Abstract
The ability to rapidly identify at scale patients that are similar based on their electronic health records (EHRs) is fundamental for a number of clinical informatics applications, such as clinical decision support, cohort selection, treatment recommendation, among others.
The effective representation of EHR data is paramount to effective computational similarity methods. Such representation would take into account the complex properties of EHR data including temporality and multivariaty. Of critical importance for this is the modelling of: (i) compound information – multiple medical events for a patient occur in order and may be at the same time, (ii) clinical patterns – frequent common sequential patterns that are associated with specific sequences of clinical events. To model these, in this paper we exploit the recently proposed Temporal Tree technique to capture compound information and we further apply sequential pattern mining (SPM) with gap constraint to discover more complex clinical patterns.
The effectiveness of the proposed EHR representation method is evaluated using a real EHR dataset, MIMIC III, based on two task types within an Intensive Care Unit setting: (i) similar patients retrieval (ii) sepsis prediction and mortality prediction. The empirical results show that representation of EHRs with Temporal Tree and SPM, used in conjunction with traditional similarity measures or more complex embedding methods, delivers significant improvements in effectiveness in the considered tasks.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Electronic health records representation
- Temporal Tree
- Sequential pattern mining
- Gap constraint
- Patient similarity
1 Introduction
The availability of increasingly larger amounts of electronic health records (EHRs) has motivated the exploration of large scale artificial intelligence, data mining and machine learning methods aimed to provide effective clinical decision support [14, 22]. Key to many of these methods is the representation of such patient data available in the form of EHRs. This is prominent for example in the task of similarity computing, i.e. establishing how similar a pair of patients is based on the information recorded in their EHRs [20, 21]. The computation of such a similarity requires to take into account treatments, symptoms, laboratory reports, vital signals, among other data types. The task of similarity computing, indeed, is central to other advance applications such as automatic stratification of patients [14], analysis of patient’ clinical pathways [13], personalise healthcare [32], and identifying the relationship between diseases and co-morbidities [18]. Because of its central role, in this paper we investigate a new EHR data representation method in the context of similarity computing.
The effective representation of EHR data for similarity computing, however, is challenging due to the complex nature of EHRs. EHRs in fact are multivariate, temporal, heterogeneous, irregular, and sparse. In addition, because of these multivariate and temporal characteristics, many inherent relationships between clinical events are present in EHR data. These relationships take two forms: that of compound information, and that of clinical patterns.
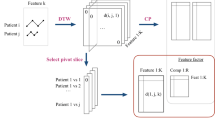
EHR information is compounded when multiple clinical events appear at the same point in time (or within a short period of time). For example, in Fig. 1a, the following patient measurements are recorded within one hourFootnote 1: <Systolic Blood Pressure(SBP):\(80{>}\), <Respiratory Rate(RR):\(18{>}\), <Glasgow Coma Scale(GCS):\(16{>}\) – because these clinical measurements are recorded within the same temporal time unit used for representation (one hour), they form a compound information.
Clinical patterns (CPs) occur when common sequential clinical events emerge over time from the EHR data. For example, Fig. 1a shows the clinical event sequence for SBP as <SBP:80, SBP:78,...\({>}\).
Clinical diagnoses are often made on the basis of observations at one point in time (compound information) and the trend of similar observations over a period of time (clinical patterns). Thus, the availability of an effective method for jointly modelling compound information and clinical patterns may be crucial for example for differential diagnosis and for reaching a deeper understanding of patient conditions – important factors when considering patient similarity or other advance clinical decision support tasks.
Previous methods have been proposed that address the above challenges, but only partially. Multivariate time series has been used to represent EHR data [6, 8], thus modelling the temporal and multivariate aspects; however this method does not consider the inherent relationships between the clinical events, such as events occurring within a short period of time [26]. Graph mining methods such as subgraph2vec [23] and deep graph [35] can be used to represent compound information through the neighbourhoods of a node but they do not allow for the representation of temporal information. Sequential Pattern Mining (SPM) methods [34, 36] or SPM with gap constraint [24] are useful to discover clinical pattens, however they only use univariate data and cannot model compound information. Recently, the Temporal Tree technique [26] has been proposed to capture compound information in EHRs; however Temporal Tree cannot identify and represent clinical patterns.
To overcome the limitations of existing methods and address the identified challenges, in this paper we propose a novel method that builds upon the recent Temporal Tree technique by integrating Sequential Pattern Mining. The proposed method works as follows: First, the Temporal Tree representation is used for capturing compound information. Then, sequences of clinical events are generated from the Temporal Tree representation. Subsequently, SPM with gap constraint is applied for discovering the complex clinical patterns. In this process, clinical patterns are generated not only from sequences of univariate observations (e.g., observations of SBP over a period of time) but also, with the help of Temporal Tree, from sequences of compound information captured across multiple levels of the hierarchical structure (e.g., combined observations of SBP, RR, GCS over a period of time). By doing so, the discovered clinical patterns encode complex relationships between clinical events due to the ability to capture multiple observations at the same time as well as frequent patterns over a period of time. We evaluate the proposed Temporal Tree with Sequential Pattern Mining for EHR representation across multiple clinical tasks where similarity computing is paramount, and consider an array of similarity measures as representative instantiations of traditional methods (Jaccard, overlap, weighted-cosine) and state-of-the-art embedding techniques (pv-dbow, pv-dm, soft-cosine).
This paper puts forward the following contributions:
-
1.
A novel method for representing EHR data that captures complex inherent relationships between clinical events. This method is based on the Temporal Tree technique and Sequential Pattern Mining with gap constraint.
-
2.
An evaluation of the proposed representation method on clinical tasks where effective similarity computing is paramount: (i) similar patient retrieval (ii) sepsis prediction and patient mortality prediction.
-
3.
A comparison of the effectiveness of the proposed representation method against state-of-the-art methods on real ICU data, showing that the proposed method provides significant improvements in effectiveness in the considered evaluation tasks.
2 Related Work
Patient Similarity. The problem of computationally establishing how similar two patients are based on their EHRs has been explored in a number of previous studies. For example, Sun et al. [31] used locally supervised metric learning to compute a patient similarity matrix. Miotto et al. [20] used unsupervised deep feature learning to derive a general purpose patient representation. Jia et al. [15] used diagnoses sets and converted the multi-label classification problem into a single-value regression problem to identify similar patients. A common drawback of these methods, however, is that they do not consider the inherent relationships between clinical events when computing similarity.
Other methods do tackle the problem of representing inherent relationships between clinical events. For example Wang et al. [33] first derived dynamic Bayesian networks (DBNs) from the EHRs for finding the correlation among variables, and then exploited the DBNs within a recurrent neural network architecture to generate a representation of each patient. These sequences were utilized to learn patient embeddings using med2vec [9]. However, this method does not consider compound information and is characterised by an overwhelming amount of parameters, rendering the learning process difficult and lengthy.
Patient Embeddings. Embedding techniques have been exploited to represent EHR data into lower dimensional vectors, where similar patients would be represented by similar embeddings. Zhang et al. [37], Choi et al. [10], and Glicksberg et al. [11] used word2vec [19] to construct a lower dimensional embedding, while Bajor et al. [5] used the document-level embedding approach [17] (also known as doc2vec). However, these methods do not explicitly model the inherent relationships between clinical events. The method of Pokharel et al. [26], Temporal Tree, which is at the basis of the method put forward in this paper, models compound information for representing a patient into an embedding; however it does not model clinical patterns.
Sequential Pattern Mining on EHRs. SPM [1] has found wide application for discovering frequent patterns from the EHR data. Wright et al. [34] used SPM to identify temporal relationships between drugs; these relationships were then exploited to predict the next prescribed medications. Similarly, Rjeily et al. [27] applied SPM, specifically Compact Prediction Tree plus (CPT+), for identifying heart failure patients. These previous examples, however, rely on SPM using univariate data only. In addition, they do not use the gap constraint in their SPM, which is important to identify clinical events that occur in close time proximity: these events are in fact likely to be more meaningful than distantly occurring events. Batal et al. [7] used pattern mining on multivariate temporal data for identifying patients who can have potential risk of heparin-induced thrombocytopenia. However they did not consider compound information and gap constraint.
3 Temporal Tree with Sequential Pattern Mining
In this paper we investigate the application of the Temporal Tree technique [26] to capture compound information and further propose to extend this method using SPM with gap constraint with the aim of discover and model more complex clinical patterns. Next, we detail the Temporal Tree technique (Sect. 3.1) and subsequently the use of SPM to generate clinical patterns (Sect. 3.2).
3.1 Temporal Tree
A Temporal Tree [26] is a temporal hierarchical structural network which is constructed based on the temporal co-occurrence of clinical events, and it allows us to represent the compound information present in EHR data. A Temporal Tree is constructed for each patient. An example of a simple Temporal Tree is shown in Fig. 1b. Each branch from a root node of a SubTree represents an event type such as laboratory events, prescriptions, etc. In this paper, we use a single event type, i.e., quick Sequential Organ Failure(qSOFA) variables (see Sect. 4.2 for more details), but multiple event types are possible.
Compound information in the Temporal Tree is generated based on the local neighbourhood relationships between clinical events and is represented in a hierarchical form. Here, the leaf nodes represent the actual clinical events that appear at the respective timestamps and non-leaf nodes represent the compound information which is generated by the relabelling process. For relabelling, the Weisfeiler-Lehman graph kernels re-labelling method [29] is used. Note that during the relabelling process, the label of a parent node is generated from its children nodes by sorting them first and then concatenating them e.g., in Fig. 1b GCSN is generated from GCN and N rather than NGCS.
Generation of Compound Information Sequences. Compound information sequences are generated from a Temporal Tree by Breath First Search (BFS) traversal. To avoid the unmeaningful labels and to capture the clinical patterns for each variables as well as compound information separately, we modify the original formulation of Temporal Tree [26] when generating a clinical sequence as follows. (1) We only consider qSOFA variables, hence we have only one branch from the root node of level 0. As a result, the labels of level 0 and level 1 are the same and thus the labels of level 0 are ignored when generating the sequence. Similarly, we also ignore the level 3 because if we generate the sequences from level 3, then they only contain the repetition of the same level (e.g., SBP, RR, GCS, A, N) which is not meaningful for distinguishing patients. (2) We generate many sequences from level 1 and level 2 as indicated by the horizontal doted line in Fig. 1b – this is unlike in the original Temporal Tree where one sequence for each level was generated. For example, in Fig. 1b we generate three different sequences from level 2 (for each variable: SBP, RR, GCS). Then, for each patient, all the generated sequences are concatenated to form a single clinical event sequence.
3.2 Clinical Patterns
Clinical Sequence. Each patient is considered as a sequence of compound information as described in Sect. 3.1. Formally, let \(\varSigma \) be a set of symbols (compound information) and \(|\varSigma |\) denote its cardinality. A clinical sequence S is defined as a temporally ordered list of clinical events and is written as S = \(\{e_{1},e_{2},\ldots ,e_{l}\}\) where \(e_{i}\) \(\epsilon \) \(\varSigma \) is the symbol at position i. D = \(\{S_{1},S_{2},\ldots ,S_{N}\}\) is a dataset of N sequences.
Subsequence. Let \(S_{1}\) = \(\{e_{1},e_{2},\ldots ,e_{m}\}\) and \(S_{2}\) = \(\{\acute{e}_{1},\acute{e}_{2},\ldots ,\acute{e}_{n}\}\) be two sequences over \(\varSigma \). Then, \(S_{1}\) is a subsequence of \(S_{2}\) (denoted by \(S_{1}\) \(\subseteq \) \(S_{2}\) and also referred to as \(S_{2}\) contains \(S_{1}\)) if there exists a one-to-one mapping \(\phi \) : [1, m] \(\rightarrow \) [1, n], such that \(S_{1}[i]\) = \(S_{2}[\phi (i)]\) and for any two positions i, j in \(S_{1}\), \(i<j\) \(\Rightarrow \) \(\phi (i)\) < \(\phi (j)\) [36].
General SPM instantiations are set to discover all present patterns without modelling the gap between the symbols. We argue however that in the case of clinical event sequences, this gap does matter. During a patient’s stay in ICU, for example, a patient condition is very unstable and one of the key goals of an ICU doctor is to bring the patient to a stable condition. Treatments are thus provided based on the immediate condition of a patient being observed; further observations are made thereof, adjusting or changing the treatment regime. Thus, the closer two clinical events are (close gap), the more meaningful and strong their relationship is. To model this, we consider sequential patterns under gap constraint satisfaction.
Gap Constraint. A gap (denoted by \(\varDelta \)) is a positive integer, \(\varDelta \) \({>}\) 0. Let a clinical sequence be S = \(\{\acute{e}_{1},\acute{e}_{2},\ldots ,\acute{e}_{n}\}\) and an occurrence o \(=\) \(\{i_{1},i{2},\ldots ,i_{n}\}\) of a subsequence X of S. If \(i_{k+1}\) \(\le \) \(i_{k}\) + \(\varDelta \) \((\forall i_{k} \epsilon [i,n-1])\), then o satisfies the \(\varDelta \)-gap constraint. If there is at least one occurrence o of X that satisfies the \(\varDelta \)-gap constraint, then X satisfies the \(\varDelta \)-gap constraint [24].
SPM with Gap Constraint. Given a clinical sequential dataset D, a gap constraint \(\varDelta \) and a minimum support threshold (denoted by \(\delta \in [0,1]\)), sequential pattern discovery deals with finding all the subsequences(X), along with their corresponding supports (\(\sigma \)), such that \(\sigma (X,\varDelta ) \ge \delta \).
Example 1. Figure 2 shows an example of generating the clinical pattens for each patient using Temporal Tree and SPM with gap constraint.
(a) An example of compound information; (b) sequence of clinical events without applying Temporal Tree; (c) clinical sequence generated by Temporal Tree using BFS traversal; (d) from table c, ten clinical patterns are discovered using SPM with gap constraint, given that \(\varDelta \) = 1 and \(\delta \) = 0.6; (e) sequence of clinical patterns for each patient.
Generating the clinical patterns has the following advantages: (1) they capture the inherent complex temporal and multivariate relationships between clinical events. For example, X9 is a pattern of a patient having normal GCS and then abnormal GCS. In real data, we observe a large number of more complex patterns than the example X9. (2) They increase the accuracy of the representation. (3) By considering both singleton clinical events and clinical patterns, we can increase the vocabulary size which results in better feature representation.
Example 2. Listing 1.1 shows an example of discovered clinical patterns using a real dataset. Here, X101 represents that a patient has normal GCS and RR, but abnormal SBP at the same time. Likewise X974 represents a clinical pattern of the type: abnormal RR, abnormal RR, abnormal RR, normal RR.

4 Evaluation Methodology
4.1 Dataset and Patient Cohort Selection
We use a publicly available de-identified real ICU dataset, MIMIC III [16], to evaluate the proposed approach. We consider each ICU admission as referring to a unique patient. Patients were selected according to the following criteria: (i) adults (patients aged 16 years or more), (ii) have at least one value recorded for each qSOFA variable (see Sect. 4.2 for more details), (iii) have been admitted to ICU for the first time – re-admitted patients are excluded because it is likely a patient is re-admitted for the same condition, and thus the data would show a high correlation, (iv) top-3 most frequent firstFootnote 2 diagnoses only. This is because the use of all diagnoses available would result in a largely sparse similarity matrix to be used for evaluation. To avoid this, we filter the patients using such a criteria. Note that this is not a limitation of the proposed method, but an empirical setting chosen to maintain reliability in the evaluation. The filtered subset contains a total of 5, 274 patients. Note the dataset presents a bias towards survival patients (did not die during hospital stay); mortality: 642, survival: 4, 632. Similarly, the dataset contains 1, 783 patients that have developed sepsis and 3, 491 with no sepsis.
4.2 Features Selection
The features are selected based on the qSOFA criteria. The qSOFA score [4] is a simplified version of Sequential Organ Failure Assessment (SOFA) Score as an initial way to assess patients at high risk of poor outcome with respect to infection/sepsis. The advantage of qSOFA is its simplicity compared to SOFA which requires numerous lab tests, more time and is more expensive. qSOFA can be repeated serially and it can be applied outside the ICU setting as an initial way to identify patients at risk. qSOFA uses three variables to test the abnormality of organs according to the following criteria: Low Blood Pressure (SBP \(\le \)100 mmHg), High Respiratory Rate (RR \(\ge \) 22 breaths/min) and Altered Mentation (GCS \(\le \) 13) [28].
We set the time intervals to one hour and if more than one event is found within an interval, we take the average value of the events. Further, missing data is restored using linear interpolation. Numeric values are converted into categorical values (\(A{:}\ Abnormal\), \(N{:}\ Normal\)) using the qSOFA criteria. We also set time intervals for Temporal Tree to one hour because the clinical events that occur in close temporal proximity often have a stronger relationship than events that occur far apart (at least during an admission in ICU).
4.3 Baselines
In the experiments, we apply the proposed representation to both traditional (jaccard, overlap, weighted-cosine) as well as state-of-the-art embedding based similarity methods (pv-dbow, pv-dm, soft-cosine), see Table 1.
-
Jaccard: Similarity is computed based on the number of common clinical patterns shared by two patients (each patient is represented as a set of clinical patterns) over the size of the union for two patients.
-
Overlap: Similarity is computed based on number of common clinical patterns shared by two patients (each patient is represented as a set of clinical patterns) over the size of the smaller set of the two patients.
-
Weighted-cosine: The clinical patterns are weighted according to term frequency (tf)-inverse document frequency(idf). Then, the similarity is calculated based on the cosine angle between the two patient vectors.
-
PV-DM [17]: State-of-the-art embedding based method where concatenation (or average) of a sequence vector along with surrounding CPs is used to predict a target CP. The sequence vector represents the missing information from the current context and can act as a memory of the topic (in the original paper, topic refers to topic of a paragraph) of the sequence of CPs. Note that each patient is represented as a sequence of CPs as described in Sect. 3.2.
-
PV-DBOW [17]: State-of-the-art embedding based method where the sequence vector is trained to predict the CPs in a small window. Unlike the PV-DM, this model ignores the surrounding CPs in the input, but force the model to predict CPs which are randomly sampled from the sequence of CPs in the output. Note that each patient is represented as a sequence of CPs as described in Sect. 3.2.
-
Soft-cosine [30]: The tf-idf schema is used for assigning weights to clinical patterns and pv-dbow is used for calculating the similarity between clinical patterns. We have used the optimized soft-cosine formula as shown in Table 1 to reduce computational complexity.
As for comparison, we consider the following EHRs representations (i) raw features - clinical events without considering Temporal Tree and the modelling of clinical patterns (ii) Temporal Tree [26] - originally proposed without modelling of clinical patterns (iii) Temporal Tree with SPM - the proposed representation method in this paper where Temporal Tree with modelling of clinical patterns are considered. We apply all the above measures to these three considered representations.
4.4 Evaluation Tasks
The effectiveness of the proposed representation method is evaluated in two contexts: (i) similar patients retrieval (ii) prediction models.
Similar Patients Retrieval. We cast the similarity computing problem into an information retrieval problem where the task is to retrieve the patients that are similar to a query patient. For this, we follow the previous similar works by Gottlieb et al. [12] and Pokharel et al. [26]. Similar to them, we use the International Classification of Diseases, Ninth Revision (ICD-9) codes as gold standard to measure patient similarity. The gold standard similarity between two patients is calculated based on the number of diagnoses shared along with the respective ranking of diagnoses. This is achieve by using the SimIndex function [25]. For a query patient, the ranking of similar patients retrieved by the system is evaluated in terms of the following parameters: (1) Mean Square Error (MSE): computes the error made by the system when retrieving similar patients, compared to the gold standard. (2) Normalized Discounted Cumulative Gain (nDCG): Discounted Cumulative Gain (DCG) computes a weighted sum of the degree of relevancy while ranking the retrieved similar patients by the system. And the nDCG is DCG normalized by the ideal DCG - in our case, ideal DCG is DCG measure from patient similarity matrix which is obtained by using gold standard. (3) Precision: we follow the work of Gottlieb et al. [12] where they only consider the top two diagnoses (highest priority) and don’t use simIndex function. So, relevance of the query patient is defined as follows: if a retrieved patient contains any of the two diagnosis of the query patient, then the patient is considered as relevant to the query patient. We restrict the number of retrieved patients for a given patient query to \(k=1,5,10,20\).
Prediction Models. Intuitively, similar patients are likely to exhibit similar mortality and sepsis risks – thus effective representation methods would exhibit similar features for similar patients and thus be effective for prediction tasks such as (i) sepsis prediction (ii) in ICU patient mortality prediction. We use these two important ICU tasks as a down-stream application of the proposed EHR data representation method.
For sepsis prediction, we consider the sepsis information (sepsis or not-sepsis) for a patient as the class labels, thus becoming a binary classification problem. For obtaining the sepsis information, we follow the work of Angus et al. [3] which is common practice in hospitals for sepsis patient identification.
For mortality prediction, we consider the mortality information (survive or not-survive) for a patient at the end of their ICU stay as the class labels, thus becoming a binary classification problem.
For both prediction tasks, we use k-Nearest Neighbourhood (kNN) for classification as it is an intuitive similarity-based approach that can directly rely on the representations studied in this paper. We evaluate the classification effectiveness according to \(f1\_micro\), \(f1\_macro\) and Area Under the Receiver Operating Characteristic Curve (AUC). We apply 5-folds cross validation: in each fold (training:four portions, testing:one portion), the training dataset is further divided into sub-training and sub-validation dataset with 80:20 ratio to determine the value of k (which gives the maximum accuracy); k is varied in the range [0,20] with step 1; thus obtaining k is used for evaluation by using training and testing dataset. The whole process is repeated five times and the effectiveness is averaged to weed out bias due to the random partition of the training data.
5 Analysis of Empirical Results
5.1 Experimental Criteria Setup
Three main parameters govern our experiments: the minimum threshold (\(\delta \)), the gap constraint (\(\varDelta \)) for discovering clinical patterns, and the number of embedding dimensions (ed). We set \(\delta = 0.05\) which is a common value for this parameter, \(\varDelta = 2\) because in the case of ICU, the clinical events that are close to each other in time are more meaningful than others, and \(ed = 50\) following Altszyler et al. [2] who suggested 50 dimensions are appropriate for a medium-size dataset like the one we consider.
5.2 Similar Patients Retrieval Task
Tables 2, 3 and 4 report the performance of different methods for similar patient retrieval in terms of nDCG, Precision and MSE. Note that all differences between methods are statistically significant (t-test with Bonferroni correction). In the rest of the paper, the suffixes indicate the following strategies: \(\_raw\): without use of Temporal Tree and clinical patterns, \(\_tt\): with use of Temporal Tree only, \(\_tt\_spm\): with use of Temporal Tree with clinical patterns.
The following observations can be made based on the empirical results:
-
1.
In general, all similarity methods obtain higher effectiveness when representing EHRs with Temporal Tree and even better effectiveness when using in addition SPM.
-
2.
Surprisingly, jaccard performs better than embedding based methods when EHRs are represented with Temporal Tree and SPM.
-
3.
The soft-cosine method gives mixed result when using Temporal Tree and SPM.
-
4.
The best effectiveness is obtained by jaccard (in terms of nDCG for \(k=5,10,20\), precision for \(k=1,5,10,20\)) and overlap (in terms of in terms of nDCG for \(k=1\)) when applying Temporal Tree and SPM. In the case of MSE, the best effectiveness is obtained by jaccard (for \(k=1\)) and soft-cosine (for \(k=5\)) with Temporal Tree only and soft-cosine with raw features (for \(k=10,20\)).
-
5.
Overall, Temporal Tree with SPM is found to be highly effective for representing EHRs (note, for MSE, the lower the better).
5.3 Prediction Tasks
Tables 5 and 6 reports the effectiveness of prediction methods in terms of \(f1\_micro\), \(f1\_macro\) and AUC for sepsis prediction and mortality prediction, respectively. From the results, the following observations can be made:
-
1.
The most effective methods have been those that used when Temporal Tree and Temporal Tree with SPM for representing EHRs.
-
2.
pv-dm, an embedding based method, gives mixed results when using Temporal Tree with SPM.
-
3.
Weighted cosine is the most effective method for sepsis prediction, while jaccard (for \(f1\_micro\)) and pv-dbow (for \(f1\_macro\) and AUC) are the most effective for the task of mortality prediction. All these methods perform when Temporal Tree with SPM is used to represent EHRs.
6 Conclusion and Future Work
In this paper we introduce a novel method for representing patient EHR data based on Temporal Tree with gap constraint with sequential pattern mining. The ability to holistically represent EHR data is paramount to effective patient similarity computation, which forms the basis of many methods in clinical decision support. Key to our method is the modelling of complex clinical patterns which exist within EHRs.
To demonstrate the proposed method, we perform an empirical evaluation that exploits our method within traditional as well as recent embedding based techniques for patient similarity computation. The empirical results show that the proposed method, Temporal Tree with sequential pattern mining with gap constraint, is an effective representation to be exploited when computing patient similarity from EHR. In future work, we plan to extend our method by investigating its capabilities in making the representation and the similarity computation explainable so as to increase clinicians’ understanding of the results produced by the computational methods.
Notes
- 1.
Assume one hour is the temporal time unit used for representation.
- 2.
Each patient may have multiple diagnoses: we only consider the first diagnosis when filtering the data to create the subset for evaluation. The used primary icd9_code are: “41401”,“0389” and “51881”.
References
Agrawal, R., Srikant, R.: Mining sequential patterns. In: Proceedings of the 11th International Conference on Data Engineering, pp. 3–14. IEEE (1995)
Altszyler, E., Ribeiro, S., Sigman, M., Slezak, D.F.: The interpretation of dream meaning: resolving ambiguity using latent semantic analysis in a small corpus of text. Conscious. Cogn. 56, 178–187 (2017)
Angus, D.C., Linde-Zwirble, W.T., Lidicker, J., Clermont, G., Carcillo, J., Pinsky, M.R.: Epidemiology of severe sepsis in the united states: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29(7), 1303–1310 (2001). Society of Critical Care Medicine
Angus, D.C., et al.: A framework for the development and interpretation of different sepsis definitions and clinical criteria. Crit. Care Med. 44(3), e113 (2016)
Bajor, J.M., Mesa, D.A., Osterman, T.J., Lasko, T.A.: Embedding complexity in the data representation instead of in the model: A case study using heterogeneous medical data. arXiv preprint arXiv:1802.04233 (2018)
Batal, I., Fradkin, D., Harrison, J., Moerchen, F., Hauskrecht, M.: Mining recent temporal patterns for event detection in multivariate time series data. In: Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 280–288 (2012)
Batal, I., Valizadegan, H., Cooper, G.F., Hauskrecht, M.: A pattern mining approach for classifying multivariate temporal data. In: 2011 IEEE International Conference on Bioinformatics and Biomedicine, pp. 358–365. IEEE (2011)
Batal, I., Valizadegan, H., Cooper, G.F., Hauskrecht, M.: A temporal pattern mining approach for classifying electronic health record data. ACM Trans. Intell. Syst. Technol. (TIST) 4(4), 63 (2013)
Choi, E., et al.: Multi-layer representation learning for medical concepts. In: Proceedings of the 22nd ACM SIGKDD, pp. 1495–1504 (2016)
Choi, Y., Chiu, C.Y.I., Sontag, D.: Learning low-dimensional representations of medical concepts. AMIA Jt. Summits Transl. Sci. Proc. 2016, 41 (2016)
Glicksberg, B.S., et al.: Automated disease cohort selection using word embeddings from electronic health records. In: PSB, pp. 145–156. World Scientific (2018)
Gottlieb, A., Stein, G.Y., Ruppin, E., Altman, R.B., Sharan, R.: A method for inferring medical diagnoses from patient similarities. BMC Med. 11(1), 194 (2013)
Huang, Z., Dong, W., Duan, H., Li, H.: Similarity measure between patient traces for clinical pathway analysis: problem, method, and applications. IEEE J. Biomed. Health Inform. 18(1), 4–14 (2014)
Jensen, P.B., Jensen, L.J., Brunak, S.: Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 13(6), 395 (2012)
Jia, Z., Zeng, X., Duan, H., Lu, X., Li, H.: A patient-similarity-based model for diagnostic prediction. Int. J. Med. Inform. 135, 104073 (2020)
Johnson, A.E., et al.: MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016)
Le, Q., Mikolov, T.: Distributed representations of sentences and documents. In: International Conference on Machine Learning, pp. 1188–1196 (2014)
Li, L., et al.: Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci. Transl. Med. 7(311), 311ra174 (2015)
Mikolov, T., Sutskever, I., Chen, K., Corrado, G.S., Dean, J.: Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems, pp. 3111–3119 (2013)
Miotto, R., Li, L., Kidd, B.A., Dudley, J.T.: Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 6, 26094 (2016)
Miotto, R., Weng, C.: Case-based reasoning using electronic health records efficiently identifies eligible patients for clinical trials. J. Am. Med. Inform. Assoc. 22(e1), e141–e150 (2015)
Musen, M.A., Middleton, B., Greenes, R.A.: Clinical decision-support systems. In: Shortliffe, E.H., Cimino, J.J. (eds.) Biomedical Informatics, pp. 643–674. Springer, London (2014). https://doi.org/10.1007/978-1-4471-4474-8_22
Narayanan, A., Chandramohan, M., Chen, L., Liu, Y., Saminathan, S.: subgraph2vec: Learning distributed representations of rooted sub-graphs from large graphs. arXiv preprint arXiv:1606.08928 (2016)
Nguyen, D., Luo, W., Nguyen, T.D., Venkatesh, S., Phung, D.: Sqn2Vec: learning sequence representation via sequential patterns with a gap constraint. In: Berlingerio, M., Bonchi, F., Gärtner, T., Hurley, N., Ifrim, G. (eds.) ECML PKDD 2018. LNCS (LNAI), vol. 11052, pp. 569–584. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-10928-8_34
Pokharel, S., Li, X., Zhao, X., Adhikari, A., Li, Y.: Similarity computing on electronic health records (2018)
Pokharel, S., Zuccon, G., Li, X., Utomo, C.P., Li, Y.: Temporal tree representation for similarity computation between medical patients. Artif. Intell. Med. 108, 101900 (2020)
Rjeily, C.B., Badr, G., Al Hassani, A.H., Andres, E.: Predicting heart failure class using a sequence prediction algorithm. In: 2017 4th International Conference on Advances in Biomedical Engineering (ICABME), pp. 1–4. IEEE (2017)
Seymour, C.W., et al.: Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315(8), 762–774 (2016)
Shervashidze, N., Schweitzer, P., van Leeuwen, E.J., Mehlhorn, K., Borgwardt, K.M.: Weisfeiler-lehman graph kernels. J. Mach. Learn. Res. 12, 2539–2561 (2011)
Sidorov, G., Gelbukh, A., Gómez-Adorno, H., Pinto, D.: Soft similarity and soft cosine measure: similarity of features in vector space model. Computación y Sistemas 18(3), 491–504 (2014)
Sun, J., Wang, F., Hu, J., Edabollahi, S.: Supervised patient similarity measure of heterogeneous patient records. ACM SIGKDD Expl. Newsl. 14(1), 16–24 (2012)
Utomo, C.P., Kurniawati, H., Li, X., Pokharel, S.: Personalised medicine in critical care using Bayesian reinforcement learning. In: Li, J., Wang, S., Qin, S., Li, X., Wang, S. (eds.) ADMA 2019. LNCS (LNAI), vol. 11888, pp. 648–657. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-35231-8_47
Wang, Y., Chen, W., Pi, D., Boots, R.: Graph augmented triplet architecture for fine-grained patient similarity. World Wide Web 23(5), 2739–2752 (2020). https://doi.org/10.1007/s11280-020-00794-y
Wright, A.P., Wright, A.T., McCoy, A.B., Sittig, D.F.: The use of sequential pattern mining to predict next prescribed medications. J. Biomed. Inform. 53, 73–80 (2015)
Yanardag, P., Vishwanathan, S.: Deep graph kernels. In: Proceedings of the 21th ACM SIGKDD, pp. 1365–1374. ACM (2015)
Zaki, M.J., Meira Jr., W., Meira, W.: Data Mining and Analysis: Fundamental Concepts and Algorithms. Cambridge University Press, Cambridge (2014)
Zhang, J., Kowsari, K., Harrison, J.H., Lobo, J.M., Barnes, L.E.: Patient2Vec: a personalized interpretable deep representation of the longitudinal electronic health record. IEEE Access 6, 65333–65346 (2018)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Pokharel, S., Zuccon, G., Li, Y. (2020). Representing EHRs with Temporal Tree and Sequential Pattern Mining for Similarity Computing. In: Yang, X., Wang, CD., Islam, M.S., Zhang, Z. (eds) Advanced Data Mining and Applications. ADMA 2020. Lecture Notes in Computer Science(), vol 12447. Springer, Cham. https://doi.org/10.1007/978-3-030-65390-3_18
Download citation
DOI: https://doi.org/10.1007/978-3-030-65390-3_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-65389-7
Online ISBN: 978-3-030-65390-3
eBook Packages: Computer ScienceComputer Science (R0)