, Kim Y-K, Gho YS (2013) Immunization with Escherichia coli outer membrane vesicles protects bacteria-induced lethality via Th1 and Th17 cell responses. J Immunol 190(8):4092–4102. https://doi.org/10.4049/jimmunol.1200742
Klieve AV, Yokoyama MT, Forster RJ, Ouwerkerk D, Bain PA, Mawhinney EL (2005) Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl Environ Microbiol 71(8):4248–4253. https://doi.org/10.1128/aem.71.8.4248-4253.2005
Klimentova J, Pavkova I, Horcickova L, Bavlovic J, Kofroňová O, Benada O, Stulik J (2019) Francisella tularensis subsp. holarctica releases differentially loaded outer membrane vesicles under various stress conditions. Front Microbiol 10:2304
Klimentová J, Stulík J (2015) Methods of isolation and purification of outer membrane vesicles from Gram-negative bacteria. Microbiol Res 170:1–9. https://doi.org/10.1016/j.micres.2014.09.006
Knox K, Vesk M, Work E (1966) Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J Bacteriol 92(4):1206–1217
Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, Stanton BA (2016) A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog 12(6):e1005672. https://doi.org/10.1371/journal.ppat.1005672
Kolling GL, Matthews KR (1999) Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol 65(5):1843–1848
Kosgodage US, Matewele P, Mastroianni G, Kraev I, Brotherton D, Awamaria B, Nicholas AP, Lange S, Inal JM (2019) Peptidylarginine deiminase inhibitors reduce bacterial membrane vesicle release and sensitize bacteria to antibiotic treatment. Front Cell Infect Microbiol 9(227). https://doi.org/10.3389/fcimb.2019.00227
Kulkarni HM, Nagaraj R, Jagannadham MV (2015) Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol Res 181:1–7. https://doi.org/10.1016/j.micres.2015.07.008
Kulp A, Kuehn MJ (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. https://doi.org/10.1146/annurev.micro.091208.073413
Kunsmann L, Rüter C, Bauwens A, Greune L, Glüder M, Kemper B, Fruth A, Wai SN, He X, Lloubes R (2015) Virulence from vesicles: Novel mechanisms of host cell injury by Escherichia coli O104: H4 outbreak strain. Sci Rep 5:13252
Lapinet JA, Scapini P, Calzetti F, Pérez O, Cassatella MA (2000) Gene expression and production of tumor necrosis factor alpha, interleukin-1beta (IL-1beta), IL-8, macrophage inflammatory protein 1alpha (MIP-1alpha), MIP-1beta, and gamma interferon-inducible protein 10 by human neutrophils stimulated with group B meningococcal outer membrane vesicles. Infect Immun 68(12):6917–6923. https://doi.org/10.1128/iai.68.12.6917-6923.2000
Lee E-Y, Bang JY, Park GW, Choi D-S, Kang JS, Kim H-J, Park K-S, Lee J-O, Kim Y-K, Kwon K-H, Kim K-P, Gho YS (2007) Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7(17):3143–3153. https://doi.org/10.1002/pmic.200700196
Lee E-Y, Choi D-Y, Kim D-K, Kim J-W, Park JO, Kim S, Kim S-H, Desiderio DM, Kim Y-K, Kim K-P, Gho YS (2009) Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9(24):5425–5436. https://doi.org/10.1002/pmic.200900338
Lee J, Lee E-Y, Kim S-H, Kim D-K, Park K-S, Kim KP, Kim Y-K, Roh T-Y, Gho YS (2013) Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother 57(6):2589–2595. https://doi.org/10.1128/AAC.00522-12
Lee Y, Park JY, Lee EH, Yang J, Jeong BR, Kim YK, Seoh JY, Lee S, Han PL, Kim EJ (2017) Rapid Assessment of Microbiota Changes in Individuals with Autism Spectrum Disorder Using Bacteria-derived Membrane Vesicles in Urine. Exp Neurobiol 26(5):307–317. https://doi.org/10.5607/en.2017.26.5.307
Lekmeechai S, Su YC, Brant M, Alvarado-Kristensson M, Vallström A, Obi I, Arnqvist A, Riesbeck K (2018) Helicobacter pylori outer membrane vesicles protect the pathogen from reactive oxygen species of the respiratory burst. Front Microbiol 9:1837. https://doi.org/10.3389/fmicb.2018.01837
Li Z, Clarke AJ, Beveridge TJ (1998) Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol 180(20):5478–5483
Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B (2017) Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol 17(1):66. https://doi.org/10.1186/s12866-017-0977-7
Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT (2014) Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196(13):2355–2366. https://doi.org/10.1128/jb.01493-14
Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X (2017) A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. https://doi.org/10.1038/ncomms14888
Lindholm M, Metsäniitty M, Granström E, Oscarsson J (2020) Outer membrane vesicle-mediated serum protection in Aggregatibacter actinomycetemcomitans. J Oral Microbiol 12(1):1747857. https://doi.org/10.1080/20002297.2020.1747857
Lindmark B, Rompikuntal PK, Vaitkevicius K, Song T, Mizunoe Y, Uhlin BE, Guerry P, Wai SN (2009) Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol 9(1):1–10. https://doi.org/10.1186/1471-2180-9-220
Manning AJ, Kuehn MJ (2011) Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. https://doi.org/10.1186/1471-2180-11-258
Martinez J, Fernandez JS, Liu C, Hoard A, Mendoza A, Nakanouchi J, Rodman N, Courville R, Tuttobene MR, Lopez C, Gonzalez LJ, Shahrestani P, Papp-Wallace KM, Vila AJ, Tolmasky ME, Bonomo RA, Sieira R, Ramirez MS (2019) Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci Rep 9(1):17251. https://doi.org/10.1038/s41598-019-53847-2
Mashburn LM, Jett AM, Akins DR, Whiteley M (2005) Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187(2):554–566. https://doi.org/10.1128/jb.187.2.554-566.2005
Mashburn LM, Whiteley M (2005) Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437(7057):422–425. https://doi.org/10.1038/nature03925
Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M (2008) Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69(2):491–502. https://doi.org/10.1111/j.1365-2958.2008.06302.x
Mayer F, Gottschalk G (2003) The bacterial cytoskeleton and its putative role in membrane vesicle formation observed in a Gram-positive bacterium producing starch-degrading enzymes. J Mol Microbiol Biotechnol 6(3–4):127–132. https://doi.org/10.1159/000077243
Mayrand D, Grenier D (1989) Biological activities of outer membrane vesicles. Can J Microbiol 35(6):607–613. https://doi.org/10.1139/m89-097
Mazmanian SK, Round JL, Kasper DL (2008) A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453(7195):620–625. https://doi.org/10.1038/nature07008
McBroom AJ, Kuehn MJ (2007) Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 63(2):545–558. https://doi.org/10.1111/j.1365-2958.2006.05522.x
Metruccio MME, Evans DJ, Gabriel MM, Kadurugamuwa JL, Fleiszig SMJ (2016) Pseudomonas aeruginosa outer membrane vesicles triggered by human mucosal fluid and lysozyme can prime host tissue surfaces for bacterial adhesion. Front Microbiol 7:871. https://doi.org/10.3389/fmicb.2016.00871
Mezhenskaya D, Isakova-Sivak I, Rudenko L (2019) M2e-based universal influenza vaccines: a historical overview and new approaches to development. J Biomed Sci 26(1):76. https://doi.org/10.1186/s12929-019-0572-3
Mitra S, Sinha R, Mitobe J, Koley H (2016) Development of a cost-effective vaccine candidate with outer membrane vesicles of a tolA-disrupted Shigella boydii strain. Vaccine 34(15):1839–1846. https://doi.org/10.1016/j.vaccine.2016.02.018
Moon DC, Choi CH, Lee JH, Choi C-W, Kim H-Y, Park JS, Kim SI, Lee JC (2012) Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol 50(1):155–160
Ñahui Palomino RA, Vanpouille C, Laghi L, Parolin C, Melikov K, Backlund P, Vitali B, Margolis L (2019) Extracellular vesicles from symbiotic vaginal lactobacilli inhibit HIV-1 infection of human tissues. Nat Commun 10(1):5656. https://doi.org/10.1038/s41467-019-13468-9
Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, de Mazière A, Klumperman J, Schlatter M, Delamarre L, Mellman I (2014) Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509(7499):240–244. https://doi.org/10.1038/nature13133
Nakao R, Myint SL, Wai SN, Uhlin BE (2018) Enhanced biofilm formation and membrane vesicle release by Escherichia coli expressing a commonly occurring plasmid gene, kil. Front Microbiol 9:2605. https://doi.org/10.3389/fmicb.2018.02605
Nakao R, Takashiba S, Kosono S, Yoshida M, Watanabe H, Ohnishi M, Senpuku H (2014) Effect of Porphyromonas gingivalis outer membrane vesicles on gingipain-mediated detachment of cultured oral epithelial cells and immune responses. Microbes and infection / Institut Pasteur 16(1):6–16. https://doi.org/10.1016/j.micinf.2013.10.005
Namork E, Brandtzaeg P (2002) Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet (London, England) 360(9347):1741. https://doi.org/10.1016/s0140-6736(02)11721-1
O’Donoghue EJ, Krachler AM (2016) Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol 18(11):1508–1517. https://doi.org/10.1111/cmi.12655
O’Donoghue EJ, Sirisaengtaksin N, Browning DF, Bielska E, Hadis M, Fernandez-Trillo F, Alderwick L, Jabbari S, Krachler AM (2017) Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLoS Pathog 13(11):e1006760. https://doi.org/10.1371/journal.ppat.1006760
Obana N, Nakao R, Nagayama K, Nakamura K, Senpuku H, Nomura N (2017) Immunoactive clostridial membrane vesicle production is regulated by a sporulation factor. Infect Immun 85(5):e00096–e00017. https://doi.org/10.1128/iai.00096-17
Ochoa-Repáraz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH (2010) A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol 3(5):487–495. https://doi.org/10.1038/mi.2010.29
Ojima Y, Sawabe T, Konami K, Azuma M (2020) Construction of hypervesiculation Escherichia coli strains and application for secretory protein production. Biotechnol Bioeng 117(3):701–709. https://doi.org/10.1002/bit.27239
Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martín-Peña R, González-Reyes JA, Jiménez-Munguía I, Gómez-Gascón L, Fernández J, Luque-García JL, García-Lidón C, Estévez H, Pachón J, Obando I, Casadevall A, L-a P, Rodríguez-Ortega MJ (2014) Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteome 106:46–60. https://doi.org/10.1016/j.jprot.2014.04.023
Oster P, Lennon D, O’Hallahan J, Mulholland K, Reid S, Martin D (2005) MeNZB: a safe and highly immunogenic tailor-made vaccine against the New Zealand Neisseria meningitidis serogroup B disease epidemic strain. Vaccine 23(17–18):2191–2196. https://doi.org/10.1016/j.vaccine.2005.01.063
Oster P, O'Hallahan J, Aaberge I, Tilman S, Ypma E, Martin D (2007) Immunogenicity and safety of a strain-specific MenB OMV vaccine delivered to under 5-year olds in New Zealand. Vaccine 25(16):3075–3079. https://doi.org/10.1016/j.vaccine.2007.01.023
Papayannopoulos V (2018) Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 18(2):134–147. https://doi.org/10.1038/nri.2017.105
Park JY, Choi J, Lee Y, Lee JE, Lee EH, Kwon HJ, Yang J, Jeong BR, Kim YK, Han PL (2017) Metagenome Analysis of Bodily Microbiota in a Mouse Model of Alzheimer Disease Using Bacteria-derived Membrane Vesicles in Blood. Exp Neurobiol 26(6):369–379. https://doi.org/10.5607/en.2017.26.6.369
Parker H, Chitcholtan K, Hampton MB, Keenan JI (2010) Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun 78(12):5054–5061. https://doi.org/10.1128/iai.00299-10
Patten DA, Hussein E, Davies SP, Humphreys PN, Collett A (2017) Commensal-derived OMVs elicit a mild proinflammatory response in intestinal epithelial cells. Microbiology 163(5):702–711
Pérez-Cruz C, Cañas MA, Giménez R, Badia J, Mercade E, Baldomà L, Aguilera L (2016) Membrane Vesicles Released by a hypervesiculating Escherichia coli Nissle 1917 tolR Mutant Are Highly Heterogeneous and Show Reduced Capacity for Epithelial Cell Interaction and Entry. PLoS One 11(12):e0169186. https://doi.org/10.1371/journal.pone.0169186
Petousis-Harris H, Paynter J, Morgan J, Saxton P, McArdle B, Goodyear-Smith F, Black S (2017) Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: a retrospective case-control study. Lancet (London, England) 390(10102):1603–1610. https://doi.org/10.1016/s0140-6736(17)31449-6
Pettit RK, Judd RC (1992a) Characterization of naturally elaborated blebs from serum-susceptible and serum-resistant strains of Neisseria gonorrhoeae. Mol Microbiol 6(6):723–728. https://doi.org/10.1111/j.1365-2958.1992.tb01521.x
Pettit RK, Judd RC (1992b) The interaction of naturally elaborated blebs from serum-susceptible and serum-resistant strains of Neisseria gonorrhoeae with normal human serum. Mol Microbiol 6(6):729–734. https://doi.org/10.1111/j.1365-2958.1992.tb01522.x
Pieters J (2008) Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe 3(6):399–407
Pollak CN, Delpino MV, Fossati CA, Baldi PC (2012) Outer membrane vesicles from Brucella abortus promote bacterial internalization by human monocytes and modulate their innate immune response. PLoS One 7(11):e50214. https://doi.org/10.1371/journal.pone.0050214
Post DM, Zhang D, Eastvold JS, Teghanemt A, Gibson BW, Weiss JP (2005) Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J Biol Chem 280(46):38383–38394. https://doi.org/10.1074/jbc.M508063200
Prados-Rosales R, Weinrick BC, Piqué DG, Jacobs WR, Casadevall A, Rodriguez GM (2014) Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol 196(6):1250–1256. https://doi.org/10.1128/jb.01090-13
Pritsch M, Ben-Khaled N, Chaloupka M, Kobold S, Berens-Riha N, Peter A, Liegl G, Schubert S, Hoelscher M, Löscher T, Wieser A (2016) Comparison of intranasal outer membrane vesicles with Cholera toxin and injected MF59C.1 as adjuvants for malaria transmission blocking antigens AnAPN1 and Pfs48/45. J Immunol Res 2016:e3576028. https://doi.org/10.1155/2016/3576028
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4):373–383. https://doi.org/10.1083/jcb.201211138
Rappazzo CG, Watkins HC, Guarino CM, Chau A, Lopez JL, DeLisa MP, Leifer CA, Whittaker GR, Putnam D (2016) Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 34(10):1252–1258. https://doi.org/10.1016/j.vaccine.2016.01.028
Ren D, Nelson KL, Uchakin PN, Smith AL, Gu XX, Daines DA (2012) Characterization of extended co-culture of non-typeable Haemophilus influenzae with primary human respiratory tissues. Exp Biol Med (Maywood) 237(5):540–547. https://doi.org/10.1258/ebm.2012.011377
Renelli M, Matias V, Lo RY, Beveridge TJ (2004) DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150(7):2161–2169. https://doi.org/10.1099/mic.0.26841-0
Repoila F, Darfeuille F (2009) Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell 101(2):117–131. https://doi.org/10.1042/bc20070137
Resch U, Tsatsaronis JA, Le Rhun A, Stubiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Wai SN, Charpentier E (2016) A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. MBio 7 (6):e00207–e00216. doi:https://doi.org/10.1128/mBio.00207-16
Reyes-Robles T, Dillard RS, Cairns LS, Silva-Valenzuela CA, Housman M, Ali A, Wright ER, Camilli A (2018) Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol 200(15):e00792–e00717. https://doi.org/10.1128/JB.00792-17
Rivera J, Cordero RJB, Nakouzi AS, Frases S (2010) Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 107(44):19002–19007. https://doi.org/10.1073/pnas.1008843107
Roier S, Blume T, Klug L, Wagner GE, Elhenawy W, Zangger K, Prassl R, Reidl J, Daum G, Feldman MF, Schild S (2015) A basis for vaccine development: comparative characterization of Haemophilus influenzae outer membrane vesicles. Int J Med Microbiol 305(3):298–309. https://doi.org/10.1016/j.ijmm.2014.12.005
Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, Daum G, Reidl J, Feldman MF, Schild S (2016) A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. https://doi.org/10.1038/ncomms10515
Romeu B, Lastre M, García L, Cedré B, Mandariote A, Fariñas M, Oliva R, Pérez O (2014) Combined meningococcal serogroup A and W135 outer-membrane vesicles activate cell-mediated immunity and long-term memory responses against non-covalent capsular polysaccharide A. Immunol Res 58(1):75–85. https://doi.org/10.1007/s12026-013-8427-6
Rompikuntal PK, Thay B, Khan MK, Alanko J, Penttinen A-M, Asikainen S, Wai SN, Oscarsson J (2012) Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infect Immun 80(1):31–42. https://doi.org/10.1128/IAI.06069-11
Roszkowiak J, Jajor P, Guła G, Gubernator J, Żak A, Drulis-Kawa Z, Augustyniak D (2019) Interspecies outer membrane vesicles (OMVs) modulate the sensitivity of pathogenic bacteria and pathogenic yeasts to cationic peptides and serum complement. Int J Mol Sci 20(22). https://doi.org/10.3390/ijms20225577
Round JL, Mazmanian SK (2010) Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci 107(27):12204–12209
Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G (2011) Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55(7):3084–3090. https://doi.org/10.1128/aac.00929-10
Salvachúa D, Werner AZ, Pardo I, Michalska M, Black BA, Donohoe BS, Haugen SJ, Katahira R, Notonier S, Ramirez KJ, Amore A, Purvine SO, Zink EM, Abraham PE, Giannone RJ, Poudel S, Laible PD, Hettich RL, Beckham GT (2020) Outer membrane vesicles catabolize lignin-derived aromatic compounds in Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 117(17):9302–9310. https://doi.org/10.1073/pnas.1921073117
Salverda ML, Meinderts SM, Hamstra HJ, Wagemakers A, Hovius JW, van der Ark A, Stork M, van der Ley P (2016) Surface display of a borrelial lipoprotein on meningococcal outer membrane vesicles. Vaccine 34(8):1025–1033. https://doi.org/10.1016/j.vaccine.2016.01.019
Schaar V, de Vries SP, Perez Vidakovics ML, Bootsma HJ, Larsson L, Hermans PW, Bjartell A, Mörgelin M, Riesbeck K (2011) Multicomponent Moraxella catarrhalis outer membrane vesicles induce an inflammatory response and are internalized by human epithelial cells. Cell Microbiol 13(3):432–449. https://doi.org/10.1111/j.1462-5822.2010.01546.x
Schager AE, Dominguez-Medina CC, Necchi F, Micoli F, Goh YS, Goodall M, Flores-Langarica A, Bobat S, Cook CNL, Arcuri M, Marini A, King LDW, Morris FC, Anderson G, Toellner KM, Henderson IR, López-Macías C, MacLennan CA, Cunningham AF (2018) IgG Responses to Porins and Lipopolysaccharide within an Outer Membrane-Based Vaccine against Nontyphoidal Salmonella Develop at Discordant Rates. MBio 9(2). https://doi.org/10.1128/mBio.02379-17
Schetters STT, Jong WSP, Horrevorts SK, Kruijssen LJW, Engels S, Stolk D, Daleke-Schermerhorn MH, Garcia-Vallejo J, Houben D, Unger WWJ, den Haan JMM, Luirink J, van Kooyk Y (2019) Outer membrane vesicles engineered to express membrane-bound antigen program dendritic cells for cross-presentation to CD8(+) T cells. Acta Biomater 91:248–257. https://doi.org/10.1016/j.actbio.2019.04.033
Schild S, Nelson EJ, Camilli A (2008) Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect Immun 76(10):4554–4563. https://doi.org/10.1128/iai.00532-08
Schlatterer K, Beck C, Hanzelmann D, Lebtig M, Fehrenbacher B, Schaller M, Ebner P, Nega M, Otto M, Kretschmer D, Peschel A (2018) The mechanism behind bacterial lipoprotein release: phenol-soluble modulins mediate Toll-like receptor 2 activation via extracellular vesicle release from Staphylococcus aureus. MBio 9(6). https://doi.org/10.1128/mBio.01851-18
Schooling SR, Beveridge TJ (2006) Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188(16):5945–5957. https://doi.org/10.1128/jb.00257-06
Schooling SR, Hubley A, Beveridge TJ (2009) Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol 191(13):4097–4102. https://doi.org/10.1128/jb.00717-08
Schrempf H, Koebsch I, Walter S, Engelhardt H, Meschke H (2011) Extracellular Streptomyces vesicles: amphorae for survival and defence. Microb Biotechnol 4(2):286–299. https://doi.org/10.1111/j.1751-7915.2011.00251.x
Schroder K, Zhou R, Tschopp J (2010) The NLRP3 inflammasome: a sensor for metabolic danger? Science 327(5963):296–300. https://doi.org/10.1126/science.1184003
Schulz E, Goes A, Garcia R, Panter F, Koch M, Müller R, Fuhrmann K, Fuhrmann G (2018) Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. J Control Release 290:46–55. https://doi.org/10.1016/j.jconrel.2018.09.030
Schwechheimer C, Kuehn MJ (2015) Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13(10):605–619. https://doi.org/10.1038/nrmicro3525
Schwechheimer C, Kulp A, Kuehn MJ (2014) Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol 14(1):324
Shan Q, Dwyer M, Rahman S, Gadjeva M (2014) Distinct susceptibilities of corneal Pseudomonas aeruginosa clinical isolates to neutrophil extracellular trap-mediated immunity. Infect Immun 82(10):4135–4143. https://doi.org/10.1128/iai.02169-14
Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK (2012) Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12(4):509–520. https://doi.org/10.1016/j.chom.2012.08.004
Sinha R, Koley H, Nag D, Mitra S, Mukhopadhyay AK, Chattopadhyay B (2015) Pentavalent outer membrane vesicles of Vibrio cholerae induce adaptive immune response and protective efficacy in both adult and passive suckling mice models. Microb Infect 17(3):215–227. https://doi.org/10.1016/j.micinf.2014.10.011
Sisquella X, Ofir-Birin Y, Pimentel MA, Cheng L, Abou Karam P, Sampaio NG, Penington JS, Connolly D, Giladi T, Scicluna BJ, Sharples RA, Waltmann A, Avni D, Schwartz E, Schofield L, Porat Z, Hansen DS, Papenfuss AT, Eriksson EM, Gerlic M, Hill AF, Bowie AG, Regev-Rudzki N (2017) Malaria parasite DNA-harbouring vesicles activate cytosolic immune sensors. Nat Commun 8(1):1985. https://doi.org/10.1038/s41467-017-02083-1
Sjöström AE, Sandblad L, Uhlin BE, Wai SN (2015) Membrane vesicle-mediated release of bacterial RNA. Sci Rep 5:15329. https://doi.org/10.1038/srep15329
Stentz R, Carvalho AL, Jones EJ, Carding SR (2018) Fantastic voyage: the journey of intestinal microbiota-derived microvesicles through the body. Biochem Soc Trans. https://doi.org/10.1042/bst20180114
Stentz R, Horn N, Cross K, Salt L, Brearley C, Livermore DM, Carding SR (2015) Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J Antimicrob Chemother 70(3):701–709. https://doi.org/10.1093/jac/dku466
Stephens DS, Edwards KM, Morris F, McGee ZA (1982) Pili and outer membrane appendages on Neisseria meningitidis in the cerebrospinal fluid of an infant. J Infect Dis 146(4):568. https://doi.org/10.1093/infdis/146.4.568
Stevenson TC, Cywes-Bentley C, Moeller TD, Weyant KB, Putnam D, Chang YF, Jones BD, Pier GB, DeLisa MP (2018) Immunization with outer membrane vesicles displaying conserved surface polysaccharide antigen elicits broadly antimicrobial antibodies. Proc Natl Acad Sci U S A 115(14):E3106–e3115. https://doi.org/10.1073/pnas.1718341115
Subramanian P, Pirbadian S, El-Naggar MY, Jensen GJ (2018) Ultrastructure of Shewanella oneidensis MR-1 nanowires revealed by electron cryotomography. Proc Natl Acad Sci 115(14):E3246. https://doi.org/10.1073/pnas.1718810115
Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y (1978) Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol Gen Genet 167(1):1–9. https://doi.org/10.1007/bf00270315
Taheri N, Fällman M, Wai SN, Fahlgren A (2019) Accumulation of virulence-associated proteins in Campylobacter jejuni outer membrane vesicles at human body temperature. J Proteome 195:33–40
Tan TT, Morgelin M, Forsgren A, Riesbeck K (2007) Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 195(11):1661–1670. https://doi.org/10.1086/517611
Tashiro Y, Hasegawa Y, Shintani M, Takaki K, Ohkuma M, Kimbara K, Futamata H (2017) Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target cells. Front Microbiol 8(571). https://doi.org/10.3389/fmicb.2017.00571
Thanassi DG, Hultgren SJ (2000) Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Opin Cell Biol 12(4):420–430. https://doi.org/10.1016/s0955-0674(00)00111-3
Thay B, Damm A, Kufer TA, Wai SN, Oscarsson J (2014) Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-kappaB activation. Infect Immun 82(10):4034–4046. https://doi.org/10.1128/iai.01980-14
Thay B, Wai SN, Oscarsson J (2013) Staphylococcus aureus alpha-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One 8(1):e54661. https://doi.org/10.1371/journal.pone.0054661
Thompson SS, Naidu YM, Pestka JJ (1985) Ultrastructural localization of an extracellular protease in Pseudomonas fragi by using the peroxidase-antiperoxidase reaction. Appl Environ Microbiol 50(4):1038–1042
Toyofuku M, Cárcamo-Oyarce G, Yamamoto T, Eisenstein F, Hsiao C-C, Kurosawa M, Gademann K, Pilhofer M, Nomura N, Eberl L (2017) Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun 8(1):481. https://doi.org/10.1038/s41467-017-00492-w
Tran F, Boedicker JQ (2017) Genetic cargo and bacterial species set the rate of vesicle-mediated horizontal gene transfer. Sci Rep 7(1):8813. https://doi.org/10.1038/s41598-017-07447-7
Treanor JJ (2015) Prospects for broadly protective influenza vaccines. Vaccine 33(Suppl 4):D39–D45. https://doi.org/10.1016/j.vaccine.2015.08.053
Tulkens J, De Wever O, Hendrix A (2020) Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nat Protoc 15(1):40–67. https://doi.org/10.1038/s41596-019-0236-5
Turkina MV, Olofsson A, Magnusson K-E, Arnqvist A, Vikström E (2015) Helicobacter pylori vesicles carrying CagA localize in the vicinity of cell–cell contacts and induce histone H1 binding to ATP in epithelial cells. FEMS Microbiol Lett 362(11):fnv076–fnv076. https://doi.org/10.1093/femsle/fnv076
Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Carcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB (2016) Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. https://doi.org/10.1038/ncomms11220
Turner L, Bitto NJ, Steer DL, Lo C, D’Costa K, Ramm G, Shambrook M, Hill AF, Ferrero RL, Kaparakis-Liaskos M (2018) Helicobacter pylori outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Front Immunol 9(1466). https://doi.org/10.3389/fimmu.2018.01466
Turner L, Praszkier J, Hutton ML, Steer D, Ramm G, Kaparakis-Liaskos M, Ferrero RL (2015) Increased outer membrane vesicle formation in a Helicobacter pylori tolB mutant. Helicobacter 20(4):269–283. https://doi.org/10.1111/hel.12196
van de Waterbeemd B, Zomer G, Kaaijk P, Ruiterkamp N, Wijffels RH, van den Dobbelsteen GP, van der Pol LA (2013) Improved production process for native outer membrane vesicle vaccine against Neisseria meningitidis. PLoS One 8(5):e65157. https://doi.org/10.1371/journal.pone.0065157
Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam VA (2016) Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell 165(5):1106–1119. https://doi.org/10.1016/j.cell.2016.04.015
Vasilyeva NV, Tsfasman IM, Suzina NE, Stepnaya OA, Kulaev IS (2008) Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp. XL1 into the medium by means of outer membrane vesicles. FEBS J 275(15):3827–3835. https://doi.org/10.1111/j.1742-4658.2008.06530.x
Vdovikova S, Luhr M, Szalai P, Nygård Skalman L, Francis MK, Lundmark R, Engedal N, Johansson J, Wai SN (2017) A novel role of Listeria monocytogenes membrane vesicles in inhibition of autophagy and cell death. Front Cell Infect Microbiol 7:154. https://doi.org/10.3389/fcimb.2017.00154
Vidakovics ML, Jendholm J, Morgelin M, Mansson A, Larsson C, Cardell LO, Riesbeck K (2010) B cell activation by outer membrane vesicles - a novel virulence mechanism. PLoS Pathog 6(1):e1000724. https://doi.org/10.1371/journal.ppat.1000724
Wagner T, Joshi B, Janice J, Askarian F, Škalko-Basnet N, Hagestad OC, Mekhlif A, Wai SN, Hegstad K, Johannessen M (2018) Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J Proteome 187:28–38. https://doi.org/10.1016/j.jprot.2018.05.017
Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE (2003) Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115(1):25–35
Wang X, Eagen WJ, Lee JC (2020) Orchestration of human macrophage NLRP3 inflammasome activation by Staphylococcus aureus extracellular vesicles. Proceedings of the National Academy of Sciences:201915829. doi:https://doi.org/10.1073/pnas.1915829117
Wang Z, Lazinski DW, Camilli A (2017) Immunity Provided by an Outer Membrane Vesicle Cholera Vaccine Is Due to O-Antigen-Specific Antibodies Inhibiting Bacterial Motility. Infect Immun 85(1). https://doi.org/10.1128/iai.00626-16
Wang X, Thompson CD, Weidenmaier C, Lee JC (2018) Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat Commun 9(1):1379. https://doi.org/10.1038/s41467-018-03847-z
Watkins HC, Rappazzo CG, Higgins JS, Sun X, Brock N, Chau A, Misra A, Cannizzo JPB, King MR, Maines TR, Leifer CA, Whittaker GR, DeLisa MP, Putnam D (2017) Safe recombinant outer membrane vesicles that display M2e elicit heterologous influenza protection. Mol Ther 25(4):989–1002. https://doi.org/10.1016/j.ymthe.2017.01.010
Whitchurch CB (2002) Extracellular DNA required for bacterial biofilm formation. Science 295(5559):1487–1487. https://doi.org/10.1126/science.295.5559.1487
Winter J, Letley D, Rhead J, Atherton J, Robinson K (2014) Helicobacter pylori membrane vesicles stimulate innate pro- and anti-inflammatory responses and induce apoptosis in Jurkat T cells. Infect Immun 82(4):1372–1381. https://doi.org/10.1128/iai.01443-13
Yamasaki-Yashiki S, Miyoshi Y, Nakayama T, Kunisawa J, Katakura Y (2019) IgA-enhancing effects of membrane vesicles derived from Lactobacillus sakei subsp. sakei NBRC15893. Biosci Microbiota Food Health 38(1):23–29. https://doi.org/10.12938/bmfh.18-015
Yara D, Stentz R, Wileman T, Schüller S (2019) The impact of the colonic milieu on enterohaemorrhagic E. coli outer membrane vesicle production. Access. Microbiology 1(1A)
Yaron S, Kolling GL, Simon L, Matthews KR (2000) Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol 66(10):4414–4420
Yonezawa H, Osaki T, Fukutomi T, Hanawa T, Kurata S, Zaman C, Hojo F, Kamiya S (2017) Diversification of the AlpB outer membrane protein of Helicobacter pylori affects biofilm formation and cellular adhesion. J Bacteriol 199(6). https://doi.org/10.1128/jb.00729-16
Yoo JW, Irvine DJ, Discher DE, Mitragotri S (2011) Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 10(7):521–535. https://doi.org/10.1038/nrd3499
Yoo JY, Rho M, You YA, Kwon EJ, Kim MH, Kym S, Jee YK, Kim YK, Kim YJ (2016) 16S rRNA gene-based metagenomic analysis reveals differences in bacteria-derived extracellular vesicles in the urine of pregnant and non-pregnant women. Exp Mol Med 48(2):e208. https://doi.org/10.1038/emm.2015.110
Yun SH, Park EC, Lee S-Y, Lee H, Choi C-W, Yi Y-S, Ro H-J, Lee JC, Jun S, Kim H-Y, Kim G-H, Kim SI (2018) Antibiotic treatment modulates protein components of cytotoxic outer membrane vesicles of multidrug-resistant clinical strain, Acinetobacter baumannii DU202. Clin Proteomics 15(1):28. https://doi.org/10.1186/s12014-018-9204-2
Zavan L, Bitto NJ, Johnston EL, Greening DW, Kaparakis-Liaskos M (2019) Helicobacter pylori growth stage determines the size, protein composition, and preferential cargo packaging of outer membrane vesicles. Proteomics 19(1–2):e1800209. https://doi.org/10.1002/pmic.201800209
Zavan L, Bitto NJ, Kaparakis-Liaskos M (2020) Introduction, History, and Discovery of Bacterial Membrane Vesicles. In: Kaparakis-Liaskos M, Kufer TA (eds) Bacterial Membrane Vesicles. Springer, Switzerland, pp 1–21
Zhan X, Stamova B, Jin LW, DeCarli C, Phinney B, Sharp FR (2016) Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 87(22):2324–2332. https://doi.org/10.1212/wnl.0000000000003391
Zhang H, Zhang Y, Song Z, Li R, Ruan H, Liu Q, Huang X (2020) sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL-8 secretion in human cells. Int J Med Microbiol 310(1):151356. https://doi.org/10.1016/j.ijmm.2019.151356
Zhao K, Deng X, He C, Yue B, Wu M (2013) Pseudomonas aeruginosa outer membrane vesicles modulate host immune responses by targeting the Toll-like receptor 4 signaling pathway. Infect Immun 81(12):4509–4518. https://doi.org/10.1128/iai.01008-13
Zielke RA, Wierzbicki IH, Weber JV, Gafken PR, Sikora AE (2014) Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Molecular & cellular proteomics: MCP 13(5):1299–1317. https://doi.org/10.1074/mcp.M113.029538
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Gilmore, W.J., Bitto, N.J., Kaparakis-Liaskos, M. (2021). Pathogenesis Mediated by Bacterial Membrane Vesicles. In: Mathivanan, S., Fonseka, P., Nedeva, C., Atukorala, I. (eds) New Frontiers: Extracellular Vesicles. Subcellular Biochemistry, vol 97. Springer, Cham. https://doi.org/10.1007/978-3-030-67171-6_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-67171-6_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-67170-9
Online ISBN: 978-3-030-67171-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)