Abstract
Acoustic bone shadow information in ultrasound (US) is important during imaging bones in US-guided orthopedic procedures. In this work, an end to end deep learning-based method is proposed to segment the bone shadow region from US data. In particular, we decompose the bone shadow segmentation task into two subtasks, coarse bone shadow enhancement (BSE) and horizontal bone interval mask (HBIM) estimation. Outputs from two subtasks are processed by a masking operation to generate the final bone shadow segmentation. To better leverage the mutual information in different tasks, our model features a shared encoder as deep feature extractor for both subtasks and two multi-scale pyramid pooling decoders. Additionally, we propose a conditional shape discriminator to regularize the shape of the output segmentation map. The proposed method is validated on 814 in vivo US scans obtained from knee, femur, distal radius and tibia bones. Validation against expert annotation achieved statistically significant improvements in segmentation of bone shadow regions compared to the state-of-the-art method.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
In order to provide a safe alternative to intra-operative fluoroscopy, ultrasound (US)has been investigated as an alternative intra-operative imaging modality in various orthopedic procedures [4]. US provides real-time, safe, and 2D/3D imaging. However, low signal-to-noise (SNR) ratio, limited field of view, and various imaging artifacts have hindered the wide spread use of US in computer assisted orthopedic surgery (CAOS) applications. Furthermore, regions corresponding to bone boundaries appear several millimeters (mm) in thickness due to the width of the US beam further complicating the interpretation of the collected US data. In order to alleviate some of these difficulties, various groups have proposed bone segmentation or enhancement methods [4].
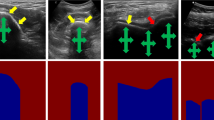
In the context of bone imaging, using US, bone boundaries have the highest intensity in the image followed by a region with low intensity values denoted as the shadow region. Shadow region is the result of a high acoustic impedance mismatch between the soft tissue and the bone boundary resulting in most of the US signal being reflected back to the transducer surface. In order to improve the accuracy and robustness of bone segmentation, several groups have incorporated bone shadow information into their framework [4]. Bone shadow information can also be used in order to guide the orthopedic surgeon to a standardized diagnostic viewing plane with minimal artifacts. Most recently, bone shadow information was also incoporated into deep learning-based bone segmentation methods [9, 10]. In [10], the authors have proposed a simultaneous bone enhancement, classification and segmentation framework based on deep learning. The bone enhancement stage [10] uses bone shadow image features extracted using the method proposed in [3]. The bone shadow enhancement method, proposed in [3], is based on the construction of a signal transmission map from the local phase bone image features. Although the method improves general appearance of the bone shadow region, it produces suboptimal bone shadow enhancement results and can not run in real-time (Fig. 1).
(a) B-mode US image of in vivo femur. Thick yellow arrows point to the bone shadow region. Red arrows point to the bone surface response. Green arrows point to soft tissue interface resembling bone response. (b) Bone shadow enhanced image obtained using [3]. (c) Gold standard bone shadow obtained by expert manual segmentation. In both (a) and (c) regions corresponding to soft tissue are displayed with black color coding, regions corresponding to bone shadow are displayed with gray/white color coding. (Color figure online)
In this work, our goal is to improve the bone shadow segmentation by proposing a deep learning-based method which yields better performance over other methods. The motivation and contribution of the proposed method are as follows:
-
Because of US imaging principle and anatomy of bone structures, bone shadows share some common shape profiles. In Fig. 1(c), the gold standard bone shadow will ideally have sharp horizontal cut-off for non-bone area and certain bone surfaces on top. Thus we propose an adversarial network to implicitly impose the shape regularization.
-
Expert manual annotations of medical images are expensive and time consuming. We leverage the bone shadow image features extracted using the method proposed in [3] and use it as surrogate ground truth to not only provide additional supervision on intermediate results, but also enable the semi-supervised learning for US bone shadow segmentation.
-
By using only left and right boundary of bone, one can create a horizontal bone interval mask and apply it on bone shadow enhanced image, Fig. 1(b), to output bone shadow segmentation results that are close to ground truth. We propose a subnetwork that estimate the bone regions horizontally by only learning from manually annotated bone landmarks which has lower annotation cost than full segmentation. This could lead to larger scale dataset for training.
2 Proposed Method
In the proposed method, two subnetworks are first trained separately to produce a coarse bone shadow enhancement (BSE) and horizontal bone interval mask (HBIM). After obtaining both coarse BSE and HBIM, a masking operation is used to generate the final bone shadow. As a result, we provide a joint trainable end-to-end deep learning model for robust bone shadow segmentation. The proposed CNN model consists of one shared encoder and two independent multi-scale decoders for coarse BSE and HBIM estimation. To further regularize the shape of the output bone shadow, we introduce a conditional shape discriminator which can guide the training of bone shadow segmentation network by adding the adversarial loss on the shape information. Figure 2 provides an overview of our framework.
2.1 Conditional Shape Discriminator
Unlike other semantic segmentation tasks, bone shadow segmentation is different in many ways. One major difference of the output segmentation map is the general shape. The type of bones (knee, fibia, femur, etc.), view planes (longitudinal and transverse), and most importantly the orientation of the US transducer with respect to the imaged bone anatomy would affect the bone shadow shape individually.
To ensure specific shape on the estimated bone shadows by a CNN, a conditional shape discriminator D is added in the training stage and designed following a conditional Generative Adversarial Network (cGAN) framework [6]. It takes both the input image and its corresponding bone shadow segmentation (segmentation from proposed network or ground truth) to identify if the segmentation is ground truth on the basis of binary images. From the perspective of segmentation network, it regularizes N output \(\hat{Y}\) using the binary cross entropy loss:
where \(X_i\) is input image and \(Y_i\) is the corresponding ground truth. Because, for binary segmentation task, the output segmentation is binary which varies in different shapes, this adversarial loss can effectively enforce the output segmentation map to follow a reasonable shape even with different types of bones and view planes.
2.2 Coarse Bone Shadow Enhancement
One of the main challenges in deep learning-based medical image analysis is the generalization ability of the trained model due to the lack of large amounts of manually annotated data. However, recent studies have shown that, by training the model through semi supervised learning on automatic annotated or weakly labelled data, the model gains better generalization ability and improves the overall performance even for different imaging modalities [2].
In this work, we propose to use Bone Shadow Enhancement (BSE) method, proposed in [3], to filter the US image and generate a coarse estimation of the bone shadow regions. BSE image signal at position (x, y) is computed by modeling the interaction of the US signal within the tissue as scattering and attenuation information using:
where \(CM_{LP}(x,y)\) is the confidence map image obtained by modeling the propagation of US signal inside the tissue taking into account bone features present in local phase bone image LP(x, y) [3]. \(US_{A}(x,y)\) maximizes the visibility of high intensity bone features inside a local region and satisfies the constraint that the mean intensity of the local region is less than the echogenicity of the tissue confining the bone [3]. Tissue attenuation coefficient is represented by \(\delta \). \(\rho \) is a constant related to tissue echogenicity confining the bone surface, and \(\epsilon \) is a small constant used to avoid the division by zero [3].
2.3 Horizontal Bone Interval Mask
As shown in Fig. 1(b) and (c), the previously defined BSE image can be regarded as a coarse estimation of bone shadow regions. While the sharp boundary of the bone surface is usually well preserved, it can also have high confidence shadows leaking into non-bone regions horizontally. To solve this shadow leakage problem, image processing technique that can remove shadows corresponding to non-bone structure while keeping the bone shadow needs to be applied on the BSE image. From the observation that the shadow leakage usually happens below the bone surface and expands horizontally, a Horizontal Bone Interval Mask (HBIM) is proposed to mask out the non-bone shadows. Given a US image X(m, n) of size \(N\times M\), its corresponding BSE image BSE(m, n) and the manually segmented bone shadow Y(m, n), HBIM is defined as follows:
HBIM can be seen as a vector in which 1 indicates the presence of bone surface along corresponding vertical line in US image. Thus we can derive the final fine bone shadow segmentation \(\hat{Y}\) using HBIM as follows,
As a result, one is able to calculate a high quality bone shadow segmentation using only the input US image and the horizontal location information of the bone in the US image. Moreover, as will be shown later, this leads to a much more robust and predictable bone shadow segmentation than a simple end-to-end training scheme.
2.4 Network Structure
The proposed framework features three tasks: (1) Coarse BSE estimation, (2) HBIM estimation, and (3) final bone shadow segmentation. Noticeably, with three different tasks, our proposed framework is a multi-task learning (MTL) model.
We view the first two tasks as intermediate tasks that are highly correlated with the final task. In the proposed method, we use a ResNet50 [5] pretrained on ImageNet [1] as the shared encoder to take the advantage of very deep neural network. The first convolutional layer is modified to take a single channel input. While the ResNet50 encoder is shared across all tasks for deep feature extraction, each of the intermediate tasks has its own decoder. As noted in U-Net [8], the key part of precise pixel-wise prediction for biomedical image segmentation task is to make good use of the multi-scale features. In our network, we adopt the decoder that was first proposed in [11]. For HBIM estimation, the desired output is a one-dimensional row vector. In order to achieve that, we changed the pyramid pooling to (1, 1), (1, 2), (1, 3), (1, 6) and add another average pooling layer between the input deep feature and concatenation to align the feature size.
Finally, we complete the final bone shadow segmentation by using the estimated HBIM and BSE of previous two tasks following Eq. 4. Given the proposed MTL model for these three tasks, it turns out helpful to extract comprehensive image features by sharing a shared encoder and then branching out for task-specific losses for each task. To further maximize the synergy across all the tasks, we propose a combined loss function containing four task-specific losses: \( L = L_{BSE} + L_{HBIM} + L_{B} + \lambda L_{AD}\), where \(L_{HBIM}\) and \(L_B\) are binary cross entropy loss of estimated HBIM and bone shadow, and \(L_{BSE}\) is the \(L_1\) loss of estimated BSE. \(L_{AD}\) represents the adversarial loss (loss from the discriminator D) as defined in Eq. 1 with weight \(\lambda \). As for the structure of the discriminator D, we follow the structure that was proposed in [7].
3 Dataset and Training
After obtaining the institutional review board (IRB) approval, a total of 814 different US images, from 20 healthy volunteers, were collected using SonixTouch US machine (Analogic Corporation, Peabody, MA, USA). The scanned anatomical bone surfaces include knee, femur, radius, and tibia. All bone shadows of the collected data were manually annotated by an expert ultrasonographer in the preprocessing stage. The BSE images were obtained using the filter parameters defined in [3] and the HBIMs were obtained using Eq. 3 with bone shadow annotations. The datasets were randomly separated on the subject level into training and testing sets by an 60%/40% split (573/241 in images level). Any subject with data included in the training set were excluded from the testing set. During preprocessing, the images were resampled into 0.15 mm isotropic resolution, and resized to \(256\times 256\).
The coarse BSE and HBIM estimation tasks are trained first with a batch size of 32 for 100 epochs in which only \(L_{BSE}\) and \(L_{HBIM}\) are used to train the network. The base network is optimized by the Adam optimizer with a learning rate of \(10^{-4}\). A joint training using all four losses is applied afterwards with a batch size of 32 for 50 epochs with \(\lambda =0.1\). During testing, the image can be forwarded though the network for all tasks by one shot. The experiments are performed on a Linux workstation equipped with an Intel 3.50 GHz CPU and a 12 GB NVidia Titan Xp GPU using the PyTorch framework. The average running time of our model for single testing image is around 0.03 s which makes real-time application possible.
4 Experimental Results
4.1 Bone Shadow Segmentation
We compare the performance of our method with that of the following four methods: Unet [8], PSPnet [11], PSPGAN and PSPnet-MTL. PSPGAN denotes the method that combines the proposed conditional shape discriminator in Sect. 2 and PSPnet. PSPnet-MTL is the multi-task version of PSPnet without conditional shape discriminator. The comparison between PSPGAN, PSPnet-MTL and PSPGAN-MTL is for the purpose of ablation study. For all the compared methods, parameters are set as suggested in their corresponding papers and trained using the same training dataset as used to train our network.
The Dice coefficient, mean Intersection over Union (mIoU) and pixel-wise accuracy (Acc.) are used to measure the segmentation performance of different methods. Average results of all test scans are shown in Table 1. As can be seen from this table, in all three metrics, our method provides the best performance compared to the other methods. Going directly from PSPGAN to PSPGAN-MTL provides implicit data augmentation and bone shadow prior for the tasks with limited data, thus results in a much more robust and accurate bone shadow segmentation. By adding proposed conditional shape discriminator, both PSPGAN and PSPGAN-MTL can outperform their counterparts, PSPnet, PSPnet-MTL. These experiments clearly show the significance of each component of proposed method, integrating coarse BSE estimation and HBIM for bone shadow segmentation and conditional shape discriminator.
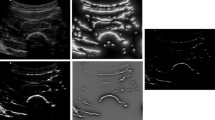
Apart from the quantitative comparison of Dice, mean IoU and pixel accuracy, we also compared our method PSPGAN-MTL with others qualitatively by visual inspection. The segmentation results corresponding to different methods and the intermediate outputs of PSPGAN-MTL are shown in Fig. 3. The more shape alike PSPGAN result shows the effect of the proposed conditional shape discriminator comparing to PSPnet.
For the final experiment of bone shadow segmentation, we compare two methods: PSPGAN-MTL and PSPGAN, in term of their ability to correctly segment spine (multiple bones) which is not present in the dataset. From the results shown in Fig. 4, it is clear that with the help of the proposed multi-task bone shadow segmentation, PSPGAN-MTL suffers no mis-segmentation and provides a more complete segmentation compared with PSPGAN.
4.2 Bone Surface Localization
One main application of bone shadow segmentation is bone surface localization from bone shadow in which accurate and robust localization is important for the improved guidance in US-based CAOS procedures. In this experiment, we applied raycasting method to perform bone surface localization from bone shadows.
The Average Euclidean Distance (AED) results (mean+std) in Table 1 show that the proposed PSPGAN-MTL outperforms the other methods on test scans by a large margin. Note that the bone surface localization experiment was carried out using previous bone shadow segmentation results for all methods. Therefore, the networks are not trained specificly on the bone surface localization task. A further paired t-test between PSPGAN-MTL and PSPGAN at a 5% significance level with p-value of 0.0009 clearly indicates that the improvements of our method are statistically significant.
5 Conclusion
In this paper, we proposed an end-to-end deep learning framework that enabled robust and accurate bone shadow segmentation for bone ultrasound examination. The main novelty lies in (1) the introduction of conditional shape discriminator to shape specific image segmentation problem, (2) the design of two subtasks, coarse bone shadow enhancement and horizontal bone interval mask to improve the performance of each task and (3) the integration of the highly-related homogeneous tasks into a single unified bone shadow segmentation network. Formulating the network with a single powerful encoder based on Resnet50 and two pyramid pooling decoders, the proposed network brings strong synergy across all tasks when extracting shared deep features. Future work will include more extensive validation and extension to 3D data for processing volumetric US scans.
References
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. IEEE (2009)
Ganaye, P.-A., Sdika, M., Benoit-Cattin, H.: Semi-supervised learning for segmentation under semantic constraint. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11072, pp. 595–602. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00931-1_68
Hacihaliloglu, I.: Enhancement of bone shadow region using local phase-based ultrasound transmission maps. Int. J. Comput. Assist. Radiol. Surg. 12(6), 951–960 (2017). https://doi.org/10.1007/s11548-017-1556-y
Hacihaliloglu, I.: Ultrasound imaging and segmentation of bone surfaces: a review. Technology 5(02), 74–80 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Mirza, M., Osindero, S.: Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784 (2014)
Radford, A., Metz, L., Chintala, S.: Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Villa, M., Dardenne, G., Nasan, M., Letissier, H., Hamitouche, C., Stindel, E.: FCN-based approach for the automatic segmentation of bone surfaces in ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 13(11), 1707–1716 (2018)
Wang, P., Patel, V.M., Hacihaliloglu, I.: Simultaneous segmentation and classification of bone surfaces from ultrasound using a multi-feature guided CNN. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 134–142. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_16
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2881–2890 (2017)
Acknowledgment
This work was supported in part by 2017 North American Spine Society Young Investigator grant.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, P., Vives, M., Patel, V.M., Hacihaliloglu, I. (2020). Robust Bone Shadow Segmentation from 2D Ultrasound Through Task Decomposition. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12266. Springer, Cham. https://doi.org/10.1007/978-3-030-59725-2_78
Download citation
DOI: https://doi.org/10.1007/978-3-030-59725-2_78
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59724-5
Online ISBN: 978-3-030-59725-2
eBook Packages: Computer ScienceComputer Science (R0)