Abstract
Multi-modal medical image fusion plays a significant role in clinical applications like noninvasive diagnosis and image-guided surgery. However, designing an efficient image fusion technique is still a challenging task. In this paper, we propose an improved multi-modal medical image fusion method to enhance the visual quality and contrast of the fused image. To achieve this work, the registered source images are firstly decomposed into low-frequency (LF) and several high-frequency (HF) sub-images via non-subsampled shearlet transform (NSST). Afterward, LF sub-images are combined using the proposed weight local features fusion rule based on local energy and standard deviation, while HF sub-images are fused based on the novel sum-modified-laplacien (NSML) technique. Finally, inversed NSST is applied to reconstruct the fused image. Furthermore, the proposed method is extended to color multi-modal image fusion that effectively restrains color distortion and enhances spatial and spectral resolutions. To evaluate the performance, various experiments conducted on different datasets of gray-scale and color images. Experimental results show that the proposed scheme achieves better performance than other state-of-art proposed algorithms in both visual effects and objective criteria.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Nowadays, multi-modal medical image fusion has been emerging as a crucial area for clinical diagnosis and analysis. The target of multi-modal image fusion is to integrate complementary information of multi-modal source images into one comprehensive image, with the aim to improve visual quality, preserves more content information and decrease computational task [1]. In general, modern medical imaging modalities are available to guide doctors and radiologist in specific medical applications. These modalities are broadly classified into structural and functional [1]. Magnetic resonance imaging (MRI) and computed tomography (CT) reflect the structural information of an organ with high spatial resolution, therefore represented structural modalities. Where, functional MRI (fMRI), positron emission tomography (PET) and single-photon emission CT (SPECT) images give functional information with low spatial resolution, so grouped as functional modalities. Based on [1, 2], it is hard to obtain accurate information about specified organ from a single modality. For example, MRIs provide detailed information about pathological soft tissues, while CT images can clearly give the information of bone structures. Likewise, PET images can be utilized for the quantitative and dynamic detection of metabolic substances in the human body, while SPECT images show clinically variations in metabolism. Therefore, we need to create an effective multi-modal fusion method to facilitate the aided diagnosis and treatment planning. In the literature, image fusion algorithms are mainly developed at three levels: pixel [2], feature [3] and decision [4]. Usually, pixel-level fusion is used in the medical field because of its several advantages [2]. It is divided into spatial-domain and transform-domain. The spatial-domain fusion methods like principal component analysis (PCA) [5] and intensity-hue-saturation (IHS) [6] are broadly suitable for mono-modal image fusion, but they often suffer from block or region artifacts and spectral distortion. In the transform domain, different kinds of transforms have achieved great success, such as pyramid transform [7], wavelet transform [5], curvelet transform [8], contourlet transform [9] and shearlet transform [10]. Recently, Easley et al. [11] have proposed the NSST transform that has been successfully adopted in the medical fusion field.
On the other hand, the design of fusion rules for decomposed coefficients is the key factor that influences the image fusion quality. So far, a variety of fusion methods have been proposed. In the transform domain, sparse representation (SR) [11] and fuzzy logical [12] have successfully used for medical image fusion. In the similarity, pulse-coupled neuronal network (PCNN) and its modified versions have been widely adopted for medical fusion domain [13]. We have proposed a simplified PCNN model for fusing MRI and PET images in [14]. However, the major limitation of these models is time-consuming due to several parameters and complicated function mechanisms [14, 15]. In a similar vein, deep learning is a recent machine learning used for image fusion, but it not been widely applied in the medical fusion field due to the high time-consuming limitation and the significant demand for computational power [16]. Recently, sum-modified-laplacian (SML) is one of the obvious tools that can well-reflect the feature information about contours and edges of the image. At the beginning, Huang et al. [17] have employed SML for multi-focus image fusion and they achieved good results. Then, a new fusion scheme based on a novel SML (NSML) was proposed by Yin et al. [18] and used for capturing all salient features [19].
Inspired by the transform-domain algorithms, we proposed a pixel-based fusion method for multi-modal medical images. The core contribution of the proposed fusion method is the weight local features rules implementation for approximated coefficients based on local energies and standard deviation that significantly enhance visual appearance and reduce blurring effects. Besides, NSML is used for capturing all salient features from detailed coefficients. The proposed algorithm is further extended to color medical image fusion based on YIQ transformation. The rest of this paper is organized as follows. Section 2 briefly introduces related theories of NSST. Section 3 explains the proposed method in detail. Section 4 describes the extensive experimental results and discussions. Finally, Sect. 5 concludes the paper.
2 Non-subsampled Shearlet Transform
The NSST is a shift-invariant version of the shearlet transforms [11]. It has several advantages like shift-invariance, multi-directionality and computational simplicity. In two dimensions (2D), the affine system with composite dilation defined by [11]:
A denotes the scaling matrix. S stands for the shear matrix. j, k, m are the scale, direction and shift parameters, respectively. For each d > 0 and s \( \in \) R,
For any \( \upxi = \left( {\upxi_{1} ,\upxi_{2} } \right) \in {\mathbb{R}}^{2} \), \( \upxi_{1} \ne 0, \) the shearlet function is defined as:
Where \( \widehat{\uppsi} \) is the Fourier transform of \( \uppsi \), \( \uppsi_{1} \in {\text{C}}^{\infty } \left( {\text{R}} \right) \) and \( \uppsi_{2} \in {\text{C}}^{\infty } \left( {\text{R}} \right) \) are both wavelet and supp \( \uppsi_{1} \subset [ - 1/2, - 1/16\left] \,{ \sqcup }\, \right[1/16,1/2 \), supp \( \uppsi_{2} \subset \left[ { - 1,1} \right] \). This indicates that \( \uppsi_{0} \in {\text{C}}^{\infty } \left( {\text{R}} \right) \) and supp \( \uppsi_{0} \subset \left[ { - 1/2,1/2} \right]^{2} \). Subsequently, we suppose that:
For each \( {\text{j}} \ge 0 \), \( \uppsi_{2} \) satisfies that:
Based on several examples of \( \uppsi_{1} \) and \( \uppsi_{2} \), Eq. 4 and Eq. 5 notice that:
The discrete NSST transform can be obtained from the different equations mentioned above. More theoretical background can be found in [11, 18].
3 Proposed Fusion Method
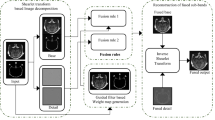
Let A and B denote the input images. In the beginning, we confirm that image registration is not related to the entire system. The input images are selected from a registered medical source. A schematic diagram of the proposed method is illustrated in Fig. 1. First, the input images were normalized and then decomposed up to three levels into LF and HF sub-bands by applying the NSST to separate the principal information and the edge details of the source image. The direction numbers from finer to coarser levels are set at [3, 4]. The ‘maxflat’ pyramidal filter is used and the size of the shearing window is set at 32, 32, and 16. After that, proposed fusion rules are adopted to integrate coefficients. Finally, inversed NSST is applied to get the fused image.
3.1 Fusion Rule of the Low Frequency Sub-images
Approximated coefficients are very important for the visual quality of the fused image. Traditional ways for fusing LF sub-bands are by taking the weighted average or the maximum of the coefficient, but they directly affect the contrast and resolution of the output image [18].Therefore, a new fusion rule is given based on local energy and standard deviation, which are frequently used for fusing salient features [14, 19].
The local energy features enable us to describe the inherent texture of the image by analyzing the grade of associated information [9]. Besides, the local texture features of an image have a strong connection with the variation of the image coefficients, as well as its neighborhood [14]. The variation can be depicted by the regional standard deviation. Let \( {\text{C}}_{\text{F}} \left( {{\text{a}},{\text{b}}} \right) \) denotes the LF coefficient located at (a, b). The proposed fusion method is described as follows:
Where \( \updelta_{\upmu} \) is the weight for the standard deviation \( {\text{D}}_{\upmu} \) while \( \upvarepsilon_{\upmu} \) is the weight of the local energy \( {\text{E}}_{\upmu} \). A and B are the input image, and \( \upmu \) = A,B. They are calculated in the 3 × 3 neighborhood as follows:
The regional standard deviation \( {\text{D}}_{\upmu} \) and the local energy \( {\text{E}}_{\upmu} \) are calculated as:
Where the template \( \upomega = \left\{ {\begin{array}{*{20}c} 1 & 2 & 1 \\ 2 & 4 & 2 \\ 1 & 2 & 1 \\ \end{array} } \right\} \times \frac{1}{16} \), and \( {\text{S}}_{\upmu} \) is calculated as follows:
3.2 Fusion Rule of the High Frequency Sub-images
For the fusion of detailed coefficients, the system utilizes NSML technique recently applied by several works [18, 19]. Indeed, NSML can well-reflect the important features and properly assesses the focused features [19]. Let \( {\text{C}}_{\text{F}}^{{{\text{l}},{\text{k}}}} \left( {{\text{i}},{\text{j}}} \right) \) stands for the HF coefficient in the position (i, j) in the lth scale and kth direction. The fused \( {\text{C}}_{\text{F}}^{{{\text{l}},{\text{k}}}} \) coefficients are selected by computing and comparing NSML. The coefficient \( {\text{C}}_{\text{F}}^{{{\text{l}},{\text{k}}}} \left( {{\text{i}},{\text{j}}} \right) \) with maximum NSML value is selected as follows:
The NSML is defined as:
Where \( {\text{MP}}^{{{\text{l}},{\text{k}}}} \left( {{\text{i}},{\text{j}}} \right) \) denotes the directional band-pass, sub-band coefficient located at the pixel (i, j) in the lth sub-band at the kth decomposition level. While computing the derivative, \( {\text{step}} \) denoted the variable spacing between pixels, typically equal to 1. P and Q are the parameters that determine the window with a size of \( \left( {2{\text{P}} + 1} \right)\left( {2{\text{Q}} + 1} \right) \), \( {\text{w}}\left( {{\text{m}},{\text{n}}} \right) \) represents the weights of the \( {\text{ML}}^{{{\text{l}},{\text{k}}}} \left( {{\text{i}} + {\text{m}},{\text{j}} + {\text{n}}} \right) \), and must satisfying the normalization rules i.e. \( \sum\nolimits_{\text{m}} {\sum\nolimits_{\text{n}} {\left( {{\text{m}},{\text{n}}} \right) = 1} } \). Therefore, we choose P = Q = 1 and the same weighed window used for fusing LF coefficients.
3.3 Color Image Fusion
In recent years, fusion of structural and functional images has been an interesting hybrid tool that brings an important revolution in the medical field [1]. Usually, color images are in RGB (Red, Green, Blue) color space, which contains almost all the basic colors that can be perceived by human vision. Nevertheless, the three colors are treated equally and their components are strongly correlated. So, it makes the RGB color space very difficult to determine what color of the image will be changed if a component changes. To overcome this problem, several color transformations, such as YIQ, HSV, and IHS are proposed. In YIQ, image data consists of three components. The first component luminance (Y) represents the grayscale information, while the last two components hue (I) and saturation (Q) denote the chrominance information.
In this study, the color inputs are transformed from RGB to YIQ, which provides better spectral and spatial characteristics and reduces color distortion. The proposed scheme for color image fusion is illustrated in Fig. 2 and described as follows: First, the input PET/SPECT image perfectly registered and normalized in advance is transformed into the YIQ independent components. Second, the proposed gray fusion method is applied. The final output is obtained using the inversed YIQ by exploiting the new intensity and the original I and Q components of the color image.
4 Experimental Results and Discussion
To determine the overall performance of the proposed fusion method, extensive experiments are conducted on different types of pre-registered dataset images. All used images were frequently taken from the Website of Medical School of Harvard University “http://www.med.harvard.edu/AANLIB/” and from the fusion website “http://www.imagefusion.org”. In this study, five objective image quality metrics are adopted [20]: (i) Entropy (E) that evaluates the quantity of information in the fused image. (ii) Feature Mutual Information (FMI) that measures the amount of feature information. (iii) Fusion Quality index (QAB/F) that gives valuable information about edge preservation. (iv) Standard Deviation (SD) that reflects the contrast information in the fused images. (v) Structural Similarity Index Measure (SSIM), which determines the structural similarity between two images.
4.1 Experiment-1: Gray-Scale Image Fusion
For the first section of experiments, four different datasets of CT and MRI images of the size 256 × 256 are selected as shown in Fig. 3. To verify the effectiveness of the proposed approach, the following existing state-of-art fusion methods are considered: Method.1 [12], Method.2 [21], Method.3 [18], Method.4 [13], Method.5 [22] and Method.6 [19].
Visual Analysis. Fusion results produced by the above techniques are shown in Fig. 4. Images (a) in every set are clearly noted blurred and having brightness issues and high noise. From images (b) and (c), a loss of contours and edges information can easily be noted as compared to images (g). The implementation of the modified MPCNN model in the MSVD domain gives better results, as can be shown by images (d). Method.5 provided clear results (see images (e)), but due to direct averaging of the LF coefficients, it gives less distinction or low contrast in some superposed positions. Hence, Method.6 produces superior results with good contrast. Moreover, by comparing all these images at once, it can easily notice that the images obtained from the proposed method are clearer (see images (g)), almost all the salient features are clearly visible. This is mainly because of the NSST decomposition because of the advantage of high computational efficiency. Where, the good contrast and high resolution are preserved due to the proposed fusion rule for LF coefficients and the utilization of NSML for focusing and capturing high features from the HF coefficients.
Quantitative Analysis. Table 1, 2, 3, 4 provide quantitative performance. It should be noted that a larger measure implies better quality. First, through the significant results obtained for E given in Table 1, it can be clearly concluded that the proposed method gains the highest E for almost all the datasets, which shows that maximum structural information is present in the fused image. Second, the FMI results listed in Table 2 shows that the proposed method gives higher FMI for all datasets. It shows that maximum information about the edge strength, texture and contrast from the source images is retained in the fused image. Third, almost all datasets preserved the highest values of the QAB/F (Table 3) except for dataset-1 in Method. 6, which indicates that more edge details are provided by our algorithm. Finally, results obtained for SD are given Table 4. The proposed technique produces higher contrast for all sets except dataSet-1 in Method.6. From these results, it can be observed that the proposed method provides an efficient fusion tool compared with other mainstream algorithms.
4.2 Experiment-2: Color Image Fusion
For this section of experiments, five data-sets of MRI, PET and SPECT images having a size of 256 × 256 are selected as shown in Fig. 5.The following state-of-art schemes are selected to verify the effectiveness of the proposed color fusion method: Scheme.1 [12], Scheme.2 [15], Scheme.3 [23], Scheme.4 [14] and Scheme.5 [19].
Visual Analysis. Fused results are given in Fig. 6. Images (a) produced by Scheme.1 preserve both structural and functional information, but the approach works in grayscale space. From images (b), it can be observed that most of the information of brain structures in the non-functional area is lost. Furthermore, these images suffer from color distortion and contrast reduction. We expect more satisfactory results from Scheme.3 (see images (c)) because of the advantages of implementing fusion rules on every channel separately after each local Laplacian filtering (LLF) decomposition level. Although, the images obtained through this scheme contain some noise and are showing deficiency in edge strength. In general, more visual superiority is retained by Scheme.4 and Scheme.5, where the fused outputs have good resolution and contrast as well (see images (d)) and images (e)). Compare to these existing methods, it can be easily recognized in the images obtained by the proposed scheme (see images (f)) that problem of color and contrast reduction is highly improved.
Quantitative Analysis. In order to support the visual quality of the proposed scheme, the quantitative matrices FMI, QAB/F, SSIM and SD for the previously mentioned schemes are also computed. Results obtained from these matrices for all the five sets are plotted in Fig. 7, 8, 9, 10. In general, the proposed scheme gives the highest values for almost every set. Furthermore, our fused images show more visual superiority and effectiveness in contrast with excellent spatial and spectral resolutions. Hence, from the visual and quantitative analysis, we conclude that the proposed algorithm has successfully injected the anatomical information of the high-resolution MRI image into the metabolic information of the PET/SPECT image.
Computational Efficiency. In pursuit of the clear comparison, time costs of the compared algorithms are illustrated in Table 5. It can be observed that methods [21] and [14] consume lot more time (83 s and 180 s) compare to others. This is because these methods are based on learning algorithms like SR and PCNN. The method [23] provides advantageous results, but it was taking also important time 135 s due to the LLF decomposition process. The proposed method is taking less time about 6 s to produce one fusion image of size 256 × 256 from two input images on the platform implemented in Matlab2018b on a PC with Intel core2 Duo CPU and 4 GB of RAM. To conclude, the proposed fusion method is light-weight and efficient. Thus it can be dedicated to real time-aided diagnosis and treatment planning systems without consuming too many computational resources.
Although the proposed method achieves better performance, it has two limitations. Firstly, the proposed method is designed to fuse the pre-registered images, thus adding an image registration module might enable the proposed system to deal with the unregistered datasets. Second, the proposed fusion method needs more ground truth data to prove more efficiency for large databases. In future works, the proposed method can be extended to other medical aided diagnostic applications like segmentation and classification. Specifically, it will be dedicated for classifying low-grade and high-grade gliomas based on radiomics analysis [24]. Indeed, glioma classification before surgery is of the utmost important in clinical decision making and prognosis prediction. Therefore, an aided diagnosis framework composed of two parts, fusion and classification, will be proposed to assist radiologists in glioma classification.
5 Conclusion
The purpose of this study was to propose a multi-modal image fusion method based on an adopted weight local features and NSML fusion rules in the NSST domain. First, NSST has been applied on pre-registered source images. Subsequently, we design an adapted weight local features fusion rule for fusing the approximated sub-images, while detailed coefficients are combined via NSML technique. Furthermore, the proposed method is extended for functional and anatomical image fusion by transforming color images to YIQ color space, which produces fused images with high spatial and spectral resolutions. Overall, extensive experiment results have proved the effectiveness of the proposed method. In future work, the proposed fusion method will be adopted for an accurate glioma classification based on radiomics analysis.
References
Du, F., et al.: An overview of multi-modal medical image fusion. Neurocomputing 215, 3–20 (2016)
Li, S., et al.: Pixel-level image fusion: a survey of the state of the art. Inform. Fusion 33, 100–112 (2017)
Shahdoosti, H.R., Tabatabaei, Z.: MRI and PET/SPECT image fusion at feature level using ant colony based segmentation. Biomed. Sign. Process. Control 47, 63–74 (2019)
Mangai, U.G., et al.: A survey of decision fusion and feature fusion strategies for pattern classification. IETE Tech. Rev. 27(4), 293–307 (2010)
Reena Benjamin, J., Jayasree, T.: Improved medical image fusion based on cascaded PCA and shift invariant wavelet transforms. Int. J. Comput. Assist. Radiol. Surg. 13(2), 229–240 (2017). https://doi.org/10.1007/s11548-017-1692-4
He, C., et al.: Multimodal medical image fusion based on IHS and PCA. Procedia Eng. 7, 280–285 (2010)
Wang, W., Chang, F.: A multi-focus image fusion method based on laplacian pyramid. JCP 6(12), 2559–2566 (2011)
Ali, F.E., et al.: Curvelet fusion of MR and CT images. Prog. Electromagn. Res. 3, 215–224 (2008)
Yang, L., Guo, B.L., Ni, W.: Multimodality medical image fusion based on multiscale geometric analysis of contourlet transform. Neurocomputing 72(1–3), 203–211 (2008)
Miao, Q.G., Shi, C., Xu, P.F., Yang, M., Shi, Y.B.: A novel algorithm of image fusion using shearlets. Opt. Commun. 284(6), 1540–1547 (2011)
Easley, G., Labate, D., Lim, W.Q.: Sparse directional image representations using the discrete shearlet transform. Appl. Comput. Harmonic Anal. 25(1), 25–46 (2008)
Manchanda, M., Sharma, R.: An improved multimodal medical image fusion algorithm based on fuzzy transform. J. Vis. Commun. Image Represent. 51, 76–94 (2018)
Ouerghi, H., Mourali, O., Zagrouba, E.: Multimodal medical image fusion using modified PCNN based on linking strength estimation by MSVD transform. Int. J. Comput. Commun. Eng. 6(3), 201–211 (2017)
Ouerghi, H., Mourali, O., Zagrouba, E.: Non-subsampled shearlet transform based MRI and PET brain image fusion using simplified pulse coupled neural network and weight local features in YIQ colour space. IET Image Proc. 12(10), 1873–1880 (2018)
Ganasala, P., Kumar, V.: Feature-motivated simplified adaptive PCNN-based medical image fusion algorithm in NSST domain. J. Digit. Imaging 29(1), 73–85 (2016)
Zhang, Y., et al.: IFCNN: A general image fusion framework based on convolutional neural network. Inform. Fusion 54, 99–118 (2020)
Huang, W., Jing, Z.: Evaluation of focus measures in multi-focus image fusion. Pattern Recogn. Lett. 28(4), 493–500 (2007)
Yin, M., et al.: A novel image fusion algorithm based on nonsubsampled shearlet transform. Optik 125(10), 2274–2282 (2014)
Ullah, H., et al.: Multi-modality medical images fusion based on local-features fuzzy sets and novel sum-modified-Laplacian in non-subsampled shearlet transform domain. Biomed. Signal Process. Control 57, 101724 (2020)
Jagalingam, P., Hegde, A.V.: A review of quality metrics for fused image. Aquatic Procedia 4. Icwrcoe 133–142 (2015)
Mohammed, A., Nisha, A.KL., Sathidevi, P.S.: A novel medical image fusion scheme employing sparse representation and dual PCNN in the NSCT domain. In: IEEE Region 10 Conference (TENCON). pp. 2147–2151. IEEE, Singapore (2016)
Liu, X., Mei, W., Du, H.: Multi-modality medical image fusion based on image decomposition framework and nonsubsampled shearlet transform. Biomed. Signal Process. Control 40, 343–350 (2018)
Du, J., Li, W., Xiao, B.: Anatomical-functional image fusion by information of interest in local Laplacian filtering domain. IEEE Trans. Image Process. 26(12), 5855–5866 (2017)
Lotan, E., et al.: State of the art: Machine learning applications in glioma imaging. Am. J. Roentgenol. 212(1), 26–37 (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Ouerghi, H., Mourali, O., Zagrouba, E. (2020). Multi-modal Image Fusion Based on Weight Local Features and Novel Sum-Modified-Laplacian in Non-subsampled Shearlet Transform Domain. In: Bebis, G., et al. Advances in Visual Computing. ISVC 2020. Lecture Notes in Computer Science(), vol 12510. Springer, Cham. https://doi.org/10.1007/978-3-030-64559-5_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-64559-5_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-64558-8
Online ISBN: 978-3-030-64559-5
eBook Packages: Computer ScienceComputer Science (R0)