Abstract
Magnetic resonance imaging (MRI) is the primary clinical tool to examine inflammatory brain lesions in Multiple Sclerosis (MS). Disease progression and inflammatory activities are examined by longitudinal image analysis to support diagnosis and treatment decision. Automated lesion segmentation methods based on deep convolutional neural networks (CNN) have been proposed, but are not yet applied in the clinical setting. Typical CNNs working on cross-sectional single time-point data have several limitations: changes to the image characteristics between single examinations due to scanner and protocol variations have an impact on the segmentation output, while at the same time the additional temporal correlation using pre-examinations is disregarded.
In this work, we investigate approaches to overcome these limitations. Within a CNN architectural design, we propose convolutional Long Short-Term Memory (C-LSTM) networks to incorporate the temporal dimension. To reduce scanner- and protocol dependent variations between single MRI exams, we propose a histogram normalization technique as pre-processing step. The ISBI 2015 challenge data was used for network training and cross-validation.
We demonstrate that the combination of the longitudinal normalization and CNN architecture increases the performance and the inter-time-point stability of the lesion segmentation. In the combined solution, the dice coefficient was increased and made more consistent for each subject. The proposed methods can therefore be used to increase the performance and stability of fully automated lesion segmentation applications in the clinical routine or in clinical trials.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system (CNS) that produces demyelination and axonal/neuronal damage [9]. The demyelinating process is associated with persistent inflammation throughout the CNS and, as a result, the demyelinated lesion, also known as plaque, is the main pathological feature of MS [1, 10]. Due to its high sensitivity, magnetic resonance imaging (MRI) is extensively used for diagnosis and monitoring of MS [29]. However, manual segmentations by trained experts are tedious, time-consuming and often lack reproducibility [24]. For this reason, automated MS lesion segmentation techniques have been developed during the last years [29], and particularly convolutional neural networks (CNN) have gained popularity for this task, after proving their effectiveness in other neuroimaging tasks [26].
From the perspective of how the data is used to train a model, MS lesion segmentation algorithms can be classified as either longitudinal or cross-sectional. Longitudinal approaches make use of the time information provided by subsequent scans (known as time-points or visits) of the same patient. In cross-sectional approaches, all scans, even if belonging to the same patient, are treated as independent scans and no time information is considered. Only few CNN-based approaches can be found in the literature that tackle the problem of MS lesion segmentation in a longitudinal manner.
The first CNN-based longitudinal method found in the literature was proposed in [6]. Although the CNN is used only for classifying candidates extracted using intensity and atlas information, the method employs different time-points to perform the task. Another longitudinal method is described in [19], where the goal is to detect lesion load change using CNNs. To achieve this, a hybrid between the U-Net [23] and the Dense-Net [16] was used as basis. A special type of loss function generates probabilities that are used together with the segmentation masks for obtaining information about lesion change. Following this idea of detecting changes, a CNN is proposed in [27] for detecting new T2-weighted (T2-w) lesions. This architecture consists of a first block based on Voxelmorph [4] to learn deformation fields and register the baseline image to the follow-up images, and a second block to perform the segmentation of new lesions using the results of the first block.
One important issue when using longitudinal data is the normalization across time-points, or longitudinal normalization. The goal is to increase the similarity, in terms of image intensity regarding tissue classes, of the different time-points, without modifying the structures whose changes are due to pathological conditions. MS lesions are an example of those structures, as they can persist, change or disappear in time [25]. A statistical normalization method is proposed in [28], in which all histograms are centered using statistical measures obtained from the white matter voxels. Other methods based on histograms use landmarks to perform the normalization across different time-points [21]. Another longitudinal normalization method is presented in [25]. In this case voxel changes in time are modeled mathematically depending on temporal intensity variations and the lesion priors are used for keeping the lesion voxels unchanged.
Following the idea that including longitudinal information in CNNs can produce better segmentation results [6], this work proposes an effective pipeline based on the Chi-Square metric and convolutional long short-term memory (C-LSTM) [15] networks, for the exploitation of longitudinal MRI data for segmentation of MS lesions. To the best of our knowledge, this is the first MS lesion segmentation longitudinal method that combines traditional CNNs with convolutional recurrent neural networks (C-RNN).
2 Method
2.1 Longitudinal Normalization
A simple yet effective MRI longitudinal normalization based on the Chi-Square metric \( \chi ^2 \) is proposed. The Chi-Square test is commonly used for analyzing the difference between observed and expected distributions [30], but in this case only the metric is used to measure and maximize the similarity between the histograms of volumes of the same modality for different patients and different time-points. Equation 1 shows the Chi-Metric as a means of comparison of two histograms \(H_a\) and \(H_b\), for voxel intensities I.
Let s, t and m represent subject, time-point and modality, respectively. For each modality m a reference volume \(V_{\hat{s}\hat{t}}^{(m)}\) is selected to normalize the other volumes \(V_{st}^{(m)}\) of that modality, with \( s \ne \hat{s}, t\ne \hat{t}\). For each \(V_{st}^{(m)}\) an optimal scalar \(\theta _{st}^{(m)}\) is found using Eq. 2, where \(H_{\hat{s}\hat{t}}^{(m)}\) and \(H_{st}^{(m)}\) are the histograms of the normal appearing white matter (NAWM) of \(V_{\hat{s}\hat{t}}^{(m)}\) and \(V_{st}^{(m)}\), respectively. The normalized images are the result of the product \(\theta _{st}^{(m)} V_{st}^{(m)}\). To obtain the NAWM masks, the Computational Anatomy Toolbox (CAT12) [13] applied within the Statistical Parametric Mapping (SPM12) [22] toolkit was used.


2.2 Network Architecture
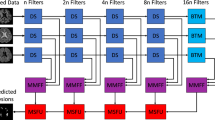
In order to exploit time information, an extension of the architecture presented in [20] is proposed for the segmentation of MS lesions from longitudinal multi-modal brain images. This architecture is a hybrid between the well known U-Net and the Convolutional Long Short-Term Memory (C-LSTM) network. Figure 1 shows the architecture. The input patches have size (T, M, H, W, D), where T and M represent the number of selected time-points to process for each sample and the number of modalities, respectively. H, W and D represent the height, width and depth of the patches in each volume. The patches are extracted from the volumes that have been previously pre-processed. Just like in the U-Net architecture, there is an encoder for extracting hierarchical features, but these are extracted for each time-point separately. These features are then combined, at the deepest level and for all time-points, by a C-LSTM. After the features are processed by the first C-LSTM, a decoder upsamples them so that the input dimensions can be reached again. At the output of the decoder, a second C-LSTM combines again the features of the different time-points. Finally, the feature maps corresponding to a specific time-point (e.g. the one in the middle, if T is odd) are selected and a last convolution takes place to reduce the number of maps to 2, one for each class, lesion or non-lesion.
U-Net ConvLSTM architecture. Patch dimensions are included in gray text, where T and M denote the number of selected time-points and number of modalities, respectively. H, W and D denote the spatial dimensions of the patches in the volumes. Dashed lines denote skip connections by copying and concatenation.
2.3 Post-processing
After a segmentation is produced, a post-processing step is performed to exclude potential false-positive (FP) detected lesions. This is achieved by imposing a minimal lesion size of 3 \(\mathrm{mm}^3\), as it has been found to improve the performance of MS lesion segmentation methods [12].
3 Experimental Setup
3.1 Dataset
One of the most popular datasets for MS lesion segmentation is the one provided by the longitudinal MS lesion segmentation challenge, which was part of the International Symposium on Biomedical Imaging (ISBI) in 2015 [17] and continues to be publicly available. The dataset, acquired with a 3T scanner, is subdivided into training (5 subjects) and testing (14 subjects) sets. Only the training set contains lesion segmentation masks generated by two different expert raters. Each subject contains between 4 and 6 time-points, each of which consists of T1-weighted (T1-w) magnetization prepared rapid gradient echo (MPRAGE), T2-weighted (T2-w) and Proton Density weighted (PDw) produced with double spin echo (DSE) and Fluid Attenuated Inversion Recovery (FLAIR) images. The average time between subsequent time-points is 1 year [8, 17].
Both the original and the pre-processed images are available for use. For this work, the pre-processed images were used. The pre-processing steps are: first N4 correction, skull-stripping, dura stripping, second N4 correction and registration to an isotropic MNI template. The proposed longitudinal normalization was applied on the images that resulted from this pre-processing procedure.
3.2 Longitudinal Normalization Configuration
One important step for the longitudinal normalization is the white matter segmentation. The segmentation was performed on each volume using CAT with the default parameters. For finding the values of \(\theta _{st}^{(m)}\), the first time-point of subject 01 was selected as reference for each modality and the Nelder–Mead Simplex method [11] was employed for the minimization of the distance function.
For comparison purposes, the min-max normalization (Eq. 3) was used as default normalization. This type of normalization, in which intensity values are mapped to the interval [0, 1], is one of the methods that can be used as pre-processing in MRI data [5, 7, 14]. In Eq. 3 \(I_{orig}\), \(I_{min}\) and \(I_{max}\) represent the original, minimum and maximum intensities of a volume, respectively, and \(I_{norm}\) is the assigned intensity.
3.3 Training and Implementation Details
After having normalized the pre-processed images, a leave-one-out (subject-wise) cross-validation was performed on the training set. For each fold, from the 4 subjects not used for testing, one was used for validation and 3 for training. The model was trained using 32 \(\times \) 32 \(\times \) 32 patches with step size 16 \(\times \) 16 \(\times \) 16, and extracted only from the brain region using a brain mask generated by thresholding the pre-processed volumes. All four modalities were used (\(M=4\)) and three time-points were taken for each training sample (\(T=3\)). Sampling in time was performed using padding by repeating the first and last time-points. Training was performed using the Adam optimizer [18] for a maximum of 200 epochs with an early stopping condition of 20 epochs, and a batch size of 16. To deal with the class imbalance (more normal tissue as compared to lesion tissue), the dice loss function was used. All models were trained with only one segmentation mask (mask1) of the two available, and the validation subjects were randomly chosen a first time obtaining the order 03, 03, 04, 05, 02, and then set to be the same for all experiments. No data augmentation was performed in order to increase the comparability between different experiments.
The proposed model was implemented in PyTorch, using a GPU NVIDIA Tesla T4.
3.4 Evaluation
Four experiments were carried out for determining the advantages of the normalization procedure and the longitudinal architecture: the cross-sectional version of the model (without ConvLSTM blocks and without considering the time dimension), which is a normal 3D U-Net, for both min-max and the proposed normalization; and the proposed model (Fig. 1) also for both types of normalization.
In order to evaluate the performance of the method, the Dice score (DSC), lesion-wise false positive rate (LFPR) and lesion-wise true positive rate (LTPR) were used. The DSC is computed according to Eq. 4, where TP, FP and FN denote number of true positive, false positive, and false negative voxels, respectively. The LFPR is the number of lesions in the produced segmentation that do not overlap with a lesion in the ground truth, divided by the total number of lesions in the produced segmentation. The LTPR is computed as the number of lesions in the ground truth that overlap with a lesion in the produced segmentation, divided by the total number of lesions in the ground truth [2].
4 Results
The results of the normalization process are presented in Fig. 2 for all pre-processed training subjects of the dataset.
Table 1 shows the mean values of the metrics for the four evaluated cases, computed for all time-points of all subjects. An increase is noticed in the DSC and LTPR, particularly, when the longitudinal approach is combined with the proposed normalization. Figure 3 shows an example of a segmentation result for which a higher DSC in the longitudinal approach denotes a better segmentation quality with respect to the cross-sectional (3D U-Net) version.
Example of resulting segmentation masks from cross-validation experiment for patient 01, slice 89 from the ISBI training dataset for cross-sectional and longitudinal models. Masks are shown on FLAIR images and t denotes time-point index. The standard cross-sectional approach (second column) leads to unsegmented lesions especially on \(t=2\). Both proposed techniques improve the segmentation when applied individually (third and fourth columns), while the combined method (last column) yields the best results as compared to the ground truth.
5 Discussion and Conclusion
We have proposed a supervised longitudinal pipeline for MS lesion segmentation from MRI images. The approach combines a whole-volume longitudinal normalization scheme with a patch-based 3D CNN architecture that exploits time information. The method was evaluated on data from the ISBI 2015 challenge, obtaining an improvement in the DSC and LTPR with respect to a cross-sectional architecture and with respect to a simple normalization algorithm. Time padding allowed to reduce the impact of the reduction of samples that occurs when the time dimension is considered. Both intra- and inter-subject standard deviations were reduced in comparison to the cross-sectional approach for the DSC metric, which denotes more consistent segmentations in time. However, the LFPR increased for the proposed pipeline, which can be explained, to a certain extent, by the fact that in several cases (e.g. Fig. 3), the normalization and the C-RNN-based architecture allow to segment lesions that were not segmented for example in the cross-sectional case. These new lesions can represent a single lesion in the ground truth and therefore the LFPR is expected to increase. Ensembling several models could be used for reducing the amount of false positive lesions, while keeping high DSC and LTPR values. When compared to other approaches like for instance [7] in which the DSC for the same cross-validation was 0.684, or to [2] with a DSC of 0.698, our DSC (0.699) is similar to these values, but it is lower than the best reported result, which is 0.765 [3]. Considering the absence of data augmentation in our experiments, our approach offers promising results.
The proposed CNN architecture requires a higher training time (about 1.5x the time of the cross-sectional approach), which can be seen as a limitation, but the improvement in both the time consistency and values of segmentation metrics justifies the increase in training time. The increase in training time also strongly depends on the implementation of the C-LSTM, which in our case has not yet been optimized for time efficiency.
In comparison to other normalization methods such as histogram matching, the proposed method allows to preserve the basic shape of the histograms, which prevents from loosing information about the lesions and other structures. The optimization of the similarity reduces problems that peak/landmark based methods can show when the histograms differ too much before normalization. We chose an approach using a pre-segmented WM mask, assuming that normalizing the surrounding tissue value of white matter lesions optimally supports the detection of the pathological lesions. This approach relies on a rough segmentation of the white matter before applying the CNN. However, the normalization can also be applied on the original histograms, at the cost of a higher influence of the lesion volume in the quality of the normalization.
The longitudinal normalization pre-processing method increased the robustness of a trained network in respect to the histogram variations of the input data, which were present in the ISBI 2015 training data. Thus, it is a promising technique to be applied also on MRI data from various sources, e.g. in the context of multi-center trials. Future work of our group will therefore include the validation of the algorithm on heterogeneous real-life data.
Diagnosis and treatment decision based on lesion inspection on MRI data is a central aspect in MS. The clinical workflow also contains the comparison to pre-examinations to assess inflammatory activity. This process is tedious when looking at up to above 100 slices in high resolution imaging, at least four modalities and several pre-examinations. Still, common solutions for automated lesion segmentation do not rely on neural networks and are not typically applied in the clinical setting. Thus, our work is highly relevant as it investigates ways to improve the state-of-the art regarding the important aspect of longitudinal analysis, in order to make longitudinal lesion segmentation applicable and reliable in clinical MS neuroimaging.
References
Arnon, R., Miller, A.: Translational NeuroImmunology in Multiple Sclerosis, 1st edn. Academic Press Inc., London (2016)
Aslani, S., Dayan, M., Murino, V., Sona, D.: Deep 2D encoder-decoder convolutional neural network for multiple sclerosis lesion segmentation in brain MRI. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11383, pp. 132–141. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11723-8_13
Aslani, S., et al.: Multi-branch convolutional neural network for multiple sclerosis lesion segmentation. NeuroImage 196, 1–15 (2019). https://doi.org/10.1016/j.neuroimage.2019.03.068
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38(8), 1788–1800 (2019)
Baur, C., Wiestler, B., Albarqouni, S., Navab, N.: Deep autoencoding models for unsupervised anomaly segmentation in brain MR images. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11383, pp. 161–169. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11723-8_16
Birenbaum, A., Greenspan, H.: Longitudinal multiple sclerosis lesion segmentation using multi-view convolutional neural networks 10008, 58–67 (2016). https://doi.org/10.1007/978-3-319-46976-8_7
Brosch, T., Tang, L.Y., Yoo, Y., Li, D.K., Traboulsee, A., Tam, R.: Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging 35(5), 1229–1239 (2016). https://doi.org/10.1109/TMI.2016.2528821
Carass, A., et al.: Longitudinal multiple sclerosis lesion segmentation: resource and challenge. NeuroImage 148, 77–102 (2017). https://doi.org/10.1016/j.neuroimage.2016.12.064
Cohen, J.A., Rae-Grant, A.: Handbook of Multiple Sclerosis, 1st edn. Springer Healthcare, London (2012). https://doi.org/10.1007/978-1-907673-50-4
Compston, A., et al.: McAlpine’s Multiple Sclerosis, 4th edn. Churchill Livingstone. Elsevier Inc. (2005)
Dennis Jr., J.E., Woods, D.J.: Optimization on microcomputers. The Nelder-Mead simplex algorithm. Technical report (1985)
Fartaria, M.J., et al.: Partial volume-aware assessment of multiple sclerosis lesions. NeuroImage Clin. 18, 245–253 (2018)
Gaser, C., Dahnke, R.: Cat-a computational anatomy toolbox for the analysis of structural MRI data. HBM 2016, 336–348 (2016)
Ghafoorian, M., Bram, P.: Convolutional neural networks for MS lesion segmentation, method description of DIAG team. In: Proceedings of the 2015 Longitudinal Multiple Sclerosis Lesion Segmentation Challenge, pp. 1–2 (2015)
Hochreiter, S., Schmidhuber, J.: Long short-term memory. Neural Comput. 9(8), 1735–1780 (1997)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017 2017, vol. 2017 pp. 2261–2269 (2017). https://doi.org/10.1109/CVPR.2017.243
IACL: The 2015 longitudinal MS lesion segmentation challenge (2018). http://iacl.ece.jhu.edu/MSChallenge. Accessed 12 May 2020
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
McKinley, R., et al.: Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence. NeuroImage Clin. 25, 102104 (2020). https://doi.org/10.1016/j.nicl.2019.102104
Novikov, A.A., Major, D., Wimmer, M., Lenis, D., Buehler, K.: Deep sequential segmentation of organs in volumetric medical scans. IEEE Trans. Med. Imaging 38(5), 1207–1215 (2018)
Nyul, L.G., Udupa, J.K., Zhang, X.: New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 19(2), 143–150 (2000)
Penny, W.D., Friston, K.J., Ashburner, J.T., Kiebel, S.J., Nichols, T.E.: Statistical Parametric Mapping: the Analysis of Functional Brain Images. Elsevier (2011)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer Assisted Intervention, vol. 9351, pp. 234–241 (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Roy, S., Butman, J.A., Reich, D.S., Calabresi, P.A., Pham, D.L.: Multiple sclerosis lesion segmentation from brain MRI via fully convolutional neural networks (2013) (2018). http://arxiv.org/abs/1803.09172
Roy, S., et al.: Longitudinal intensity normalization in the presence of multiple sclerosis lesions. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, pp. 1384–1387. IEEE (2013)
Salem, M., et al.: Multiple sclerosis lesion synthesis in MRI using an encoder-decoder U-NET. IEEE Access 7, 25171–25184 (2019). https://doi.org/10.1109/ACCESS.2019.2900198
Salem, M., et al.: A fully convolutional neural network for new T2-w lesion detection in multiple sclerosis. NeuroImage Clin. 25, 102149 (2020). https://doi.org/10.1016/j.nicl.2019.102149
Shinohara, R.T., et al.: Statistical normalization techniques for magnetic resonance imaging. NeuroImage Clin. 6, 9–19 (2014)
Valverde, S., et al.: Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage 155, 159–168 (2017). https://doi.org/10.1016/j.neuroimage.2017.04.034
Weaver, K.F., Morales, V.C., Dunn, S.L., Godde, K., Weaver, P.F.: An Introduction to Statistical Analysis in Research: with Applications in the Biological and Life Sciences. Wiley, Hoboken (2017)
Acknowledgment
Sergio Tascon-Morales was supported by the Education, Audiovisual and Culture Executive Agency (EACEA) as part of the Erasmus Mundus Joint Master degree in Medical Imaging and Applications (MAIA).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Tascon-Morales, S. et al. (2020). Multiple Sclerosis Lesion Segmentation Using Longitudinal Normalization and Convolutional Recurrent Neural Networks. In: Kia, S.M., et al. Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology. MLCN RNO-AI 2020 2020. Lecture Notes in Computer Science(), vol 12449. Springer, Cham. https://doi.org/10.1007/978-3-030-66843-3_15
Download citation
DOI: https://doi.org/10.1007/978-3-030-66843-3_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66842-6
Online ISBN: 978-3-030-66843-3
eBook Packages: Computer ScienceComputer Science (R0)