Abstract
Identifying the mechanisms driving the distribution, diversity, and structure of parasite assemblages is critical to understand host–parasite evolution, community dynamics, and disease transmission risk. However, despite their global distribution, the broad-scale environmental factors that can affect avian haemosporidian transmission remain only partially understood across avian communities in the Neotropics. With the recent technological advances in satellite imagery, computer modeling, and molecular biology, we are now capable of studying infectious diseases in an integrated fashion over diverse spatial scales. From this perspective, ecological niche modeling (ENM) and species distribution modeling (SDM) represent useful tools to study vector-borne diseases, emphasizing the role of environmental factors in constraining their geographic distributions. Herein, we present a review of studies that have implemented modeling approaches, particularly correlative methods commonly used in ENM and SDM, to assess questions of either parasites, vectors, or host species in avian malaria. We identify that most commonly approached topics include the description of geographic distributions (biogeography), population demography, and structure of the host communities (ecology), and in low proportion, other important topics include climate change effects and potential risk for invasions. We observed that most studies were performed from local-to-regional scales and were concentrated mainly on vectors, followed by a combination of parasites and hosts. The correlative algorithm used was mainly Maxent; however, other statistical analyses included spatial regressions, smoothing procedures, and more conventional multivariate regressions developed chiefly on environmental dimensions. To date, applications of these approaches to the understanding of the geography and ecology of vector-borne diseases are in early stages. Diverse challenges related to theoretical and empirical advances, as well as the need for more (organized) data, still remain poorly explored. We present an adaptation of the Biotic-Abiotic-Mobility (BAM) framework to describe new potential arrangements in the context of this complex epidemiological/epizootiological systems. We hope this review can be useful to provide the basic knowledge and guidance for modeling of ecological niches on avian haemosporidian systems.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Climate variability
- Ecological niche
- Environmental factors
- Geographic distribution patterns
- Spatial analyses
- Vector-borne diseases
7.1 Introduction
In a rapidly changing world with many newly emerging and geographically expanding pathogens and parasites, we must investigate factors implicated in the distribution of such organisms (Doussang et al. 2019). Infectious diseases are increasingly important, as they contribute to declining populations and mortality events of wildlife species (Jones et al. 2008; Ganser et al. 2016). Changes in climatic patterns will likely further impact in the distribution of disease vectors, increasing their frequency, expanding their geographic distribution and, consequently, affecting the ecological integrity of ecosystems (Atkinson et al. 2014; Fortini et al. 2015, 2017; see Chaps. 6, 10, 11, 13 and 14) or particular species (Fortini et al. 2017). For example, several cases of climate projections estimate a range loss higher than 50% for most species in the absence of effective vector controls, or increased disease resistance (e.g., Fortini et al. 2017). Likewise, previous studies have established a link between the deforestation patterns and the abundance of Anopheles darlingi, one of the most important malaria vectors in the Neotropics (e.g., Vittor et al. 2009, Herrera et al. 2012; see Chap. 6 for a review of vector ecology concerning avian haemosporidians of tropical regions). Indeed, there has been in recent years an increased interest in the development of accurate spatial predictions integrating environmental conditions conducive to pathogen proliferation (e.g., Daszak et al. 2000; Woolhouse and Gowtage-Sequeria 2005; Sehgal et al. 2011; Moens and Pérez-Tris 2016; see Chap. 14 for anthropogenic effects on vector-borne parasites). This information is also relevant to understand the evolution and ecology of parasites, as well as to determine hotspots of potential emerging infectious diseases (Daszak et al. 2000).
Despite an accelerated focus on describing host specificity for a multitude of parasites (e.g., Hellgren et al. 2009; Clark et al. 2018; Doña et al. 2018; Park et al. 2018; see Chap. 11), there are few empirical studies accounting for the environmental dependency by considering the host–parasite contact areas or understanding the distribution patterns of vectors and parasites (Canard et al. 2014). Despite the variety of theoretical and methodological approaches that have been recently applied to the analysis of the distribution of diverse disease vectors (e.g., Escobar et al. 2016; Alkishe et al. 2017; Altamiranda-Saavedra et al. 2017), little information is available regarding the broad-scale environmental factors that can affect (and predict) the distribution and transmission of many vector-borne diseases (Pérez-Tris and Bensch 2005; Sehgal et al. 2011; see also Chap. 9 for an application of macroecology and networks to antagonistic interactions). This certainly seems to be the case for the haemosporidian parasites across avian communities in the Neotropics (Foley et al. 2010a; Galen and Witt 2014).
Ecological niche modeling (ENM) and species distribution modeling (SDM) are useful tools to predict the potential distribution of species (including parasites, vectors, and hosts) based on the relation between environmental variables associated with the sites where the species have been observed. This approach produces suitability maps that allow us to predict spatial predictions about the potential distribution of the target phenomenon or species (Peterson et al. 2011; Peterson 2014), as has been demonstrated in infectious diseases of birds (e.g., Ageep et al. 2009; Doussang et al. 2019). This approach also allows the visualization of how natural landscapes and climatic variables are associated with parasite transmission (Fuller et al. 2012a, 2012b), particularly in largely unsampled regions. Predictive maps that explain the potential distribution of these diseases can be used as early warning surveillance systems and as guides for management decisions (Ganser et al. 2016). On the other hand, the recent technological advances in satellite imagery, computer capacities, and molecular biology for lineage identifications, allow the study of infectious diseases over different spatial scales, by modeling environmental factors associated with vectors, hosts, and parasites (Kitron 1998; Sehgal et al. 2011; Eisen and Eisen 2011; Atkinson et al. 2014; Altamiranda-Saavedra et al. 2017) (Box 7.1).
Box 7.1 The General Diagram About the Implementation of ENM Approach (Modified from Martínez-Meyer 2005)

Both ENM and SDM are generated using two types of information (input data): (a) occurrence/absence records of species to be modeled and (b) descriptive variables that will define the species’ niche in “environmental” space (E-space), which correspond to those conditions where a species can potentially be distributed in “geographic” space (G-space). Standard ways to obtain occurrence data is by recording geographic coordinates during fieldwork, bibliographic sources, and/or by retrieving information from digitized collections and open digital gazetteers like the Global Biodiversity Information Facility (GBIF). Likewise, the selection of environmental data to include as part of the models requires choosing an adequate number and that these variables are associated with the most important information for the species or natural entity analyzed; these, in turn, should correspond with the objectives of the study. There are mainly three types of variables that are commonly used: climatic and bioclimatic (i.e., variables derived from monthly temperature and rainfall values in order to generate more biologically meaningful variables), topographic-edaphic, and remote sensing-derived variables. Most often, models rely on environmental variables that are more stable in relatively short periods of time and that are not directly modified or affected by the organism being modeled, which are called scenopoetic variables; instead, there are fine resolution and coupled variables to the demographic processes of the organisms being modeled which are known as bionomic variables. These represent two broad kinds of variables that can be used to classify the types of ecological niches being modeled (Peterson et al. 2011).
Once the information on the presences and variables has been defined, the most appropriate modeling technique should be selected. It is important to emphasize that there is no single best algorithm for all modeling purposes, and that choosing the right one may depend on the configuration of the analysis and type of data (i.e., presence-only, presence-absence, or presence-background information; Qiao et al. 2015). Several types of models (including statistical approaches) and algorithms can be used to perform ENMs, such as: Generalized Linear Models (GLM), Generalized Additive Models (GAM), Random Forest (RF), Boosted regression trees (BRT), BIOCLIM, GARP, and Maxent, as well as one relatively new approach to obtain consensus models (i.e., ensemble prediction). The selected modeling technique or algorithm will establish a relationship between the presence or absence of information and the range of values of the set of variables where these points are located. This relationship is usually called the adjustment of the model or classification rule, which allows us to define the environmental space where suitability conditions for species could be found.
The final step in the generation of ENM and SDM is the projection of the defined suitability conditions on geographical space to define the potential distribution areas on a map. This continuous output can be converted to a binary prediction after imposing a threshold over the suitability values above and below which it is assumed that suitable conditions exist or not, respectively. Models need to be evaluated statistically and geographically to test whether there is reliability. The process of model testing allows calculating indicators of model performance, such as the percentage of positives and negatives (i.e., “real” absences and presences of species) that are correctly predicted by the models; such values are typically summarized in what is called the confusion matrix. Finally, a particular calibration of an ENM can be used to explore the relative magnitude of environmental variables (commonly known as model transferences) in time (e.g., future climate conditions) and space (e.g., different world regions). This procedure has been very useful for assessing the effects of climate change and invasive risk on species and ecosystems.
Glossary for Box 7.1
-
Absence records: Datasets containing “records” of places where sampling has occurred but the species has not been documented. A locality where a species has been reported as absent, or assumed to be, despite sampling efforts (but note that the species may inhabit these sites, if sampling is present but inadequate).
-
Algorithm: A specific sequence of instructions for solving a problem or developing a task. It usually refers to the software used to calibrate ENMs.Bionomic variables: Variables of fine spatial and temporal resolutions that are typically coupled with the demographic processes of the species or entity being modeled (e.g., species interactions).
-
Confusion matrix: A matrix relating rows summarizing distinct combinations of predicted presence (via a binary prediction) versus absence of a species (from occurrence records of the species, as well as absence, pseudoabsence, or background data), which are commonly used to calculate the omission error rate and commission error rate (including both true and apparent commission error).
-
Distribution area: The geographical space that has been accessible to a species and where abiotic conditions and ecological interactions favor the individuals’ presence (with intrinsic growth rate greater than zero) at different scales.
-
Ecological niche modeling (ENM): Estimation of the different niches (fundamental, existing, potential, and occupied), particularly those defined using scenopoetic conditions. In practice, it is carried out via estimation of abiotically suitable conditions from observations of the presence of a species; such models can be used to estimate different distributional areas (the abiotically suitable area, potential distributional area, and occupied distributional area) by stating assumptions about factors in B and M, the latter area being the goal of species distribution modeling (SDM).
-
Ensemble prediction: A consensus prediction of a niche or a distributional area made by combining results of different methods, alternative parameterizations of the same method, or multiple iterations of stochastic methods, to generate a composite value of suitability.
-
Environmental data: Values for environmental variables (generally scenopoetic variables) used in ecological niche modeling. Typically, these variables must be a coincident raster grid for the study region employed in model calibration.
-
Environmental space (E-space): A multi-dimensional space described by environmental variables and defined by “n” dimensional units or their transformations.
-
GBIF—the Global Biodiversity Information Facility—is an international network and research infrastructure funded by the world’s governments and aimed at providing anyone, anywhere, open access to data about all types of life on Earth. This includes a database on geographic records for all types of organisms from different sources (including museums, herbaria, and studies, among others).
-
Geographic space (G-space): The space defined by latitude and longitude where environmental conditions and species are found.
-
Model transferences: The application of a model (calibrated in one region) to another place in geography (G-space) and/or to another period (e.g., climate change conditions).
-
Model: A simplified representation of some aspects of nature for the purpose of research.
-
Occurrence record: Records of species’ presence, especially voucher specimens in natural history museums and herbaria, but also including observational records from visual observations and auditory records (e.g., of birds, amphibians, bats).
-
Scenopoetic variables (or conditions): Variables that are not consumed or affected by individuals of a species, which are typically limiting species distributions and metabolic requirements and are available at coarse resolutions (e.g., temperature and precipitation).
-
Species distribution modeling (SDM): Application of niche theory to questions about real spatial distributions of species, typically in the present and obtained via estimation of the occupied distributional area from occurrence information for a species. It is supported by information of its relationship to environmental characteristics, along with their correlations with dispersal limitation and biotic interactions.
-
Species niche: It is herein defined as the sum of all the environmental factors (including biotic and abiotic) of an “n” dimensional hyperspace acting on the organism distribution.
-
Suitability: The degree to which the environment is appropriate for the species in question.
In the particular case of vector-borne parasites, several authors have suggested that vectors and hosts may promote parasite diversification and permit the coexistence of a larger range of parasite species (Krasnov et al. 2007; Poulin 2011; Clark et al. 2014; see Chaps. 11 and 12 for a thorough discussion on avian haemosporidian diversification). While much recent literature has focused on the spread of invasive vector species (such as Aedes aegypti, A. albopictus, and A. atropalpus), more studies are needed to understand vector–host–parasites distributions along climate gradients (Murray et al. 2015), as well as their relationship among different spatial scales. Although distributions of avian haemosporidian parasites can vary at macro and local scales (Wood et al. 2007; Cosgrove et al. 2008; Doussang et al. 2019), several uncertainties remain related to the role of the environment when vector and host distributions are considered at such scales. For example, analyzing how the environment influences the prevalence and diversity of haemosporidian parasites, including their interaction with hosts and vectors, will help to the understanding and prediction of their distributional and diversity patterns, including community assemblage and disease transmission risks (Pérez-Tris and Bensch 2005; Sehgal et al. 2011; Eisen and Eisen 2011; Fuller et al. 2012a, 2012b; Atkinson et al. 2014; van Hoesel et al. 2019). This information is particularly essential when considering the effect of rapid reduction of native habitats and their conversion to agriculture, livestock, and mining uses (Atkinson et al. 2014; Altamiranda-Saavedra et al. 2017).
A growing body of ENM/SDM studies on human malaria vectors have improved the understanding of the ecology and biogeography of this pathogen system, including the identification of suitable areas and environments (e.g., Foley et al. 2008, 2010b; Lambin et al. 2010; Sinka et al. 2010; Fuller et al. 2012a; Altamiranda-Saavedra et al. 2017). For example, specific data and models might be well suited for understanding the assembly of vector–hosts communities in a particular region, while being limited for generalizing management decisions across taxonomic groups in several regions (Wood et al. 2007; Cosgrove et al. 2008; Doussang et al. 2019). This means that appropriateness of a given dataset and modeling strategy needs to be analyzed based upon the type of question being addressed; therefore, best-practice standards and guidelines should be followed to support the evaluation, policy recommendations, and decisions (see, e.g., Araújo et al. 2019).
In Chaps. 5 and 6, the authors have reviewed current knowledge on the present taxonomic status, life cycle, and ecology of the dipteran vectors associated with avian haemosporidians. Herein, we present a review of studies focused on spatial and environmental questions assessed under correlative ecological approaches, including ENM and SDM and other statistical methodologies. Thus, we provide a general view on avian haemosporidian studies, based on the following questions: (i) How have different modeling approaches been implemented considering natural landscapes and climatic variables to understand parasite transmission? (ii) Which are the best-practice standards in ENM and SDM approaches? and (iii) What are current challenges and the future opportunities in modeling avian haemosporidians? From the reviewed literature, we observed a poor knowledge related to theoretical and empirical advances, as well as the need for more (organized) data. Additionally, we present an adjustment of the Biotic-Abiotic-Mobility (BAM) framework (see Soberón and Peterson 2005) to describe an alternative potential arrangement within this framework, based on this complex epidemiological system.
7.2 Historical Implementation of ENM and SDM Approaches in Avian Malaria Studies
To analyze the current state of knowledge of ENM for these vector-borne pathogens, we performed a review of research articles on avian malaria. Literature search criteria included the keywords “avian malaria AND biogeography”, “avian malaria AND ecological niche model*”, “avian malaria AND species distribution model*”, “avian malaria AND Neotropics”, “modeling/modelling avian haemosporidians” including some of the cited references within articles found based on these keywords. We found 59 articles published between 2006 and early 2019. Next, we compiled all the information from these articles in a table including the following information: (a) year; (b) entity of study (i.e., parasites, hosts, vectors, and combinations of them); (c) geographic scale (i.e., local, national, regional, global), region and/or country; (d) theme addressed: biogeography and distribution, evolution, climate change, invasion risk, and ecology (e.g., community structure, habitat requirements, prevalence, dispersal, host range, host–parasite interaction, niche breadth); (e) algorithms (e.g., Maxent, GARP, GLM, GAM, GLMM); and (f) environmental variables used.
From our compilation of studies, we observed that research on avian malaria using ENM/SDM and other statistical methodologies has shown an increase in the last decade, where most contributions (54.2%) were published during the last 6 years (2013–early 2019). However, in comparison with studies related to other vector-borne diseases (e.g., human malaria, dengue, and chagas), avian malaria and related genera have not received much attention, probably because avian malaria is not a human pathogen that can currently represent a potential emerging infectious disease.
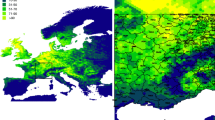
The studied entities or focal units of study (i.e., vector, host, and parasite) varied in each case (Fig. 7.1). Most studies (45.8%) focused on vectors, followed by a combination of parasite and hosts (32.2%), and few were focused exclusively on the parasite (5.1%). Even though our search was focused on cases of Neotropical avian haemosporidians, it turned out that other regions are better studied. For example, studies in countries from Asia encompass 30.7% of cases, followed by North America (20.3%, highlighting that half of those were focused exclusively in Hawaiian birds), Europe and Africa (both cases with 14.0% of studies). Studies focused in Neotropical countries (i.e., from Mexico to Argentina and Brazil, including the Caribbean islands) represented 17.4%, while only 3.5% of studies were performed in countries from Oceania. On the other hand, we observed that most studies (32.3%) were performed at local scales, followed by regional (23.7%) and national (22%) perspectives. The continental and worldwide levels of analysis represented only 15.3% and 6.7%, respectively (Fig. 7.1). This is quite relevant because different conclusions emerge from analyzing the transmission or prevalence of avian haemosporidians as scale changes (see Sect. 7.3).
The range of topics being covered varied greatly (Fig. 7.2). Most articles were centered around questions touching on some aspects of biogeography and geographic distribution (55.8% of articles), followed by studies on ecology (27.4%), climate change (12.6%), and invasion risk (4.2%). Despite the importance of each one of these topics, several articles were multidisciplinary in nature and their approach combines more than one of these topics. The most frequent combination of topics and questions were those of biogeography and geographic distributions and ecology (28.9% of cases), followed by studies including the current geographic distribution and potential effects of climate change (15.3%) (Fig. 7.2).
Regarding different modeling approaches implemented by studies, 66.7% used correlative methods, while the rest used other statistical approaches such as ModelBuilderTM or Boosted Regression Trees. For those works implementing correlative methods, 56.4% used Maxent (Phillips et al. 2006) as a tool to perform ENMs, followed by other types of statistical approaches, mainly linear models (Fig. 7.2). Finally, in terms of the environmental variables used to model either some entities (i.e., parasites, vectors, hosts) or process (e.g., levels of anthropic impacts), bioclimatic layers were the most frequently used (49.5%), followed by vegetation-related variables, such as vegetation and Normalized Difference Vegetation Index [NDVI] (13.5%), land use, and anthropic information such as human population size or livestock (16.8%). Other biological variables were used such as the host presence information (11.2%), distance layers (e.g., distance to rivers or roads; 4.4%), topographic (2.3%), and hydrology (2.3%) (Fig. 7.2). Aside from climatic variables, most studies used a combination of climate-related variables with others such as elevation and vegetation information.
7.3 Implementing Best-Practice Standards in ENM/SDM for Avian Haemosporidian Studies: A Study Case with Neotropical Human Malaria
Despite the growing body of ENM/SDM literature, and the recent demand for their use in avian haemosporidian studies, no generally agreed-upon standards for best practices yet exist for guiding the building and evaluating the adequacy of these models. Thus, to provide a general perspective about the best-practice standards applicable to a variety of available data and modeling approaches, we show such a framework with detailed guidelines for scoring key aspects of the ENM/SDM approach used in avian haemosporidian studies. For this, we analyzed the published study by Altamiranda-Saavedra et al. (2017) about the “Potential distribution of mosquito vector species in a primary malaria endemic region of Colombia” to illustrate the implementation of ENM in this chapter. Although recommendations and best-practice standards for models in biodiversity assessments exist, it is important to recognize that the criteria for judging the data and models will differ according to the particular objectives (Schwartz et al. 2012; Araújo et al. 2019). Therefore, standards showed herein do not aim to govern or guide publishing of research on ENM and/or SDM in general, but rather focus on the applicability of these methods for avian haemosporidians assessments.
Altamiranda-Saavedra et al. (2017) applied ENM methods in order to estimate the potential distribution of three endemic human malaria vector species in northern Colombia: Anopheles nuneztovari, An. albimanus, and An. darlingi. In addition, authors applied a niche overlap assessing hypotheses of niche similarity among the three vector species. The authors concentrated on evaluating the hypothesis that environmental heterogeneity is a driver for allopatric distributions of possible competing niche-related species (see Altamiranda-Saavedra et al. 2017 for a more detailed explanation), arguing that the dispersion rates and their ability to occupy diverse environmental situations may facilitate sympatry among the species of mosquitoes across environmental and geographic contexts (e.g., Laporta et al. 2011, 2015). Therefore, results may be useful for the design of malaria species-specific vector control interventions optimized for this important malaria region, especially considering the limited resources available for regular monitoring of vector species, vector-borne diseases, and control in a country like Colombia. In fact, maps based on vectors to predict the distribution of vector-borne diseases have been frequently used at broad spatial scales, with relatively fine-scale environmental factors to predict transmission dynamics of pathogens across the landscape (Pérez-Tris and Bensch 2005; Khatchikian et al. 2011).
In terms of the modeling development (Fig. 7.3), the first step was to generate predictor variables that are important in defining species’ distribution, as well as the compilation of vectors’ occurrence data. For the characterization of environmental variables, they used NDVI index obtained from the Moderate Resolution Imaging Spectroradiometer (MODIS) Terra satellite, from 2012 to 2014 and 16-day temporal resolution. The decision to use these variables to characterize the environmental variation and predict the more suitable environments for the vectors across the study region was based on the idea that spatial and temporal dynamics of vegetation could influence indirectly the mosquito reproduction and development (see Lourenco et al. 2011). For the occurrences, authors conducted sampling of vectors in or near human residences between December 2012 and March 2015, and the identification of collected vectors was performed using a morphological key and/or by PCR-RFLP-ITS2 and COI barcoding. They obtained a total of 40 localities of Urabaá – Bajo Cauca and Alto Sinuá region that were used to perform the ENM. It is important to clarify that there may be alternatives to retrieve occurrence information, such as records already available through the GBIF (https://www.gbif.org/) or VectorMap (http://vectormap.si.edu/). However, the use of alternative sources may be restricted by the availability and the quality of the information (Newbold 2010), which for cases such as malaria is scarcer than for other vector-borne human diseases.
Summary of steps and challenges in the ecological niche modeling process implemented by Altamiranda-Saavedra et al. (2017): estimation of potential distribution and test of niche similarity among three endemic human malaria vector species in northern Colombia. See text for a detailed explanation
It is important to note that authors discarded the use of alternative environmental information, such as bioclimatic variables from the WorldClim project (Hijmans et al. 2005; www.worldclim.org/) or topographic features from HYDRO1k project(USGS 2001) owing to the coarse spatial resolution available (approximately 1km2). Nevertheless, the authors specified that NDVI should properly reflect rainfall as part of the vegetation photosynthetic processes. This shows that the selection of environmental variables is an important step. In all, 69 NDVI images were used. Procedures for ENM using the large set of environmental variables have been discussed extensively, including the fact that there may exist correlations among climate variables (e.g., Graham 2003; Peterson et al. 2011). In order to reduce correlation among data layers, a principal components analysis (PCA) was performed using all images as variables. In this sense, model calibration and performance (steps 2 and 3 in Fig. 7.3) were tested for different combinations of principal components (PCs), considering only the first 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, and 55 components. Here, although authors did not discuss this point, it is important to consider that the use of PCA scores as variables in ENM represents an approach (which is not exempt from discussion) that facilitates the reduction of multicollinearity and model overfitting (Peterson et al. 2011). However, alternative methods could include the use of Pearson’s correlation coefficient, selecting only those with none or low correlation (e.g., r < 0.8), as well as the jackknife test of variable importance performed by Maxent, used frequently to identify those variables with important individual effects (Wu 1986; Elith et al. 2011).
Having collated occurrence records and environmental variables, the next step was to calibrate the models (step 2 in Fig. 7.3) to characterize the species’ ecological niche as a function of the environmental variables. This was performed using Maxent (Phillips et al. 2006), which estimates a target probability distribution by finding the probability distribution of maximum entropy (i.e., that which is most spread out, or closest to uniform), subject to a set of constraints that represent the incomplete nature of information about the target distribution. Detailed explanations for the proper implementation and interpretation of Maxent can be found elsewhere (e.g., Elith et al. 2011), and other algorithmic approaches exist that could have been used for this modeling problem, such as the Genetic Algorithm for Rule-set Production (GARP ; Stockwell 1999) and BIOCLIM (Booth et al. 2014).
A key step during the modeling process is the definition of a region for model calibration, which is particularly relevant in algorithms like Maxent where the environmental background will highly impact the results (Barve et al. 2011). The model calibration region should include a relevant area in a biogeographic context for the species being modeled. In the example, the authors specified that based on the known distribution of the vector species across Colombia and considering the entire studied endemic region for human malaria, they would set the polygon for this country as hypothesis of the accessible area (or M sensu BAM framework; Soberón and Peterson 2005; Barve et al. 2011) for the three vector species. Nevertheless, in most of the ENM literature, it is frequently observed the use of a geographical mask based on the intersection of occurrence records with Terrestrial Ecoregions (Olson et al. 2001) or the Biogeographical Provinces (Morrone 2014) to define the areas for model calibration. Such consideration is based on the assumption that these regions may define the historical accessible area for each species in geographic space. Of interest and contrary to Altamiranda-Saavedra et al.’s work, we did not observe that the analyzed studies of avian malaria using ENM/SDM applied this hypothesis of the accessible area (M) in their models, which is opposite to following a good modeling approach practice, especially when the exercise is conducted at large geographic scales. Here, we propose an adaptation of the BAM framework that considers host traits as abiotic and biotic dimensions for avian haemosporidians (Box 7.2).
Box 7.2 How Can the Biotic-Abiotic-Mobility Framework Be Incorporated in the Exploration of Avian Haemosporidians Distribution?

Initial discussions and models to represent the distribution of a species in space and then calculate the niches based on the environments were provided by Pulliam (2000) and Soberón and Peterson (2005). These last authors published a framework (known as “BAM diagram”) describing the simultaneous influence of environmental abiotic conditions (or “A”), biotic interactions (or “B”), and dispersal (or “M”) in shaping species’ geographic distributions. In general terms, the set “A” represents regions in geographic space (or “G”) where scenopoetic conditions (and existing resources) allow species’ intrinsic growth rates to be positive; while set “B” represents those geographic regions where the interacting factors (mainly biotic interactions with other species) are favorable for the presence of the species. The third set, “M” (relating to movements of individuals of the species), corresponds to the accessibility areas to the species within a given time span. The intersections of these three factors produce two components defining the “potential distributional area” (Gaston 2003) of the species: the “occupied distributional area” (Go; where the species is present [see occurrence records] representing a proxy of the species’ realized niche) and the “invadable distributional area” (Gi; where the species is absent despite the favorable conditions).
Nevertheless, it is important to highlight that delineation of “M” should be based on biological characteristics of the species under analysis and on the sampling available for that species. Each species and each geographic situation requires a more customized parameterization. Thus, the configuration of the BAM diagram for the situation under consideration and the relation of elements of the BAM diagram in environmental space become critically important. From this perspective, and considering the complex epidemiological system analyzed herein, we propose some considerations to adjust the BAM diagram for the study and definition of avian haemosporidian cases. These mainly consist in carefully interpreting the roles that vectors and hosts could represent for the abiotic, biotic and mobility sets, which have crucial theoretical and methodological implications while modeling avian haemosporidians.
As shown in the BAM diagram for the parasite case, Go depends on the biotic, abiotic, and mobility factors for both vectors and hosts (represented with white circles and lowercase letters; that is, “b” is the Biotic (B) component considering the vertebrate host and the Diptera vector, which are at the same time embedded in the abiotic (A) environment as represented by “a”). The dotted line representing “b” is smaller than “a” given that avian haemosporidians are not free-living organisms; thus, their biotic environment is restricted to the vertebrate and Diptera hosts, and consequently, the abiotic component “a” has an indirect effect on parasite occurrence via its hosts. Traditional ENM applications consider the B component to have negligent effects (the Eltonian noise hypothesis [Araújo et al. 2014]) when modeling species’ geographic distributions under the BAM framework. However, we argue that biotic interactions play a critical role in parasitic relationships in nature, so they should be considered with caution in disease ecology (Johnson et al. 2019). This is important because the congruence or amount of overlap among commonly shared factors between vectors and hosts is 1) critical at each stage of the parasite’s life cycle, its distribution, and transmission (see Rúa et al. 2005; Fuller et al. 2012b) and 2) easily affected by changes in scale.
Evidently, within A and M for both vectors and hosts, there is only a subset of areas where haemosporidian parasites could present positive intrinsic growth rates. Nevertheless, there will be areas that avian haemosporidians are not able to occupy because of present distributional constraints that cannot be overcome (e.g., elevation gradient that affects the life cycle, absence of vector or/and host). Barve et al. (2011) pointed out the crucial role of assumptions regarding M in niche modeling. These authors showed that models calibrated under different assumptions about M arrived at markedly different results, that the outcomes of model evaluations depended dramatically on which version of M was used; furthermore, the conclusions from model comparisons (Warren et al. 2008) were also dependent on assumptions regarding M. Thus, the modeling exercise for avian haemosporidians will depend on carefully thinking about the scale at which vectors and hosts are distributed, and on how abiotic, biotic, and mobility in each of these can determine the presence of the parasite.
Glossary for Box 7.2:
-
Accessibility areas (M): The biogeographic regions that individuals from a species have been capable of “testing” environmentally speaking; such regions are typically molded by factors that impede dispersal (movement) by individuals of a species (e.g., mountain chains or rivers).
-
BAM diagram: A Venn diagram that displays the joint fulfillment in geographic space (G-space) of three sets of conditions that together determine a species’ distribution: B, for biotic conditions; A, for abiotic conditions; and M, for movement of the species.
-
Biotic interactions (B): Interactions between and among species—for example, competition, mutualism, and predation.
-
Fundamental niche (FN): The set of all environmental states that permit a species to exist. Herein, we distinguish Eltonian fundamental niches from Grinnellian fundamental niches. The latter is the set of scenopoetic (non-interacting and non-linked) conditions that the species can tolerate.
-
Invadable distributional area (Gi): Corresponds to those areas in the geographic space that the species could occupy if current distributional constraints were to be overcome.
-
Occupied distributional area (Go): Those areas where the subset of the accessible region in which both scenopoetic and biotic conditions permit the species to maintain populations, and is synonymous with the “realized range” of Gaston (2003).
-
Potential distributional area: The union of the occupied distributional area and invadable distributional area for a species—that is, the regions where the abiotic and biotic conditions are suitable. (Note that much of literature uses potential distribution in a different way, however, as a synonym of what we term the abiotically suitable area).
-
Realized niche (RN): The set of all environmental states that would permit a species to exist in the presence of competitors or other negatively interacting species and restrictive factors.
In the study by Altamiranda-Saavedra et al. (2017), models were calibrated for each species, with 10 bootstrapped replicates each and the median across replicates was used as a basis for further analysis. No clamping or extrapolation options were disabled and the remaining parameters (i.e., regularization multiplier, prevalence, and features) were left as default. However, it is important to note that the calibration phase of models is critical; thus, more recent applications (such as ENMval and kuenm R packages) are exploring these parameter values in considerable detail obtaining the best models based on significance, performance, and simplicity (Muscarella et al. 2014; Cobos et al. 2019). In a first approach, to explore the robustness and predictive capabilities of the data (step 3 in Fig. 7.3), the models were generated using 50% of the locality records as training data (i.e., to calibrate the models), while the rest of data were used as testing points (i.e., for internal model evaluation). However, the final species’ models were performed using all available data. In this sense, the algorithm used localities of species records and environmental conditions to perform a certain number of iterations (500 in this case) before reaching a convergence limit. The logistic output produces a map of habitat suitability, ranging from 0 (unsuitable) to 1 (perfectly adequate; Phillips et al. 2006; Phillips and Dubik 2008). All maps were converted to binary via a conservative least presence thresholding approach (i.e., “Minimum Training Presence”), consisting of the lowest predicted value corresponding to any occurrence record of the species in the calibration dataset. It is important to note that there is no rule to set these thresholds, because its selection depends on the quality of the data used, and will vary from species to species. Detailed explanations for the proper implementation and interpretation of thresholds options in ENM could be consulted in Peterson et al. (2011) and Liu et al. (2013).
Before model predictions can be interpreted or used for any application, the predictive performance and significance need to be evaluated (step 3 in Fig. 7.3). A test using the receiver operating characteristic (ROC) curve is implemented by default in Maxent where the area under the curve (AUC) is measured with values that range from 0 to 1. However, due to the diverse critics to this test (see Lobo et al. 2008; Peterson et al. 2008 for a detailed explanation), Peterson et al. (2008) proposed the use of a modification of this test named as partial ROC. This method gives greater weight to omission errors (i.e., a false negative) and measures model performance using AUC ratios with values ranging from zero to two, where values above one indicate that models performed better than a random model ratio (AUC ratios >1.0). Bootstrap resampling was performed with 1000 iterations and with replacement of 50% of the original data points. In addition, omission rates were used as criteria to select optimal models for each species based on the evaluation of statistical significance when compared with null expectations, which was achieved by resampling 50% of the points. The partial-area ROC tests were performed using 50% of the unique occurrence data points for independent model evaluation (i.e., testing).
Finally, authors evaluated a hypothesis of niche similarity (step 4 in Fig. 7.3) among the three mosquito species following three approaches: (a) inspecting the loading values of each raw variable (16-day composite NDVI) on each of the first two principal components, and how they related to monthly rainfall averages in the study area; (b) using background similarity tests by overlaying predictions using the Schoener’s D metric, with values ranging from 0 (no overlap) to 1 (complete overlap) (see Warren et al. 2008); and (c) visualizing overall overlap based on minimum volume ellipsoids for the species in three PCA dimensions considering the Jaccard index as a numerical estimation of environmental overlap among species (see Qiao et al. 2016, 2017). These analyses allowed to obtain a better characterization of how vegetation dynamics contained in NDVI related to suitability for each species, and, at the same time, a better understanding of the dispersal capacity of these species and their ability to colonize different ecosystems across many environmental and geographic contexts.
7.4 What Are Current Challenges and the Future Opportunities in Modelling Avian Haemosporidians?
The implementation of modeling approaches in studies of limiting factors and prediction of distribution of avian haemosporidians, including the association with hosts and vectors, has seen increasing number of applications during the last years. These recent studies have been conducted to answer multiple kinds of questions, mostly to characterize current distributions and the potential spread of disease, at multiple scales across several regions and ecosystems worldwide, mostly in North America, Eurasia, and several countries of South America. This is probably a consequence of the broad applicability that ENMs possess to understand ecological requirements of species, aspects of their biogeography, predict geographic distributions, identify areas for potential risk, select areas for conservation, and forecast effects of environmental change, among others (Peterson et al. 2011; Araújo et al. 2019).
From our review, we identify six major challenges in successfully modeling of avian haemosporidians that are quite relevant for adequately assessing vector-borne parasites. The first is the proper taxonomic identification of parasites, vectors, and hosts. This is crucial not only to identify the entity being modeled (see Peterson et al. 2011), but also to be able to understand correctly the ecological and evolutionary associations and trends in the interactions among hosts, parasites, and vectors. This is more challenging perhaps for the parasite, followed by the vectors and probably less problematic for vertebrate hosts. Some studies have shown the advantage of using molecular biology techniques for this purpose (e.g., Altamiranda-Saavedra et al. 2017; see Chaps. 2 and 4 for the case of avian haemosporidians); however, they depend on having good databases derived from type specimens (e.g., COI barcodes), something that is mostly unrealistic for tropical areas particularly for vectors of nonhuman pathogens.
The second challenge is to have precise and complete information on occurrence databases (see Newbold 2010). A few efforts have been made on this aspect, mostly on the vectors (e.g., Foley et al. 2010a), but clearly there are also huge gaps on the parasites and hosts. Even if databases on birds are probably the most comprehensive among vertebrates worldwide, with highly accurate data, it is not enough to disentangle the potential distribution of avian haemosporidians. Researchers should avoid the temptation to pile occurrence data and environmental data into a niche modeling algorithm, press the button, and see what comes out (see Anderson 2015). Rather, occurrence data must be assembled carefully and comprehensively, and biases, uncertainties, and temporal characteristics must be pondered. Once the input data are assembled, and the models calibrated appropriately, outputs become considerably more rigorous.
Third and fourth challenges are the variables, and the scale and resolution that such variables better fit for the questions being asked. On this base are the conclusions and generalizations that can be made. Interestingly, the scale of analyses on which ENMs have been applied most commonly based on our review is local-to-regional, followed by larger scale analysis highlighting the broad applicability of these modeling techniques to look at the relationship between occurrence records and environmental characteristics at different scales (Overgaard et al. 2003; Foley et al. 2010b; Sinka et al. 2010; Fuller et al. 2012a; Altamiranda-Saavedra et al. 2017). This is probably because many studies aim at explaining avian malaria and its correlation with some environmental factors, which is commonly at local scales, where highest-quality data are typically available for either vectors, parasites or hosts. From this perspective, it is important to note that variables directly affecting a species’ physiology are preferred since their relationships with its geographic distribution are assumed to be stable across spatiotemporal scales (Foley et al. 2010b; Sinka et al. 2010; Fuller et al. 2012a; Anderson 2017). For instance, the slope and aspect of surface and the availability of water can be associated with anopheline habitats and their breeding sites in dry environments at local scales (Ageep et al. 2009; Fuller et al. 2012b). Recent studies showed that temperature, precipitation, and elevation can explain much of the variation in the distribution of An. albimanus in Latin America and the Caribbean (Sinka et al. 2010; Fuller et al. 2012b). This aspect is currently seeing fast advances with the incorporation of remote sensing information (Zellweger et al. 2019). As was observed in several of the publications, incorporation of high-resolution environmental surrogates, such as NDVI layers, appears to be crucial for analyzing vector-borne diseases like malaria (Foley et al. 2010b; Laporta et al. 2011; Cornuault et al. 2013a, 2013b; Ricklefs 2013; Altamiranda-Saavedra et al. 2017; Hundessa et al. 2018a, 2018b). Similarly, changes in land use and vegetation cover can also facilitate (or prevent) the spread of haemosporidian vectors (Patz et al. 2004; Vittor et al. 2009; Stresman 2010; Fecchio et al. 2018, 2019). According to Peterson (2014), ideal models of disease transmission should be based on remotely sensed datasets (e.g., Renner et al. 2016 who used laser ranging technology or LiDAR), rather than on climate data due to the lack of sufficient detail to provide genuinely helpful information in health applications (see Pérez-Rodríguez et al. 2013). Under some circumstances, no alternatives are available, but satellite imagery is invariably richer in genuine information that is measured on real-world landscapes, rather than interpolated from frighteningly sparse weather station-based data.
Another important complication in the case of avian haemosporidians is that even if we have an idea on what environmental conditions favor the transmission of the disease, we lack knowledge on the influence of several environmental factors on host communities that determine the prevalence of the parasite. In fact, the assemblage of a host or vector community does not guarantee a good prediction of parasite prevalence. Due to the complexity of the avian haemosporidian life cycles, it is difficult to draw an easy modeling framework, and even the reasoning and configuration of the BAM diagram framework (Soberón and Peterson 2005, see Box 7.2) can be challenging, because the factors within each set of conditions in B, A or M may change depending on the unit being modeled and the scale of the study. It is even further complicated because the interactions among avian haemosporidians, hosts, and vectors remain poorly understood (see Chaps. 6, 10, 11, 14, 15, and 16). Such interaction processes may be even more complex if we think about the general processes governing host specificity, in which case, we should assess both ecological and phylogenetic relationships of potential host species, in efforts to identify barriers to host range expansions (Poulin and Mouillot 2005; Hoberg and Brooks 2008; Clark et al. 2014, 2018; see Chap. 11 for an in-depth synthesis of avian haemosporidian specialization and dispersal). It seems possible to assume that this dynamic interplay may be influenced by the geography and evolutionary history of the landscape, where vector–host–parasite interactions take place (Ricklefs et al. 2004, 2014; Rivero and Gandon 2018). Since biotic interactions lie at the core of disease systems, neglecting interacting species and their role in parasite dynamics (maintenance, reproduction, and transmission) may lead to failure to forecast disease distributions (see Johnson et al. 2019). Parasite transmission is strongly influenced by interactions among infected and susceptible hosts, which can be altered by host behavior and demography (Peterson 2014; Johnson et al. 2019).
The statistical exploration of local environmental conditions linked to avian haemosporidians can be the starting point to select environmental predictors at other scales (i.e., results from local scale studies can be used to inform and parameterize coarse scale studies). For instance, globally, Haemoproteus exhibits greater lineage diversity than Plasmodium; but this pattern differs in South America, where a higher avian host diversity coupled with low Plasmodium-host specificity leads to greater lineage diversity of Plasmodium than Haemoproteus (Clark et al. 2014). However, the actual mechanism of diversification (see Chap. 12) and the broad-scale environmental factors that can affect their transmission remains only partially understood (Balls et al. 2004; Foley et al. 2010a, b; Lachish et al. 2011a, b). Opportunities exist for gaining a more comprehensive understanding of the interactions between environmental change and vector potential invasion, using different types of space-time models that can simulate environmental change or species distributions (e.g., Peterson 2009; Chaves and Koenraadt 2010).
Historical studies about ecological requirements of species and the forecasting of distribution of vector-borne disease have mainly been used at local spatial scales with relatively fine-scale environmental factors (Khatchikian et al. 2011; Ganser et al. 2016). Tools used for these analyses include spatial regressions, smoothing procedures, and more conventional multivariate regressions, all developed in “environmental” dimensions. For instance, ENM analyses of anopheline species (subgenus Nyssorhynchus) in Amazonian Brazil revealed diversification in habitat use: An. triannulatus is a generalist, whereas An. oryzalimentes and An. janconnae are specialists (Mckeon et al. 2013). ENMs were also used to predict distributions of An. bellator, An. cruzii, and An. marajoara of the Riviera Valley in southern Brazil, which revealed specific associations with land cover types (Altamiranda-Saavedra et al. 2017). Finally, low tolerance to dry environments was documented for An. darlingi; projected climate change would significantly reduce its suitable habitat mainly in Amazonian biomes, influencing both its distribution and abundance, in contrast to species of the albitarsis complex (Laporta et al. 2015).
Another challenge remains on the lack of a clear hypothesis about the areas that have been accessible (i.e., M in the BAM framework; Soberón and Peterson 2005) to the species (or entity) being modeled. This problem is not particular of avian haemosporidians, but rather an overall challenge during modeling ecological niches. However, defining the right accessibility area for model calibration in avian haemosporidians, given that it comprehends a series of interactions between hosts-parasites-vectors, complicates things. As mentioned in Box 7.2, this area is quite important because it indicates what the relevant environmental background is, and because it has huge influence on the performance of several modeling algorithms and on the significance of the model (Barve et al. 2011; Owens et al. 2013). The accessible area in the case of avian haemosporidians may change as the entity being modeled changes (i.e., parasite, host, and/or vector; Box 7.2). For example, if we focus on the parasite, this implies that its accessibility area must be restricted to some part of the accessible area of the host and some part of the accessible area of the vector. However, this accessible area may also change with the scale of analysis (Lira-Noriega et al. 2013). It is not the same to concentrate our modeling efforts at a particular landscape, as opposed to over a continental region; in the first case, most of the landscape can be assumed as accessible to either the vector or the host, but that may not be the case at the continental level. However, the definition of this accessible area will be crucial for the right interpretation of the model.
7.4.1 Future Opportunities and Directions
The literature is full of examples of research on outbreaks of a given disease, in which the relative risk of infection is assessed for a series of potential risk factors (Daszak et al. 2000; Woolhouse and Gowtage-Sequeria 2005; Sehgal et al. 2011; Peterson 2014; Escobar et al. 2016; Alkishe et al. 2017; Altamiranda-Saavedra et al. 2017). With the ecological and geographic perspectives explored in this chapter, a broader viewpoint should be possible. This perspective might be more than simply an examination of which environmental factors are important for the proper modeling of species’ niches and distributions. More in-depth studies might assess environmental correlates of key vector species’ distributional ecology, including calculation of which factors are included (or excluded) in the geographical areas from the model’s development (Peterson 2014).
Several additional steps remain to be explored in order to create better predictive maps of haemosporidians distributional patterns and transmission risk. We emphasize three crucial ones; although in all instances, good examples exist of what to do and what not to do, best practices are not always possible, feasible, or easy. First, wildlife-disease exploration requires the development of specific functionalities. One germane application is related to “time-specific” ecological niche models, which could begin to capture the essence of the temporal dynamics of species’ distributions including parasite-vectors and potential hosts (Pérez-Rodríguez et al. 2014). For these cases, occurrence data should be characterized in latitude, longitude, and time, and the occurrences would be related to environmental datasets that are similarly specific in time to produce models for a particular point in time. However, it is important to note that a major bottleneck and challenge for this field is precisely the availability of high-quality occurrence data for vector species and avian haemosporidians––unlike for the case of human malaria (Foley et al. 2010a). Likewise, these models could then, in theory, be projected to other time periods to anticipate temporal dynamics of species’ distributions. Initial explorations have been developed successfully (e.g., Peterson 2009, 2014; Tonnang et al. 2010; Pérez-Rodríguez et al. 2014; Alimi et al. 2015), but considerable additional exploration is needed.
Second, the niche specialization for a multitude of organisms is not fixed, but it is predicted to vary in response to environmental heterogeneity (Fecchio et al. 2018, 2019). A growing body of anecdotal and theoretical evidence suggests that parasites are not the exception (Hoberg and Brooks 2008; Agosta et al. 2010; Araujo et al. 2015). However, the actual mechanism of diversification and the broad-scale environmental factors that can affect their transmission remains only partially understood (Balls et al. 2004; Lachish et al. 2011a, b; Pérez-Rodríguez et al. 2014). In this sense, studies focused on the effects of climate change on avian haemosporidians, which would not be subject to the confounding patterns of human movement and economics (e.g., Gwitira et al. 2015; Ren et al. 2015; Chahad-Ehlers et al. 2018), would greatly contribute to our understanding of the impacts of changing ecological conditions on natural disease systems (Patz et al. 2004, 2008; Béguin et al. 2011; Mendenhall et al. 2013; Ren et al. 2015). It is a priority to identify which are the variables that determine and constrain distributions of disease vectors and host species, especially considering that risk of Plasmodium and Haemoproteus infection in birds is expected to increase with increasing temperatures on a global scale (Garamszegi 2011).
Finally, phylogenetic analyses are needed to reconstruct the evolutionary pathways of certain species (see Chaps. 3 and 12), and to assess whether or not current suspected hosts/reservoirs will expand in future scenarios, and whether this will result in transmission expansion (e.g., Ishtiaq et al. 2009; Svensson-Coelho and Ricklefs 2011; Mata et al. 2015). This last fact is very important considering that these changes in distribution may also affect the complex and dynamic networks of biotic interactions (Garamszegi et al. 2007; see Chap. 9). For instance, it will be relevant to analyze whether areas of high parasite prevalence are indicators of an increased abundance of vectors, increased transmission capacity, or decreased host resistance/immunity (Galen and Witt 2014; Pérez-Rodríguez et al. 2014; Zélé et al. 2014; Illera et al. 2017; Martínez et al. 2018; Pulgarín-R et al. 2018). The unresolved question that remains is whether, and to what extent, the characteristics of the landscape affect the prevalence of parasites transmitted by vectors, either directly or indirectly through the effects on hosts and/or vectors (Santiago-Alarcon et al. 2012; see Chaps. 9, 10, 13 and 14).
7.5 Conclusion
One of the major concerns is that most of the vector-borne diseases are associated with tropical environments. However, and despite that distribution limits of many haemosporidian vectors and parasites are associated with climatic conditions of temperature and precipitation, it is noteworthy that there is a poor representation of studies on avian haemosporidians in the tropics. Several studies have shown that climate variation influences the reproduction rates of parasites and the development of vectors and hosts, which in turn could affect the transmission of parasites and the exposure of parasites to new host species. Thus, the incorporation of diverse methodologies and practical considerations, such as ENM and SDM, is needed to address the diversity of questions and challenges in disease-related topics. As our literature review showed, there is an imbalance on studies addressing aspects of avian malaria, especially those including ENM and/or SDM approaches because most of them are focused only on geographic distribution patterns. Other important issues that remain poorly explored are those describing the environmental relationships at different scales (in time and space), niche shift and specialization, as well as interactions among parasites, vectors, and hosts.
Although ENM/SDM approaches to the challenge of understanding the geography and ecology of disease transmission (including avian haemosporidians) could be considered in an early stage (Peterson et al. 2011; Peterson 2014; Johnson et al. 2019), several efforts show that niche modeling has a lot to offer to the field of both public and wildlife health and epidemiology. Typical spatial applications include mapping geographic patterns of disease transmission risk, identification of risk factors (spatially or not), and assessment of populations at risk of infection. However, ENM and SDM do not capture the full complexity of the phenomenon of disease transmission because they are fitted in purely geographic dimensions, and as such, the approach unravels complex ecological and distributional phenomena into broad spatial trends.
The ideas presented in this chapter are simply examples of a complex reality. In no case is a clear and detailed analysis available that crosses all the relevant scales and resolutions. Rather, the reader is left with tidbits and suggestive indications. As highlighted, most important to the authors are the fine delimitation of a BAM diagram in which hypotheses of sets of factors affecting the distribution of haemosporidians are established, including issues of scale. It is to be hoped that, as this field develops further, more and better examples will emerge. Overall, we hope that this review and conceptual essay can be useful to provide the basic knowledge and guidance for modeling of ecological niches of avian haemosporidian systems.
References
Ageep TB, Cox J, Hassan MN et al (2009) Spatial and temporal distribution of the malaria mosquito Anopheles arabiensis in northern Sudan: influence of environmental factors and implications for vector control. Malar J 7:123–136
Agosta SJ, Janz N, Brooks DR (2010) How specialists can be generalists: resolving the “parasite paradox” and implications for emerging infectious disease. Fortschr Zool 27:151–162
Alimi OT, Fuller DO, Qualls WA et al (2015) Predicting potential ranges of primary malaria vectors and malaria in northern South America based on projected changes in climate, land cover and human population. Parasit Vectors 8:431
Alkishe AA, Peterson AT, Samy AM (2017) Climate change influences on the potential geographic distribution of the disease vector tick Ixodes ricinus. PLoS One 12:e0189092
Altamiranda-Saavedra M, Arboleda S, Parra JL et al (2017) Potential distribution of mosquito vector species in a primary malaria endemic region of Colombia. PLoS One 12:e0179093
Anderson RP (2015) El modelado de nichos y distribuciones: No es simplemente “clic, clic, clic”. Biogeografía 8:4–27
Anderson RP (2017) When and how should biotic interactions be considered in models of species niches and distributions? J Biogeogr 44:8–17
Araújo CB, Marcondes-Machado LO, Costa GC (2014) The importance of biotic interactions in species distribution models: a test of the Eltonian noise hypothesis using parrots. J Biogeogr 41:513–523
Araújo MB, Anderson RP, Márcia Barbosa A et al (2019) Standards for distribution models in biodiversity assessments. Sci Adv 5:eaat4858
Araujo SB, Braga MP, Brooks DR et al (2015) Understanding host-switching by ecological fitting. PLoS One 10:e0139225
Atkinson CT, Utzurrum RB, Lapointe DA et al (2014) Changing climate and the altitudinal range of avian malaria in the Hawaiian islands – an ongoing conservation crisis on the island of Kaua’i. Glob Change Biol 20:2426–2436
Balls MJ, Bødker R, Thomas CJ et al (2004) Effect of topography on the risk of malaria infection in the Usambara Mountains, Tanzania. Trans R Soc Trop Med Hyg 98:400–408
Barve N, Barve V, Jiménez-Valverde A et al (2011) The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol Model 222:1810–1819
Béguin A, Hales S, Rocklöv J et al (2011) The opposing effects of climate change and socio-economic development on the global distribution of malaria. Glob Environ Change 21:1209–1214
Booth TH, Nix HA, Busby JR et al (2014) BIOCLIM: the first species distribution modelling package, its early applications and relevance to most current MAXENT studies. Divers Distrib 20:1–9
Canard EF, Mouquet N, Mouillot D et al (2014) Empirical evaluation of neutral interactions in hostparasite networks. Amer Naturalist 183:468–479
Chahad-Ehlers S, Fushita AT, Lacorte GA et al (2018) Effects of habitat suitability for vectors, environmental factors and host characteristics on the spatial distribution of the diversity and prevalence of haemosporidians in waterbirds from three Brazilian wetlands. Parasit Vectors 11:276
Chaves LF, Koenraadt CJ (2010) Climate change and highland malaria: fresh air for a hot debate. Q. Rev Biol 85:27–55
Clark NJ, Clegg SM, Lima RM (2014) A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol 44:329–338
Clark NJ, Seddon JM, Slapeta J et al (2018) Parasite spread at the domestic animal–wildlife interface: anthropogenic habitat use, phylogeny and body mass drive risk of cat and dog flea (Ctenocephalides spp.) infestation in wild mammals. Parasit Vectors 11:8
Cobos ME, Peterson AT, Barve N et al (2019) Kuenm: an R package for detailed development of ecological niche models using Maxent. PeerJ 7:e6281
Cornuault J, Khimoun A, Harrigan RJ et al (2013a) The role of ecology in the geographical separation of blood parasites infecting an insular bird. J Biogeogr 40:1313–1323
Cornuault J, Warren BH, Bertrand JAM et al (2013b) Timing and number of colonizations but not diversification rates affect diversity patterns in hemosporidian lineages on a remote oceanic archipelago. Am Nat 182:820–833
Cosgrove CL, Wood MJ, Day KP et al (2008) Seasonal variation in Plasmodium prevalence in a population of blue tits Cyanistes caeruleus. J Anim Ecol 77:540–548
Daszak P, Cunningham AA, Hyatt DA (2000) Emerging infectious diseases of wildlife: threats to biodiversity and human health. Science 287:443–449
Doña J, Proctor H, Mironov S et al (2018) Host specificity, infrequent major host switching and the diversification of highly host-specific symbionts: the case of vane-dwelling feather mites. Glob Ecol Biogeogr 27:188–198
Doussang D, González-Acuña D, Torres-Fuentes LG et al (2019) Spatial distribution, prevalence and diversity of haemosporidians in the rufouscollared sparrow, Zonotrichia capensis. Parasit Vectors 12:2
Eisen L, Eisen RJ (2011) Using geographic information system and decision support systems for the prediction, prevention, and control of vector-borne diseases. Annu Rev Entomol 56:41–61
Elith J, Phillips SJ, Hastie T et al (2011) A statistical explanation of MaxEnt for ecologists. Divers Distrib 17:43–57
Escobar LE, Qiao H, Peterson AT (2016) Forecasting chikungunya spread in the Americas via data-driven empirical approaches. Parasit Vectors 9:112
Fecchio A, Bell JA, Collins MD et al (2018) Diversification by host switching and dispersal shaped the diversity and distribution of avian malaria parasites in Amazonia. Oikos 127:1233–1242
Fecchio A, Konstans K, Bell JA et al (2019) Climate variation influences host specificity in avian malaria. Ecol Lett 22:547–557
Foley DH, Rueda LM, Peterson AT et al (2008) Potential distribution of two species in the medically important Anopheles minimus complex (Diptera: Culicidae). J Med Entomol 45:852–860
Foley DH, Wilkerson RC, Birney I et al (2010a) MosquitoMap and the mal-area calculator: new web tools to relate mosquito species distribution with vector borne disease. Int J Health Geogr 9:11
Foley DH, Klein TA, Kim HC et al (2010b) Validation of ecological niche models for potential malaria vectors in the Republic of Korea. J Am Mosq Control Assoc 26:210–213
Fortini LB, Vorsino AE, Amidon FA et al (2015) Large-scale range collapse of Hawaiian forest birds under climate change and the need for 21st century conservation options. PLoS One 10:e0140389
Fortini LB, Kaise LR, Vorsino AE et al (2017) Assessing the potential of translocating vulnerable forest birds by searching for novel and enduring climatic ranges. Ecol Evol 7:9119–9130
Fuller DO, Parenti MS, Hassan AN et al (2012a) Linking land cover and species distribution models to project potential ranges of malaria vectors: an example using Anopheles arabiensis in Sudan and upper Egypt. Malar J 11:264
Fuller DO, Ahumada M, Quiñones ML et al (2012b) Near-present and future distribution of Anopheles albimanus in Mesoamerica and the Caribbean Basin modeled with climate and topographic data. Int J Health Geogr 11:13
Galen SC, Witt CC (2014) Diverse avian malaria and other haemosporidian parasites in Andean house wrens: evidence for regional co-diversification by host-switching. J Avian Biol 45:374–386
Ganser C, Gregory AJ, McNew LB et al (2016) Fine-scale distribution modeling of avian malaria vectors in north-Central Kansas. J Vector Ecol 41:114–122
Garamszegi LZ (2011) Climate change increases the risk of malaria in birds. Glob Change Biol 17:1751–1759
Garamszegi LZ, Erritzøe J, Møller AP (2007) Feeding innovations and parasitism in birds. Biol J Linn Soc 90:441–455
Gaston KJ (2003) The structure and dynamics of geographic ranges. Oxford University Press, Oxford
Graham MH (2003) Confronting multicollinearity in ecological multiple regression. Ecology 84:2809–2815
Gwitira I, Murwira A, Zengeya FM et al (2015) Modelled habitat suitability of a malaria causing vector (Anopheles arabiensis) relates well with human malaria incidences in Zimbabwe. Appl Geogr 60:130–138
Hellgren O, Pérez-Tris J, Bensch S (2009) A jack-of-all-trades and still a master of some: prevalence and host range in avian malaria and related blood parasites. Ecology 90:2840–2849
Herrera S, Quiñones ML, Quintero JP et al (2012) Prospects for malaria elimination in non-Amazonian regions of Latin America. Acta Trop 121:315–323
Hijmans RJ, Cameron SE, Parra JL et al (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Hoberg EP, Brooks DR (2008) A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host–parasite systems. J Biogeogr 35:1533–1550
Hundessa S, Williams G, Li S et al (2018a) Projecting potential spatial and temporal changes in the distribution of Plasmodium vivax and Plasmodium falciparum malaria in China with climate change. Sci Total Environ 627:1285–1293
Hundessa S, Li S, Li Liu D et al (2018b) Projecting environmental suitable areas for malaria transmission in China under climate change scenarios. Environ Res 162:203–210
Illera JC, López G, García-Padilla L et al (2017) Factors governing the prevalence and richness of avian haemosporidian communities within and between temperate mountains. PLoS One 12:e0184587
Ishtiaq F, Clegg SM, Phillimore AB et al (2009) Biogeographical patterns of blood parasite lineage diversity and avian hosts from southern Melanesian islands. J Biogeogr 37:120–132
Johnson EE, Escobar LE, Zambrana-Torrelio C (2019) An ecological framework for modeling the geography of disease transmission. Trends Ecol Evol 34:655–668
Jones KE, Patel NG, Levy MA et al (2008) Global trends in emerging infectious disease. Nature 451:990–993
Khatchikian C, Sangermano F, Kendell D et al (2011) Evaluation of species distribution model algorithms for fine-scale container-breeding mosquito risk prediction. Med Vet Entomol 25:268–275
Kitron U (1998) Landscape ecology and epidemiology of vector-borne diseases: tools for spatial analysis. J Med Entomol 35:435–445
Krasnov BR, Shenbrot GI, Khokhlova IS et al (2007) Geographical variation in the, bottom-up control of diversity: fleas and their small mammalian hosts. Glob Ecol Biogeogr 16:179–186
Lachish S, Knowles CS, Alves R et al (2011a) Fitness effects of endemic malaria infections in a wild bird population: the importance of ecological structure. J Anim Ecol 80:1196–1206
Lachish S, Knowles CS, Alves R et al (2011b) Infection dynamics of endemic malaria in a wild bird population: parasite species-dependent drivers of spatial and temporal variation in transmission rates. J Anim Ecol 80:1207–1216
Lambin EF, Tran A, Vanwambeke SO et al (2010) Pathogenic landscapes: interactions between land, people, disease vectors, and their animal hosts. Int J Health Geogr 9:54
Laporta GZ, Ramos DG, Ribeiro MC et al (2011) Habitat suitability of Anopheles vector species and association with human malaria in the Atlantic Forest in South-Eastern Brazil. Mem Inst Oswaldo Cruz 106:239–245
Laporta GZ, Linton Y-M, Wilkerson RC et al (2015) Malaria vectors in South America: current and future scenarios. Parasit Vectors 8:426
Lira-Noriega A, Soberón J, Miller CP (2013) Process-based and correlative modeling of desert mistletoe distribution: a multiscalar approach. Ecosphere 4:1–23
Liu C, White M, Newell G (2013) Selecting thresholds for the prediction of species occurrence with presence-only data. J Biogeogr 40:778–789
Lobo JM, Jiménez-Valverde A, Real R (2008) AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr 17:145–151
Lourenco PM, Sousa CA, Seixas J et al (2011) Anopheles atroparvus density modeling using MODIS NDVI in a former malarious area in Portugal. J Vector Ecol 36:279–291
Martínez J, Merino S, Badás EP et al (2018) Hemoparasites and immunological parameters in snow bunting (Plectrophenax nivalis) nestlings. Polar Biol 41:1855–1866
Martínez-Meyer E (2005) Climate change and biodiversity: some considerations in forecasting shifts in species' potential distributions. Biodivers Inform 2:42–55
Mata VA, da Silva LP, Lopes RJ et al (2015) The strait of Gibraltar poses an effective barrier to host-specialised but not to host-generalised lineages of avian Haemosporidia. Int J Parasitol 45:711–719
McKeon SN, Schlichting CD, Povoa MM et al (2013) Ecological suitability and spatial distribution of five Anopheles species in Amazonian Brazil. Am J Trop Med Hyg 88:1079–1086
Mendenhall CD, Archer HM, Brenes FO et al (2013) Balancing biodiversity with agriculture: land sharing mitigates avian malaria prevalence. Conserv Lett 6:125–131
Moens MAJ, Pérez-Tris J (2016) Discovering potential sources of emerging pathogens: South America is a reservoir of generalist avian blood parasites. Int J Parasitol 46:41–49
Morrone JJ (2014) Biogeographical regionalisation of the Neotropical region. Zootaxa 3782(1):1
Murray KA, Preston N, Allen T et al (2015) Global biogeography of human infectious diseases. Proc Natl Acad Sci USA 112:12746–12751
Muscarella R, Galante PJ, Soley-Guardia M et al (2014) ENMeval: an R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol Evol 5:1198–1205
Newbold T (2010) Applications and limitations of museum data for conservation and ecology, with particular attention to species distribution models. Prog Phys Geogr 34:3–22
Olson DM, Dinerstein E, Wikramanayake ED et al (2001) Terrestrial Ecoregions of the World: A New Map of Life on Earth. BioScience 51:933–938
Overgaard HJ, Ekbom B, Suwonkerd W et al (2003) Effect of landscape structure on anopheline mosquito density and diversity in northern Thailand: implications for malaria transmission and control. Landsc Ecol 18:605–619
Owens HL, Campbell LP, Dornak LL et al (2013) Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol Model 263:10–18
Park AW, Farrell MJ, Schmidt JP et al (2018) Characterizing the phylogenetic specialism–generalism spectrum of mammal parasites. Proc R Soc Lond [Biol] 285:20172613
Patz JA, Daszak P, Tabor GM et al (2004) Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. Environ Health Perspect 112:1092–1098
Patz JA, Olson SH, Uejio CK et al (2008) Disease emergence from global climate and land use change. Med Clin N Am 92:1473–1491
Pérez-Rodríguez A, Fernández-González S, de la Hera I et al (2013) Finding the appropriate variables to model the distribution of vector-borne parasites with different environmental preferences: climate is not enough. Glob Change Biol 19:3245–3253
Pérez-Rodríguez A, de la Hera I, Fernández-González S et al (2014) Global warming will reshuffle the areas of high prevalence and richness of three genera of avian blood parasites. Glob Change Biol 20:2406–2416
Perez-Tris J, Bensch S (2005) Dispersal increases local transmission of avian malarial parasites. Ecol Lett 8:838–8845
Peterson AT (2009) Shifting suitability for malaria vectors across Africa with warming climates. BMC Infect Dis 9:59
Peterson AT (2014) Mapping disease transmission risk: enriching models using biogeography and ecology. JHU Press
Peterson AT, Papeş M, Soberón J (2008) Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol Model 213:63–72
Peterson AT, Soberón J, Pearson RG et al (2011) Ecological niches and geographic distributions. Princeton University Press, New Jersey
Phillips SJ, Dubik M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Poulin R (2011) Evolutionary ecology of parasites. Princeton University Press, Princeton. New Jersey
Poulin R, Mouillot D (2005) Combining phylogenetic and ecological information into a new index of host specificity. J Parasitol 91:511–515
Pulgarín-R PC, Gómez JP, Robinson S et al (2018) Host species, and not environment, predicts variation in blood parasite prevalence, distribution, and diversity along a humidity gradient in northern South America. Ecol Evol 8:3800–3814
Pulliam HR (2000) On the relationship between niche and distribution. Ecol Lett 3 (4):349–361
Qiao H, Soberón J, Peterson AT (2015) No silver bullets in correlative ecological niche modelling: insights from testing among many potential algorithms for niche estimation. Methods Ecol Evol 6:1126–1136
Qiao H, Peterson AT, Campbell LP et al (2016) NicheA: creating virtual species and ecological niches in multivariate environmental scenarios. Ecography 38:805–813
Qiao H, Escobar LE, Peterson AT (2017) Accessible areas in ecological niche comparisons of invasive species: recognized but still overlooked. Sci Rep 2:1–24
Ren Z, Wang D, Hwang J et al (2015) Spatial-temporal variation and primary ecological drivers of Anopheles sinensis human biting rates in malaria epidemic-prone regions of China. PLoS One 10:e0116932
Renner SC, Lüdtke B, Kaiser S et al (2016) Forests of opportunities and mischief: disentangling the interactions between forests, parasites, and immune responses. Int J Parasitol 46:571–579
Ricklefs RE, Fallon SM, Bermingham E (2004) Evolutionary relationships, Cospeciation, and host switching in avian malaria parasites. Syst Biol 53:111–119
Ricklefs RE (2013) Habitat-independent spatial structure in populations of some forest birds in eastern North America. J Anim Ecol 82:145–154
Ricklefs RE, Outlaw DC, Svensson-Coelho M et al (2014) Species formation by host shifting in avian malaria parasites. Proc Natl Acad Sci U S A 111:14816–14821
Rivero A, Gandon S (2018) Evolutionary ecology of avian malaria: past to present. Trends Parasitol 34:712–726
Rúa GL, Quiñones ML, Vélez ID et al (2005) Laboratory estimation of the effects of increasing temperatures on the duration of gonotrophic cycle of Anopheles albimanus (Diptera: Culicidae). Mem Inst Oswaldo Cruz 100:515–520
Santiago-Alarcon D, Palinauskas V, Schaefer HM (2012) Diptera vectors of avian Haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol Rev Camb Philos Soc 87:928–964
Schwartz MW, Deiner K, Forrester T et al (2012) Perspectives on the open standards for the practice of conservation. Biol Conserv 155:169–177
Sehgal RNM, Buermann W, Harrigan RJ et al (2011) Spatially explicit predictions of blood parasites in a widely distributed African rainforest bird. Proc R Soc Lond [Biol] 278:1025–1033
Sinka ME, Rubio-Palis Y, Manguin S et al (2010) The dominant Anopheles vectors of human malaria in the Americas: occurrence data, distribution maps and bionomic précis. Parasit Vectors 3:72
Soberón J, Peterson AT (2005) Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodivers Inform 2:1–10
Stockwell D (1999) The GARP modelling system: problems and solutions to automated spatial prediction. Int J Geogr Info Sci 13 (2):143–158
Stresman GH (2010) Beyond temperature and precipitation: ecological risk factors that modify malaria transmission. Acta Trop 116:167–172
Svensson-Coelho M, Ricklefs RE (2011) Host phylogeography and beta diversity in avian haemosporidian (Plasmodiidae) assemblages of the Lesser Antilles. J Anim Ecol 80:938–946
Tonnang HEZ, Kangalawe RYM, Yanda PZ (2010) Predicting and mapping malaria under climate change scenarios: the potential redistribution of malaria vectors in Africa. Malar J 9:11
USGS (2001) HYDRO1k elevation derivate database [dataset]. Sioux Falls, SD: U.S. Geological Survey Earth Resources Observation and Science (EROS) Center. https://www.usgs.gov/centers/eros/science/usgs-eros-archive-products-overview?qt-science_center_objects=0#qt-science_center_objects. Accessed 25 December 2018
van Hoesel W, Marzal A, Magallanes S et al (2019) Management of ecosystems alters vector dynamics and haemosporidian infections. Sci Reports 9:8779
Vittor AY, Pan W, Gilman RH et al (2009) Linking deforestation to malaria in the Amazon: characterization of the breeding habitat of the principal malaria vector: Anopheles darlingi. Am J Trop Med Hyg 81:5–12
Warren DL, Glor RE, Turelli M (2008) Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62:2868–2883
Wood MJ, Cosgrove CL, Wilkin TA et al (2007) Within population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol Ecol 16:3263–3273
Woolhouse MEJ, Gowtage-Sequeria S (2005) Host range and emerging and reemerging pathogens. Emerg Infect Dis 11:1842–1847
Wu CFJ (1986) Jackknife, bootstrap and other resampling methods in regression analysis. Ann Stat 14:1261–1295
Zélé F, Vézilier J, L’Ambert G et al (2014) Dynamics of prevalence and diversity of avian malaria infections in wild Culex pipiens mosquitoes: the effects of Wolbachia, filarial nematodes and insecticide resistance. Parasit Vectors 7:437
Zellweger F, de Frenne P, Lenoir J et al (2019) Advances in microclimate ecology arising from remote sensing. Trends Ecol Evol 34:327–341
Acknowledgments
We thank the editors of this book for pushing us to write this chapter; it has been an eye-opening experience toward a complex topic in ecological modeling. The authors thank A. Townsend Peterson and Diego Santiago-Alarcon for insightful comments and inputs on earlier versions of this contribution, which were extremely helpful and greatly improved its content. We also thank the Dirección General de Asuntos del Personal Académico from Universidad Nacional Autónoma de México (DGAPA-UNAM) for a postdoctoral scholarship support to David A. Prieto-Torres.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Appendix 1. Literature Consulted for This Study but Not Cited in the Main Text
Appendix 1. Literature Consulted for This Study but Not Cited in the Main Text
-
Acheson ES, Plowright AA, Kerr JT (2015) Where have all the mosquito nets gone? Spatial modelling reveals mosquito net distributions across Tanzania do not target optimal Anopheles mosquito habitats. Malaria J 14:322
-
Akpan GE, Adepoju KA, Oladosu OR et al (2018) Dominant malaria vector species in Nigeria: Modelling potential distribution of Anopheles gambiae sensu lato and its siblings with MaxEnt. PloS ONE 13:e0204233
-
Chang KS, D.H. Yoo, Y.R. Ju et al (2016) Distribution of malaria vectors and incidence of vivax malaria at Korean army installations near the demilitarized zone, Republic of Korea. Malaria J 15:259
-
Ferraguti M, Martínez-de la Puente J, Bensch S et al (2018) Ecological determinants of avian malaria infections: An integrative analysis at landscape, mosquito and vertebrate community levels. J Anim Ecol 87:727–740
-
Ganser C, Wisely SM (2013) Patterns of spatio-temporal distribution, abundance, and diversity in a mosquito community from the eastern Smoky Hills of Kansas. J Vector Ecol 38: 229–236.
-
Gorresen PM, McMillan GP, Camp RJ et al (2009) A spatial model of bird abundance as adjusted for detection probability. Ecography 32:291–298
-
Gwitira I, Murwira A, Zengeya FM et al (2018) Application of GIS to predict malaria hotspots based on Anopheles arabiensis habitat suitability in Southern Africa. Int J Appl Earth Obs Geoinf 64:12–21
-
Hanafi-Bojd AA, Azari-Hamidian S, Hassan V et al (2011) Spatio—temporal distribution of malaria vectors (Diptera: Culicidae) across different climatic zones of Iran. Asian Pac J Trop Med 4: 498–504
-
Hanafi-Bojd AA, Sedaghat MM, Vatandoost H et al (2018) Predicting environmentally suitable areas for Anopheles superpictus Grassi (sl), Anopheles maculipennis Meigen (sl.) and Anopheles sacharovi Favre (Diptera: Culicidae) in Iran. Parasit Vectors 11:382
-
Kulkarni, MA, Desrochers RE, Kerr JT (2010) High resolution niche models of malaria vectors in northern Tanzania: a new capacity to predict malaria risk? PLoS ONE 5:e9396
-
Kulma, K, Low M, Bensch S et al (2013) Malaria infections reinforce competitive asymmetry between two Ficedula flycatchers in a recent contact zone. Mol Ecol 22:4591–4601
-
Loiseau C, Melo M, Lobato E et al (2017) Insularity effects on the assemblage of the blood parasite community of the birds from the Gulf of Guinea. J Biogeogr 44:2607–2617
-
Mohammadkhani M, Khanjani N, Bakhtiari B et al (2016) The relation between climatic factors and malaria incidence in Kerman, South East of Iran. Parasite Epidemiol Control 1:205–210
-
Padilla DP, Illera JC, González-Quevedo C et al (2017) Factors affecting the distribution of haemosporidian parasites within an oceanic island. Int J Parasitol 47:225–235
-
Padilla O, Rosas P, Moreno W et al (2017) Modeling of the ecological niches of the Anopheles spp. in Ecuador by the use of geo-informatic tools. Spat Spatiotemporal Epidemiol 21:1–11
-
Pakdad K, Hanafi-Bojd AA, Vatandoost H et al (2017) Predicting the potential distribution of main malaria vectors Anopheles stephensi, An. culicifacies sl and An. fluviatilis sl in Iran based on maximum entropy model. Acta Trop 169:93–99
-
Ricklefs RE (2013) Habitat-independent spatial structure in populations of some forest birds in eastern North America. J Anim Ecol 82:145–154
-
Samuel MD, Hobbelen PH, DeCastro F et al (2011) The dynamics, transmission, and population impacts of avian malaria in native Hawaiian birds: a modeling approach. Ecol Appl 21:2960–2973
-
Samuel MD, Woodworth BL, Atkinson CT et al (2015) Avian malaria in Hawaiian forest birds: infection and population impacts across species and elevations. Ecosphere 6:104
-
Spiegel CS, Hart PJ, Woodworth BL et al (2006) Distribution and abundance of forest birds in low-altitude habitat on Hawai’i Island: evidence for range expansion of native species. Bird Conserv Int 16:175–185
-
Zittra C, Flechl E, Kothmayer M et al (2016) Ecological characterization and molecular differentiation of Culex pipiens complex taxa and Culex torrentium in eastern Austria. Parasit Vectors 9:197
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Prieto-Torres, D.A., Rojas-Soto, O., Lira-Noriega, A. (2020). Ecological Niche Modeling and Other Tools for the Study of Avian Malaria Distribution in the Neotropics: A Short Literature Review. In: Santiago-Alarcon, D., Marzal, A. (eds) Avian Malaria and Related Parasites in the Tropics. Springer, Cham. https://doi.org/10.1007/978-3-030-51633-8_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-51633-8_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-51632-1
Online ISBN: 978-3-030-51633-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)