Abstract
Functional connectivity (FC) networks with the resting-state functional magnetic resonance imaging (rs-fMRI) help advance our understanding of brain disorders, such as Alzheimer’s disease (AD) and its prodromal stage, i.e., mild cognitive impairment (MCI). Recent studies have shown that FC networks demonstrate significant dynamic changes even in the resting state. However, previous studies typically focus on model the low-order (e.g., second-order) dynamics, without exploring the high-order dynamic properties of FC networks. In this paper, we propose to build a high-order dynamic functional connectivity network (hoDFCN) from the second-order FC networks, and define two novel measures to characterize the temporal and spatial variability of hoDFCN. Furthermore, we employ both spatial and temporal variability features for brain disease classification. Experimental results on 149 subjects with baseline resting-state functional MRI (rs-fMRI) data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) suggest the effectiveness of our proposed method in brain dementia identification.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Functional magnetic resonance imaging (fMRI) using the blood-oxygenation-level-dependent (BOLD) signal is emerging as an advanced imaging technique for the study of brain function and activity [1]. Resting-state fMRI (rs-fMRI) has great potential to serve as a biomarker for neurophysiological diseases. Brain functional connectivity (FC) networks constructed from rs-fMRI data can characterize the inter-region neural interactions of the brain, and has been successfully used to the computer-aided diagnosis of various brain diseases, e.g., Alzheimer’s disease (AD) and its prodromal stage (i.e., mild cognitive impairment, MCI) [2]. Many studies have indicated that the structure of FC networks is associated with cognitive state [3] and brain diseases [4].
In traditional methods for FC network analysis, it is implicitly assumed that the FC between distinct brain regions is constant (i.e., temporally stationary) throughout the recording period in rs-fMRI [5]. However, recent studies have suggested that the brain region correlations demonstrate significant dynamic changes even in the resting state [6], and changes in FC networks may contain a wealth of information for better understanding the brain’s functional organization [5]. Increasing evidence has shown that the dynamics of FCs are associate with cognitive brain states [7], and altered dynamics are reported in patients with brain diseases [4]. In the literature, dynamic functional connectivity networks derived from rs-fMRI have been successfully used for classification of brain diseases [8, 9]. Existing studies usually construct the low-order (i.e., second-order) dynamic FC networks (DFCN) by using sliding windows, in which the FCs are estimated using the original BOLD signals of brain regions, characterizing just the correlation of paired brain regions. In fact, studies in neuroscience have found significant high-order interactions in cortical activities [10]. However, few works explore the high-order dynamic interaction among multiple brain regions based on rs-fMRI data. Intuitively, modeling the high-order dynamics of brain FC networks can provide more prior knowledge that can be potentially used to boost the diagnosis performance.
Accordingly, in this paper, we propose to construct a high-order dynamic FC network (hoDFCN) from the conventional second-order FC networks. We further define two new measures to characterize the temporal variability and spatial variability of hoDFCN for brain disease classification. Figure 1 illustrates the framework of the proposed hoDFCN method. Specifically, we first construct a set of traditional (i.e., second-order) FC networks by computing the Pearson correlation coefficient (PCC) of BOLD signals from paired brain regions based on a non-overlapping sliding window strategy. Then, for each FC network, we further compute the correlation between two functional architectures (i.e., all FCs associated with the brain region) of brain regions. With this strategy, we can construct the high-order dynamic FC networks. Here, the obtained FCs are high-order since they are computed based on two functional architectures involving all brain regions. Then, both spatial and temporal eigenvalue features are extracted as feature representations of each subject, followed by a manifold regularized multi-task feature learning (M\(^2\)FL) for feature selection. Based on the selected features, a multi-kernel support vector machine (SVM) is employed for classification. The experimental results on 149 subjects with baseline rs-fMRI data from Alzheimer’s Disease Neuroimaging Initiative (ADNIFootnote 1) demonstrate the effectiveness of our proposed method.
2 Method
2.1 Subjects and Image Preprocessing
In this study, we use 149 subjects with rs-fMRI data from the ADNI database, including 43 late MCI (lMCI), 56 early MCI (eMCI) and 50 health controls (HCs). Data acquisition was performed as follows: the image resolution is 2.29-\(3.31\,\mathrm{mm}\) for inplane, and slice thickness is \(3.31\,\mathrm{mm}\), TE = \(30\,\mathrm{ms}\) and TR = 2.2-\(3.1\,\mathrm{s}\). The clinical and demographic information of these subjects is given in Table 1.
Following [8], we use the standard pipeline to preprocess the rs-fMRI data, including (1) discarding the first 10 rs-fMRI volumes, (2) slice timing correction, and (3) head motion correction. The brain space of fMRI scans is partitioned into 116 regions-of-interest (ROIs) using the Automated Anatomical Labeling (AAL) template [11] with a deformable registration method [12]. The band-pass filtering is performed within a frequency interval of \([0.025\,\mathrm{Hz}, 0.100\,\mathrm{Hz}]\). The BOLD signals from the gray matter tissue are extracted, and the mean time series of each ROI is calculated to construct the FC network.
2.2 High-Order Dynamic FC Network Construction
In this section, we first introduce the construction process of traditional dynamic FC networks, and then present the details of the proposed method for construction of high-order dynamic FC network.
Traditional Dynamic FC Network Construction. Based on the mean time series of ROIs, we first construct the traditional dynamic FC networks (DFCN) based on successive and non-overlapping time windows. As illustrated in Fig. 1, the PCC is used as a measure of functional connectivity between a pair of brain regions. Specifically, for each subject, we first segment the whole time series of ROIs equally into m successive and non-overlapping time windows. Then, an FC network (corresponding to an adjacency matrix) \({\mathbf{C}}^t\) \((t=1,\cdots ,m)\) is constructed by calculating the PCC between time series of paired ROIs at the \(t^{th}\) window, as follows
where corr denotes the correlation between two time series (the PCC is used in this study). Here, \(x_i^t\) and \(x_j^t\) denote segments of the BOLD signals of the \(i^{th}\) and the \(j^{th}\) ROIs within the \(t^{th}\) time window.
According to definition in Eq. 1, \({\mathbf{C}}^t(i,j)\) can only characterize the second-order interaction of paired brain regions. Therefore, given m time windows, we can generate a set of FC networks \(\mathcal {C} =\{{\mathbf{C}}^1,{\mathbf{C}}^2,\cdots ,{\mathbf{C}}^m\}\), which could implicitly characterize the dynamics of second-order FC networks.
High-Order Dynamic FC Network Construction. Based on the second-order FC network at the \(t^{th}\) time window, we proposed to construct a high-order FC network \({\mathbf{H}}^t\) by calculating the correlation between functional architectures of paired region r and q as follows
where \({\mathbf{C}}^t(r,:)=[{\mathbf{C}}^t(r,1),{\mathbf{C}}^t(r,2),\cdots ,{\mathbf{C}}^t(r,R)]\) is a functional architecture of brain region r, corresponding to the \(r^{th}\) row entity in \({\mathbf{C}}^t\), reflecting the correlation between the \(r^{th}\) \((r=1,\cdots ,R)\) region and all the other brain regions, and R is the number of brain regions.
According to Eq. 2, \({\mathbf{H}}^t(r,q)\) characterizes the high-order correlation between brain regions r and q, considering that it is calculated based on functional architectures of the regions r and q. Since such functional architectures involve the relationship between each ROI and all the other ROIs, our constructed \({\mathbf{H}}^t(r,q)\) can explicitly capture the high-order information among ROIs. Given m time windows, we can obtain a set of high-order FC networks, i.e., \(\mathcal {H}=\{{\mathbf{H}}^1, {\mathbf{H}}^2,\cdots ,{\mathbf{H}}^m \}\), characterizing the dynamics of high-order FC networks.
2.3 Temporal and Spatial Variability of hoDFCN
Recent studies have suggested that the temporal and spatial properties of FC networks may contain a wealth of information to help understand the brain networks and brain diseases [6]. Motivated by [8], we define two new metrics to characterize the temporal and spatial variability of a given brain region, and then use these metrics to assess the temporal and spatial variability of high-order dynamic FC networks. Figure 2 illustrates the construction process of temporal and spatial variability matrices associated with a specific brain region r. Specifically, given the set of high-order dynamic FC networks \(\mathcal {H}=\{{\mathbf{H}}^1, {\mathbf{H}}^2, \cdots ,{\mathbf{H}}^m \}\), to characterize the temporal changing properties of hoDFCN at a specific brain region r, we first construct a matrix \({\mathbf{E}}_r \in R^{ m\times m}\) as follows
where \(corr({\mathbf{H}}^t(r,:),{\mathbf{H}}^l(r,:))\) computes the correlation (i.e., similarity) between functional architectures of brain regions r across time windows t and l. Thus, the matrix \({\mathbf{E}}_r\) characterizes the temporal variability of functional architectures of a specific brain region r across all time windows.
Using a similar strategy, we construct another matrix \({\mathbf{S}}_r \in R^{(R-1)\times (R-1)}\) to reflect the spatial variability of a given brain region r, which is defined as
where \({\mathbf{H}}^{(:)}(r,p)=[{\mathbf{H}}^1(r,p),{\mathbf{H}}^2(r,p),\cdots ,{\mathbf{H}}^m(r,p)]^T\) is defined as the functional sequence between a pair of specific brain regions, denoting changing profile of FC between brain regions r and p within all time windows. Besides, the term \(corr({\mathbf{H}}^{(:)}(r,p),{\mathbf{H}}^{(:)}(r,q)\) defines the similarity between two functional sequence associate with region r across different brain regions. Therefore, the matrix \({\mathbf{S}}_r\) reflects the spatial variability of functional sequences associated with a specific region r across all brain regions.
To further model the dynamic changes of high-order FC networks, for each brain region r, we calculate the eigenvalues of the corresponding matrices \(\mathbf{E}_r\) and \(\mathbf{S}_r\), and select two maximum eigenvalues as features to measure the temporal variability and spatial variability of brain region r, respectively. For each type of variability, we extract a set of eigenvalue features from constructed hoDFCN of each subject. Thus, for each subject, we can obtain two sets of eigenvalue features to measure the temporal and spatial changing properties of FC networks, respectively.
2.4 Feature Learning and Classification
For each subject, we first extract two kinds of eigenvalue features (i.e., temporal variability and spatial variability) from constructed high-order FC networks to represent each subject. Since these two kinds of eigenvalue features may contain irrelevant information, we further perform manifold regularized multi-task feature learning (M\(^2\)FL) [13] to jointly select more discriminative features for brain disease classification. Finally, based on the selected features, we employ a multi-kernel support vector machine (SVM) [14] to perform brain disease classification.
3 Experiment
3.1 Experimental Setting
In this work, we perform two classification tasks: 1) lMCI vs. eMCI classification, and 2) eMCI vs. NC classification, by using a leave-one-out (LOO) cross-validation strategy. Following [8], we construct the hoDFCN using different values of m (i.e., m = \(\{ 5, 6, \cdots , 12 \}\)), and compute the average value of temporal/spatial eigenvalue features for all values of m to avoid the effect of different window lengths. In the process of multi-kernel SVM classification, we adopt a grid search strategy on training subjects to find optimal combination of multiple kernels, and use a linear SVM classifier with default parameter values for classification. To evaluate performance of different methods, we employ four metrics, i.e., accuracy (ACC), sensitivity (SEN), specificity (SPE), and the area under the receiver operating characteristic (ROC) curve (AUC).
We first compare the proposed hoDFCN method with low-order DFCN methods, including 1) the method proposed in [8] (denoted as DFCN-mean), and 2) the method that integrates temporal and spatial eigenvalue features extracted from low-order DFCN networks (denoted as DFCN-eigen). Moreover, we compare the proposed method with three hoDFCN-based methods, including 1) the method that combines temporal and spatial mean features extracted from hoDFCNs (denoted hoDFCN-mean), 2) the method using only the temporal eigenvalue features (denoted hoDFCN-temporal), and 3) the method using only the spatial eigenvalue features (denoted hoDFCN-spatial). In addition, we compare our hoDFCN with the Baseline method using clustering coefficient features from stationary FC networks.
3.2 Classification Results
The results of all seven methods are summarized in Table 2. Figure 3 plots the ROC curves of all methods. From Table 2 and Fig. 3, we can see that our proposed hoDFCN method achieves better classification performance when compared to six competing methods. For example, the proposed hoDFCN achieves the ACC values of \( 81.8\% \) and \( 81.1\% \) in two classification tasks (i.e., lMCI vs. eMCI and eMCI vs. NC), respectively, while the best accuracy of the competing methods are \(80.8\% \) and \(80.2\%\) (by hoDFCN-mean). This suggests the efficacy of our proposed method in rs-fMRI based brain disease classification.
In addition, from Table 2 and Fig. 3, we could observe that two hoDFCN-based methods (i.e., hoDFCN-mean and hoDFCN) consistently outperform the two DFCN-based methods (i.e., DFCN-mean and DFCN-eigen), suggesting the advantages of high-order FC network over traditional (i.e., second-order) FC networks. In addition, the proposed hoDFCN method consistently outperforms the competing methods that use only single eigenvalue features (i.e., hoDFCN-temporal, and hoDFCN-spatial). These results imply that the proposed spatial variability and temporal variability features contain complementary information to further improve the classification performance.
3.3 Influence of Different Combining Schemes
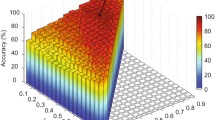
In the proposed method, we integrate temporal and spatial eigenvalue features using multi-kernel SVM technique for classification. To evaluate the effect of two kinds of eigenvalue features on performance, we test all possible combining values of two combing weights, i.e., the weight \(\beta _{TEF}\) of temporal eigenvalue features, and the weight \(\beta _{TSF}\) of spatial eigenvalue feature, with the constraint of \(\beta _{TEF}+\beta _{TSF}=1\). Figure 4 plots the obtained ACC and AUC values achieved by our hoDFCN method with different weights for spatial and temporal variability features. As can be seen from Fig. 4, one can achieve good classification performance in the inner intervals of these curves, suggesting that two kinds of network properties convey different-yet-complementary information, and thus, should be integrated for improving the classification performance. In addition, the results in Fig. 4 is inferior to the results of the proposed hoDFCN method with multi-kernel learning in Table 2. This implies that two kinds of network features should be integrated adaptively to yield better performance.
4 Conclusion
In this work, we propose a novel method to construct the high-order dynamic FC network, and also define two measures to characterize the temporal and spatial variability of FC networks for brain disease classification. Specifically, we construct the high-order FC networks based on the traditional (i.e., second-order) dynamic FC networks, by calculating the correlation between functional architectures of pairs of brain regions. Then, we construct two matrices to reflect the temporal and spatial variability of high-order FC networks, and extract two eigenvalue features to assess the dynamic properties of FC networks. Finally, we propose to select discriminative features and use a multi-kernel SVM to integrate two kinds of network features for the classification of brain diseases. The experimental results on 149 subjects with baseline rs-fMRI data from ADNI demonstrate the effectiveness of our proposed method.
Change history
29 September 2020
In a former version of this paper, the CERNET Innovation Project (NGII20190621) was missing from the Acknowledgement section. This has been corrected.
Notes
References
Cribben, I., Haraldsdottir, R., Atlas, L.Y., Wager, T.D., Lindquist, M.A.: Dynamic connectivity regression: determining state-related changes in brain connectivity. NeuroImage 61(4), 907–920 (2012)
Supekar, K., Menon, V., Rubin, D., Musen, M., Greicius, M.D.: Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput. Biol. 4(6), e1000100:1–11 (2008)
Sharp, D.J., Scott, G., Leech, R.: Network dysfunction after traumatic brain injury. Nat. Rev. Neurol. 10(3), 156–166 (2014)
Jones, D.T., et al.: Non-stationarity in the “resting brain’s” modular architecture. PloS One 7(6), e39731:1–15 (2012)
Hutchison, R.M., et al.: Dynamic functional connectivity: promise, issues, and interpretations. NeuroImage 80, 360–378 (2013)
Kudela, M., Harezlak, J., Lindquist, M.A.: Assessing uncertainty in dynamic functional connectivity. NeuroImage 149, 165–177 (2017)
Thompson, G.J., et al.: Short-time windows of correlation between large-scale functional brain networks predict vigilance intraindividually and interindividually. Hum. Brain Mapping 34(12), 3280–3298 (2013)
Jie, B., Liu, M., Shen, D.: Integration of temporal and spatial properties of dynamic connectivity networks for automatic diagnosis of brain disease. Med. Image Anal. 47, 81–94 (2018)
Wang, M., Lian, C., Yao, D., Zhang, D., Liu, M., Shen, D.: Spatial-temporal dependency modeling and network hub detection for functional MRI analysis via convolutional-recurrent network. IEEE Trans. Biomed. Eng. 67(8), 2241–2252 (2019)
Montani, F., Ince, R.A.A., Senatore, R., Arabzadeh, E., Diamond, M.E., Panzeri, S.: The impact of high-order interactions on the rate of synchronous discharge and information transmission in somatosensory cortex. Philos. Trans. 367(1901), 3297–3310 (2009)
Tzourio-Mazoyer, N., et al.: Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15(1), 273–289 (2002)
Vercauteren, T., Pennec, X., Perchant, A., Ayache, N.: Diffeomorphic demons: efficient non-parametric image registration. NeuroImage 45(1), S61–S72 (2009)
Jie, B., Zhang, D., Cheng, B., Shen, D.: Manifold regularized multitask feature learning for multimodality disease classification. Hum. Brain Mapping 36(2), 489–507 (2015)
Zhang, D., Wang, Y., Zhou, L., Yuan, H., Shen, D.: Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 55(3), 856–867 (2011)
Acknowledgment
This study was supported by NSFC (61976006, 61573023, 61703301, 61902003), Anhui-NSFC (1708085MF145, 1808085MF171), AHNU-FOYHE (gxyqZD2017010), CERNET Innovation Project (NGII20190621).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Feng, C., Jie, B., Ding, X., Zhang, D., Liu, M. (2020). Constructing High-Order Dynamic Functional Connectivity Networks from Resting-State fMRI for Brain Dementia Identification. In: Liu, M., Yan, P., Lian, C., Cao, X. (eds) Machine Learning in Medical Imaging. MLMI 2020. Lecture Notes in Computer Science(), vol 12436. Springer, Cham. https://doi.org/10.1007/978-3-030-59861-7_31
Download citation
DOI: https://doi.org/10.1007/978-3-030-59861-7_31
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59860-0
Online ISBN: 978-3-030-59861-7
eBook Packages: Computer ScienceComputer Science (R0)