Abstract
Filamentous inclusions of tau protein are found in cases of inherited and sporadic frontotemporal dementias (FTDs). Mutations in MAPT, the tau gene, cause approximately 5% of cases of FTD. They proved that dysfunction of tau protein is sufficient to cause neurodegeneration and dementia. Clinically and pathologically, cases with MAPT mutations can resemble sporadic diseases, such as Pick’s disease, globular glial tauopathy, progressive supranuclear palsy and corticobasal degeneration. The structures of tau filaments from Pick’s disease and corticobasal degeneration, determined by electron cryo-microscopy, revealed the presence of specific tau folds in each disease, with no inter-individual variation. The same was true of chronic traumatic encephalopathy.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Corticobasal degeneration
- Chronic traumatic encephalopathy
- Frontotemporal dementias
- Pick’s disease
- Tau filaments
Introduction
Ordered assembly of fewer than ten proteins into filamentous assemblies defines cases of age-related neurodegenerative diseases, including Alzheimer’s disease (AD) and Parkinson’s disease (PD). Aβ, tau, α-synuclein and TDP-43 are the best known of these proteins. For most diseases, the majority of cases are sporadic, but a small percentage is inherited in a dominant manner. Huntington’s disease and other polyglutamine repeat diseases form an exception because all cases are inherited. Chronic traumatic encephalopathy (CTE), by contrast, is probably always environmentally induced. Study of dominantly inherited forms of disease has established a causative role for ordered assembly. By extrapolation, it appears likely that inclusion formation is central to neurodegeneration in all cases of disease. Tau proteinopathies, which are characterised by the assembly of tau protein, are the most common proteinopathies of the human nervous system [1].
Frontotemporal dementias (FTDs), also known as frontotemporal lobar degenerations (FTLDs), are characterised by progressive changes in personality and/or language loss, followed by dementia [2]. Their neuroanatomical substrate is degeneration of frontal and temporal lobes of the cerebral cortex. FTDs have a genetic component that is stronger than for most other neurodegenerative diseases, with mutations in MAPT, the tau gene, GRN, the progranulin gene and C9orf72, the chromosome 9 open reading frame 72 gene, being the most common. Mutations in MAPT account for approximately 5% of cases of FTD, with an average age of onset of around 50 years and a duration of disease of approximately 10 years. Some of the clinical and neuropathological features resulting from MAPT mutations are reminiscent of sporadic tau proteinopathies, including Pick’s disease (PiD), progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), globular glial tauopathy (GGT) and chronic traumatic encephalopathy (CTE). Identification of MAPT mutations proved that dysfunction of tau protein is sufficient to cause neurodegeneration and dementia. Here, we first discuss these mutations and their effects, and then focus on sporadic PiD, CBD and CTE and their filament structures.
Tau Protein and Its Isoforms
Tau is an intrinsically disordered protein , which may have many interaction partners. It can be divided into an amino-terminal domain, a proline-rich (PXXP) region, the repeat domain and a carboxy-terminal region. The amino-terminal domain projects away from the microtubule surface and is believed to interact with components of the neuronal plasma membrane. It contains a primate-specific sequence between residues 18 and 28. The PXXP motifs in the proline-rich region are recognised by SH3 domain-containing proteins of the Src family of nonreceptor tyrosine kinases, such as Fyn [3].
The repeat region and some adjacent sequences mediate interactions between tau and microtubules. Electron cryo-microscopy (cryo-EM) has shown that each tau repeat binds to the outer microtubule surface and adopts an extended structure along protofilaments, interacting with α- and β-tubulins [4, 5]. Single-molecule tracking revealed a kiss-and-hop mechanism, with a dwell time of tau on individual microtubules of approximately 40 ms [6, 7]. Isoform differences do not influence this interaction. Despite these rapid dynamics, tau promotes microtubule assembly. It remains to be seen if microtubules are also stabilised. Tau is most abundant in the labile domain of microtubules, which has led to the suggestion that it may not stabilise microtubules, but it may enable them to have long labile domains [8, 9]. Less is known about the function of the carboxy-terminal region, which may inhibit assembly into filaments.
Despite lacking a typical low-complexity domain, full-length tau can undergo liquid-liquid phase separation through electrostatic and hydrophobic interactions [10, 11], which has been found in conjunction with amyloid aggregation, at least in vitro. Although liquid-liquid phase separation and amyloid aggregation of tau are independent processes, they may be able to influence each other.
Six tau isoforms ranging from 352 to 441 amino acids are expressed in adult human brain from a single MAPT gene [12] (Fig. 1). They differ by the presence or absence of inserts of 29 and 58 amino acids (encoded by exons 2 and 3, with exon 3 being only transcribed with exon 2) in the amino-terminal half, and the inclusion, or not, of the 31 amino acid microtubule-binding repeat, encoded by exon 10, in the carboxy-terminal half. Inclusion of exon 10 results in the production of three isoforms with four repeats (4R) and its exclusion in a further three isoforms with three repeats (3R). The repeats comprise residues 244–368, in the numbering of the 441 amino acid isoform. In adult human brain, similar levels of 3R and 4R tau are expressed [13]; the finding that a correct isoform ratio is essential for preventing neurodegeneration and dementia came as a surprise. The 2 N isoforms are underrepresented in comparison with isoforms that include exon 2 or exclude both exons 2 and 3; 2 N, 1 N and 0 N tau isoforms make up 9%, 54% and 37%, respectively. Big tau, which carries an additional large exon in the amino-terminal half, is only expressed in the peripheral nervous system.
Human brain tau isoforms . MAPT and the six tau isoforms expressed in adult human brain. MAPT consists of 14 exons (E). Alternative mRNA splicing of E2 (red), E3 (green) and E10 (yellow) gives rise to six tau isoforms (352–441 amino acids). The constitutively spliced exons (E1, E4, E5, E7, E9, E11, E12 and E13) are shown in blue. E6 and E8 (violet) are not transcribed in human brain. E4a (orange) is only expressed in the peripheral nervous system. The repeats (R1–R4) are shown, with three isoforms having four repeats (4R) and three isoforms with three repeats (3R). The core sequences of tau filaments from chronic traumatic encephalopathy (K274/S305-R379) determined by cryo-EM are underlined (in blue); the core sequences of tau filaments from Pick’s disease (K254-F378 of 3R tau) are underlined (in grey); and the core sequences of tau filaments from corticobasal degeneration (K274-E380 of 4R tau) are underlined (in cyan)
Isoform expression is not conserved between species. Thus, in adult mouse brain, 4R tau isoforms are almost exclusively present, whereas adult chicken brain expresses 3R, 4R and 5R tau isoforms [14]. However, the presence of one hyperphosphorylated 3R tau isoform lacking amino-terminal inserts is characteristic of developing vertebrates. In mice, the switch from 3R to 4R tau occurs between postnatal days 9 and 18, with tau phosphorylation decreasing over time. However, isoform switching and phosphorylation are regulated differently [15]. Adult 4R tau isoforms are better at promoting microtubule assembly and at binding to microtubules than the 3R tau isoform expressed during development.
Tau Assemblies
Full-length tau assembles into filaments [1, 16]. Negative-stain immuno-electron microscopy showed that antibodies specific for the N- and C-termini of tau decorate filaments. This was not the case of antibodies directed against R3 and R4 of tau because their epitopes are occluded in the filaments [17,18,19]. Together with biochemical studies, this work established that tau filaments consist of a core region and a fuzzy coat. Tau filaments have the biophysical characteristics of amyloid [20]. Because the region in tau that binds to microtubules also forms the filament cores, physiological function and pathological assembly may be mutually exclusive.
Phosphorylation negatively regulates the ability of tau to interact with microtubules, and filamentous tau is abnormally hyperphosphorylated [21]. It remains to be seen if phosphorylation is necessary and/or sufficient for the assembly of tau into filaments. Alternatively, a change in conformation as part of the assembly process may lead to tau hyperphosphorylation. Because tau is hydrophilic, it is not surprising that unmodified full-length protein requires cofactors, such as heparin, to assemble into filaments [22,23,24,25]. Cofactors other than heparin and/or post-translational modifications may cause the assembly of tau in human brain [26, 27].
Besides phosphorylation, other modifications may also be involved. Thus, acetylation, methylation, glycation, isomerisation, O-GlcNAcylation, nitration, sumoylation, ubiquitination and truncation of assembled tau have been described. In particular, acetylation of lysine residues has come to the fore in recent years. It reduces charge, which may play a role in filament assembly of tau. Site-specific acetylation of K280 has been shown to enhance tau aggregation, while reducing microtubule assembly [28]. Twenty-one lysine residues are present between residues 244 and 380 of tau.
In AD, CTE, tangle-only dementia and many other tauopathies, all six tau isoforms are present in disease filaments. Pick bodies of PiD are made of only 3R tau. In CBD, PSP, argyrophilic grain disease (AGD), GGT and several other diseases, 4R tau isoforms make up the filaments. The morphologies of tau filaments vary in the different diseases, even when they are made of the same isoforms.
Genetics of Microtubule-Associated Protein Tau
The relevance of tau dysfunction for neurodegeneration became clear in June 1998, when dominantly inherited mutations in MAPT were shown to cause a form of frontotemporal dementia that can be associated with parkinsonism, frontotemporal dementia and parkinsonism linked to chromosome 17 and caused by mutations in the tau gene (FTDP-17 T, also known as familial FTLD-tau) [29,30,31]. In FTDP-17 T, abundant filamentous tau inclusions are present either in nerve cells or in both nerve cells and glial cells. Aβ deposits, a defining feature of AD, are not present. This work established that a pathological pathway, leading from monomeric to assembled tau, is sufficient to cause neurodegeneration and dementia.
Sixty-five mutations in MAPT have been identified in FTDP-17 T (Fig. 2). Filamentous inclusions are composed of either 3R, 4R or 3R + 4R tau [2]. MAPT mutations are concentrated in exons 9–12 (encoding R1–R4) and the introns flanking exon 10, with a smaller number of disease-causing mutations in exon 13. Two mutations (R5H and R5L) are present in exon 1 of MAPT. Mutations can be divided into those with a primary effect at the protein level and those affecting the alternative messenger ribonucleic acid (mRNA) splicing of tau pre-mRNA.
The architecture of MAPT on chromosome 17q21.31 is characterised by two haplotypes as the result of a 900 kb inversion (H1) or noninversion (H2) polymorphism [32]. Inheritance of the H1 haplotype of MAPT is a risk factor for PSP, CBD, PD and amyotrophic lateral sclerosis (ALS), but not for PiD [33,34,35,36,37,38]. The H2 haplotype is associated with increased expression of exon 3 of MAPT in grey matter, suggesting that inclusion of exon 3 may protect against PSP, CBD, PD and ALS [39]. In experimental studies, exon 3-containing tau isoforms have been found to aggregate less than those lacking exon 3 [40].
Disease-causing mutations in MAPT have made it possible to produce transgenic rodent lines that form tau filaments and show neurodegeneration [41,42,43]. Aggregation of tau correlates with neurodegeneration [44]. Reducing aggregation and increasing degradation of aggregates are therefore therapeutic objectives. It has been reported that the removal of senescent brain cells leads to a reduction in both tau aggregates and neurodegeneration in transgenic mice [45].
Transgenic mouse lines were also essential for identification of the prion-like properties of assembled tau. Aggregation of hyperphosphorylated tau was induced following intracerebral injection of tau seeds from mice transgenic for human mutant 0N4R P301S tau into transgenic mice expressing wild-type non-aggregated 2N4R tau and, to a lesser extent, following intracerebral injection into wild-type mice [46]. Tauopathy then spread to connected brain regions, indicative of seed endocytosis, seeded aggregation, intracellular transport, and release of tau seeds. This work was complemented by studies in cells [47]. It was subsequently shown that in brain extracts from mice transgenic for human P301S tau, short filaments had the greatest seeding activity [48]. These findings may be mechanistically related to the observation that in the process leading to AD, seed-competent tau inclusions first appear in transentorhinal cortex, followed by the hippocampal formation and large parts of the neocortex [49, 50].
Conformers of assembled tau seem to exist that influence the pattern of spread in brain, reminiscent of prion strains [51,52,53]. They may explain the variety of human tauopathies. Inclusions formed and spread of pathology occurred after intracerebral injection of brain homogenates from cases of AD, tangle-only dementia, PSP, CBD and AGD into a mouse line transgenic for wild-type human 4R tau and, to a lesser extent, following intracerebral injection into non-transgenic mice [51]. PiD, the filamentous inclusions of which are made of 3R tau only, was an exception. However, seeds from PiD brain induced inclusion formation and spreading in a mouse line, expressing equal amounts of human 3R and 4R tau, in the absence of mouse tau [53].
The tau sequence and, possibly, non-tau molecular requirements for seeded aggregation in vivo remain to be defined. Tau assemblies reminiscent of those in the corresponding human diseases were observed, following the injection of brain homogenates from patients with PSP, CBD and AGD, which are 4R tau proteinopathies [51] and PiD, a 3R tau proteinopathy [53]. Although these findings are consistent with the existence of distinct tau aggregate conformers, structural information is required to prove their existence.
Neuropathological Phenotypes of FTDP-17T
Cases of FTDP-17 T are characterised by the presence of filamentous tau inclusions in nerve cells or in both nerve cells and glial cells [1, 2]. Cases with glial inclusions only have not been described. Tau inclusions are most abundant in hippocampal formation and cerebral cortex.
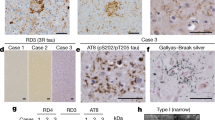
Inclusions similar to Pick bodies are often observed in the brains of individuals with mutations in exons 9, 11, 12 and 13 of MAPT. Similar to sporadic PiD, inclusions associated with mutations G272V in exon 9 and ∆K280 in exon 10 are made of 3R tau and are not phosphorylated at S262 [54,55,56]. For other mutations, such as G389R in exon 13, variable amounts of 4R tau and some phosphorylation of S262 are seen in Pick-like bodies [57] (Figs. 3 and 8). Mutation N410H in exon 13 phenocopies the tau pathology of CBD [58].
In the study mentioned earlier, tau deposits are found predominantly in neurons, whereas mutations in exon 1 and exon 10, as well as in the introns following exon 9 and exon 10, are associated with abundant neuronal and glia tau inclusions [2]. Glial pathology is in the form of coiled bodies in oligodendroglia, as well as tufted astrocytes and astrocytic plaques reminiscent of PSP and CBD. Mutations in exon 10 cause the formation of inclusions made of 4R tau; most of these mutations affect exon 10 pre-mRNA splicing, altering the ratio of 3R/4R tau. MAPT mutations P301L, P301S and P301T, the primary effects of which are at the protein level, are exceptions (Figs. 4 and 8). They continue to be important for the generation of experimental models of tauopathy and illustrate the clinical and pathological heterogeneity associated with MAPT mutations. Although most individuals with mutations P301L and P301S develop behavioural-variant FTD, cases of primary progressive aphasia have been described [59]. A P301S carrier presented with corticobasal syndrome [60]. A P301L patient had GGT, as had individuals with mutation P301T [59, 61]. GGT has emerged as a common disease associated with mutations in MAPT. Mutations in codon 301 affect only 20–25% of tau molecules, with 75–80% being wild type, arguing against a simple loss-of-function mechanism as an important disease determinant [62].
Intronic mutations in MAPT and most mutations in exon 10 affect the ratio of 3R/4R tau, which is normally 1:1, without changing the amount of total tau (Figs. 5, 6 and 8). For most mutations, this results in the relative overproduction of wild-type 4R tau and its assembly into filamentous inclusions. Tau filaments appear as twisted ribbons or half ribbons. Although these mutations often give rise to behavioural-variant FTD, cases of atypical PSP have also been described [63]. For other mutations, such as V337M (exon 12) [64] and R406W (exon 13) (Figs. 7 and 8) [65], tau inclusions resemble those of AD, and filaments are made of all six brain tau isoforms.
Negative-stain electron microscopy of tau filaments from cases of frontotemporal dementia and parkinsonism linked to chromosome 17 caused by MAPT mutations (FTDP-17 T). (1, 2), Tau filaments from a case with abundant Pick body–like inclusions and a G389R mutation. (1) Straight filaments form the majority species and (2) strongly stranded filaments are in the minority. (3–5), Tau filaments from cases with neuronal and glial inclusions and a P301L mutation or an IVS10 mutation. (3) Narrow twisted ribbons and (4) occasional rope-like filaments. (5) Wide twisted ribbons. (6, 7) Paired helical and straight tau filaments as in AD are present in cases with mutations V337M and R406W in MAPT
Structures of Tau Filaments from Pick’s Disease
PiD accounts for approximately 20% cases of FTLD-tau. Behavioural-variant frontotemporal dementia and progressive non-fluent aphasia are its most common clinical manifestations. Arnold Pick described the clinical picture and macroscopic findings in 1892 [66], and Alois Alzheimer reported the microscopic features in 1911 [67]. The presence of tau protein in Pick bodies was shown in 1985 [68, 69].
Nerve cell loss predominates in cerebral cortex (frontal > temporal > parietal), followed by hippocampal formation and amygdala, with subcortical structures being affected to variable extents [70]. The substantia nigra may be affected in some cases, while the nucleus basalis of Meynert is mostly unaffected. In frontal and anterior temporal lobes, severe circumscribed (knife-edge) atrophy is commonly seen. Microscopically, the Pick body, which consists of assembled, hyperphosphorylated 3R tau, is the pathognomonic inclusion of PiD (Fig. 9a, b) [71]. Biochemical studies have also suggested the presence of 4R tau pathology. However, this probably reflects coexisting pathologies [72] or the presence of a MAPT mutation. Pick bodies predominate in hippocampus and cerebral cortex. Fewer assemblies are present in glial cells (Fig. 9c). The glial tau pathology of PiD consists of ramified astrocytes and globular glial inclusions in oligodendrocytes. By Western blotting, assembled tau from PiD brain runs as a doublet of 60 and 64 kDa, which reveals the presence of 3R tau upon dephosphorylation [73].
By negative stain electron microscopy of sarkosyl-insoluble filaments from PiD brain, we observed narrow (Type I) and wide (Type II) tau filaments [74]. Narrow filaments had previously been described as straight, but they have a helical twist with a crossover distance of approximately 1000 Å and widths of 50–150 Å. Wide filaments have a similar crossover distance, but their widths vary from 150 to 300 Å. Immunogold negative-stain electron microscopy showed that most filaments are Type I, with a minority of Type II filaments. Filaments were not decorated by antibodies specific for R1, R3 or R4 of tau, indicating that these repeats form part of the ordered filament core.
By cryo-EM, structures of tau filaments were determined from combined frontal and temporal cortices of an individual with PiD (Fig. 10) [74]. The core of Type I filaments is made of a single protofilament that consists of residues K254-F378 of 3R tau (93 amino acids), which adopt an elongated, J-shaped, cross-β structure (Fig. 10a, c). Type II filaments are formed by the association of two Type I filaments at the distal tips of the J, where they form tight contacts through van der Waals interactions (Fig. 10b). We determined a 3.2 Å resolution map of the ordered cores of Type I filaments; the map of Type II filaments was limited to 8 Å. Each protofilament comprises nine β-strands, which are arranged into four cross-β packing stacks and are connected by turns and arcs. R1 provides two β-strands, and R3 and R4 three β-strands each. The stacks pack together in a hairpin-like fashion: β1 against one side of β8, β2 against β7, β3 against β6 and β4 against β5. The final strand, β9, is formed from the ten amino acids after R4 and packs against the other side of β8.
Structures of tau filaments from Pick’s disease . Type I and Type II tau filaments are characteristic, with Type I filaments forming the vast majority (a, b), Unsharpened cryo-EM densities of Type I (a) and Type II (b) filaments. Type I Pick filaments contain a single protofilament, whereas in Type II filaments, two identical protofilaments pack against each other symmetrically through Van der Waals interactions at the tip of the J. (c), Schematic view of the tau protofilament core of PiD. The observed nine β-strands (β1–β9) are shown as arrows
Three regions of less well-resolved density bordering the solvent-exposed faces of β4, β5 and β9 are apparent in Type I and Type II filaments. They may represent less ordered, heterogeneous and/or transiently occupied structures. The density bordering β4 is similarly located, but more extended, than that found to interact with the side chains of K317, T319 and K321 in tau filaments from AD.
Unlike tau filaments of CBD, CTE and AD, Pick body filaments are not phosphorylated at S262 [75, 76]. The reasons for this differential phosphorylation are unknown. The cryo-EM structure shows that the tight turn at G261 prevents phosphorylation of S262 in the ordered core of PiD filaments, whereas phosphorylated S262 is outside the ordered cores of tau filaments from CBD, CTE and AD. This may explain the differential phosphorylation and raises the question of whether phosphorylation at S262 may protect against PiD.
It was not previously known why only 3R tau, which lacks R2, is present in Pick body filaments. The above shows that despite sequence homology, the structure formed by K254-K274 of R1 is inaccessible to the corresponding residues from R2 (S285-S305). In support, tau filaments extracted from the brain of the patient with PiD used for cryo-EM seeded the aggregation of recombinant 3R, but not 4R, tau. Such templated misfolding may explain the selective incorporation of 3R tau in Pick body filaments.
Structures of Tau Filaments from Corticobasal Degeneration
CBD typically presents as corticobasal syndrome, which includes cortical signs, asymmetric apraxia, rigidity, myoclonus and alien limb phenomenon. It can also present as behavioural-variant FTD, Richardson’s syndrome and posterior cortical atrophy [77]. In 1925, Lhermitte et al. probably described cases of what is now known as CBD [78]. In 1968, Rebeiz et al. reported the disease as ‘corticonigral degeneration with neuronal achromasia’ [79]. The term CBD was introduced by Gibb et al. in 1989 [80]. The presence of tau protein in the inclusions of CBD was shown in 1990 [81].
Neuropathologically, CBD is characterised by asymmetric focal cortical atrophy and depigmentation of the substantia nigra. Nerve cells show diffuse cytoplasmic tau immunoreactivity, abundant neuropil threads in grey and white matter, as well as pathognomonic astrocytic plaques, mainly in affected cortical areas and in striatum (Fig. 11) [82, 83]. By Western blotting, assembled tau from CBD brains runs as a doublet of 64 kDa and 68 kDa, which consists of 4R tau upon dephosphorylation [84]. In addition, two closely related tau bands of approximately 37 kDa are typical of CBD [85].
Tau pathology in the frontal cortex of a patient with corticobasal degeneration. Tau inclusions in nerve cells and glia in grey matter (a, b), as well as oligodendrocytes in white matter (c, d) labelled by anti-tau antibodies AT8 (a, c) and RD4 (b, d). These inclusions were not labelled by RD3. Scale bars, 25 μm
By negative stain electron microscopy of sarkosyl-insoluble filaments from CBD brains, we observed narrow (Type I) and wide (Type II) tau filaments [86], in agreement with previous findings [87]. Narrow filaments have a helical twist with a crossover distance of approximately 1000 Å and widths of 80–130 Å. Wide filaments have a crossover distance of approximately 1400 Å and widths of 130–260 Å. Immunogold negative-stain electron microscopy showed that Type I and Type II filaments are present in similar amounts in some cases of CBD, with Type II filaments being more abundant in others. Filaments were not decorated by antibodies specific for R2, R3 or R4 of tau, indicating that these repeats form part of the ordered filament cores.
Structures of tau filaments were determined by cryo-EM from the frontal cortex of three individuals with CBD (Fig. 12) [86]. The core of Type I filaments is made of a single protofilament that consists of residues K274-E380 of 4R tau (107 amino acids; Fig. 12a, c). It encompasses the last residue of R1; all of R2, R3 and R4; as well as 12 amino acids after R4. In the core, there are 11 β-strands (β1–β11): three from R2 (β1–β3), three from R3 (β4–β6), four from R4 (β7–β10) and one from the sequence after R4 (β11). Each protofilament of CBD contains an additional density that is surrounded by the density of tau protein within a positively charged environment. The molecular identities of this density, as well as of those present on the outside of filament structures, remain to be identified. It has been suggested that they may correspond to post-translational modifications of tau [88]. Type II filaments consist of pairs of identical protofilaments of Type I (Fig. 12b). We obtained maps of Type I and Type II filaments at overall resolutions of 3.2 Å and 3.0 Å.
Structures of tau filaments from corticobasal degeneration. Type I and Type II tau filaments are characteristic, with Type II filaments being more numerous in some cases. (a, b), Unsharpened cryo-EM densities of Type I (a) and Type II (b) filaments. Type I filaments contain a single protofilament, whereas two symmetrically packed protofilaments are present in Type II filaments. The protofilament interface is formed by anti-parallel stacking of 343KLDFKDR349. (c), Schematic view of the tau protofilament core of CBD. The observed 11 β-strands (β1–β11) are shown as arrows. The central non-proteinaceous density is shown in blue
The 11 β-strands of each protofilament are connected by arcs and turns and form a four-layered structure. The central four layers are formed by β7, β4, β3 and β10. Strands β3 and β4 are connected by a sharp turn, whereas β7 and β10 are connected through β8 and β9, which wrap around the turn. On the other side, β2, β5 and β6 form a three-layered structure. β2 packs against one end of β5, and β6 packs against the other end. The first and the last strands, β1 and β11, pack against each other and close a hydrophilic cavity formed by residues from β2, β3, β10, β11 and the connections between β1 and β2, as well as between β2 and β3.
Each tau repeat contains a PGGG (proline-glycine-glycine-glycine) motif . In the CBD fold, that of R1 (residues 270–273) is located just outside the structured core. The PGGG motif of R2 (residues 301–304) forms a tight turn between β3 and β4, which is essential for the formation of the four-layered cross-β packing. The PGGG motif of R3 (residues 332–335) adopts an extended conformation between β6 and β7, compensating for the shorter lengths of these strands compared to the opposing β4 and β5 connected by P312. The PGGG motif of R4 (residues 364–367) adopts a similar extended conformation, forming part of the hydrophilic cavity.
In CBD Type II filaments, protofilaments are related by C2 symmetry. Their interface is formed by anti-parallel stacking of 343KLDFKDR349. Besides van der Waals interactions between the anti-parallel side chains of K347 from each protofilament, the side chain of K347 is positioned to form hydrogen bonds with the carboxyl group of D348 and the backbone carbonyl of K347 on the opposite protofilament.
CBD is characterised by abundant neuronal and glial inclusions of 4R tau. It remains to be determined if Type I and Type II filaments are differentially distributed between neuronal and glial inclusions. This notwithstanding, a single tau protofilament is characteristic of these inclusions .
Structures of Tau Filaments from Chronic Traumatic Encephalopathy
CTE is associated with repetitive head impacts or exposure to blast waves. Described as punch-drunk syndrome by Martland in 1928 [89] and dementia pugilistica by Millspaugh in 1937 [90], CTE has since been identified in former participants of other contact sports, ex-military personnel and after physical abuse. Critchley used the term in a book chapter in 1949 [91]. CTE is the best-known example of an environmentally induced neurodegenerative disease.
Clinically, CTE is characterised by behavioural, mood, cognitive and motor impairments [92]. Initial mood and behavioural changes that progress to marked cognitive impairment are often seen. For this and other reasons, we decided to include CTE in the present discussion, even though it is not generally classified under the umbrella of FTD. Motor impairments, including parkinsonism and cerebellar ataxia, have been described mostly in retired boxers.
The neuropathological concept of CTE was emphasised by Corsellis et al. in 1973, who identified generalised cerebral atrophy and widespread cortical neurofibrillary lesions in some retired boxers [93]. Antigenic similarities between the neurofibrillary lesions of CTE and Alzheimer’s disease were noted in 1988 [94]; this was followed by the description of tau inclusions using immunohistochemistry [95]. CTE is defined by an abundance of hyperphosphorylated tau in neurons, astrocytes and cell processes around small blood vessels (Figs. 13 and 14). Together with the accumulation of tau inclusions in cortical layers II and III [96], this distinguishes CTE from Alzheimer’s disease and other tauopathies. By Western blotting, assembled tau from CTE runs as major bands of 60 kDa, 64 kDa and 68 kDa, which consist of all six brain tau isoforms upon dephosphorylation [97].
By negative-stain electron microscopy of sarkosyl-insoluble material from CTE brains, it was shown that Type I tau filaments make up about 90% of filaments [98]. They differ from tau filaments of PiD, CBD and AD [74, 86, 99]. Widths are 20–25 nm and crossover spacings 65–80 nm. The remaining filaments (Type II) resemble paired helical filaments of Alzheimer’s disease; they have pronounced helical twists that result in projected widths of 15–30 nm.
Structures of tau filaments were determined by cryo-EM from the frontal cortex of three individuals with CTE (one former American football player and two ex-boxers) (Fig. 15). The core of Type I filaments is made of pairs of identical protofilaments that consist of residues K274/S305-R379 of tau (74 amino acids) (Fig. 15a). The protofilament structure (CTE fold) is similar to the C-shaped Alzheimer fold [99], but it adopts a more open conformation (Fig. 15c). Most notably, additional density—which is not present in the Alzheimer fold—is surrounded by the density of tau protein within the ordered core. Analysis of the minority Type II filaments revealed the presence of two kinds of filament, something that was not apparent by negative staining. Approximately 75% of these filaments (Type II) were composed of pairs of the same protofilament as in Type I tau filaments (including the extra density) but with a different protofilament interface. CTE Type I and Type II filaments are thus ultrastructural polymorphs that have different protofilament interfaces, but a common protofilament structure. The remaining filaments were identical to paired helical filaments of Alzheimer’s disease. This shows that cryo-EM was able to resolve what looked like paired helical filaments by negative staining into CTE Type II filaments and paired helical filaments. By cryo-EM, paired helical filaments made up 1–2% of filaments.
Structures of tau filaments from chronic traumatic encephalopathy. Type I and Type II tau filaments are characteristic, with Type I filaments forming the vast majority. (a, b), Unsharpened cryo-EM densities of Type I (a) and Type II (b) filaments. The Type I filament was resolved to 2.3 Å and the Type II filament to 3.4 Å. Both filament types show identical pairs of protofilaments. They differ in their inter-protofilament packing (ultrastructural polymorphs). In CTE Type I filaments, protofilaments pack through an anti-parallel steric zipper formed by residues 324SLGNIH329. The interface in CTE Type II filaments comprises residues 332PGGGQ336. (c), Schematic view of the tau protofilament core of CTE. The observed eight β-strands (β1–β8) are shown as arrows. The central non-proteinaceous density is shown in violet
Each CTE protofilament is C-shaped and contains eight β-strands, five of which give rise to two regions of anti-parallel β-sheets, with the other three forming a β-helix. The carboxy-terminal residues of R1 and R2 form part of the first β-strand. R3 contributes three and R4 four β-strands, with the final β-strand being formed by the 11 amino acids after the end of R4; β1 and β2 pack against β8, β3 packs against β7, with β4, β5 and β6 giving rise to the C-shaped β-helix. The CTE fold is similar to the Alzheimer fold [98, 99], with the main differences being present at the tip of the C, where the packing of β4–β6 coincides with an opening up of the C-shape, and a reversal in the orientation of residues S356 and L357. In CTE Type I filaments, two identical protofilaments pack in a staggered manner through an anti-parallel steric zipper formed by residues 324SLGNIH329. The interface in CTE Type II filaments is also staggered and comprises the same residues as the interface in Alzheimer’s disease–paired helical filaments (332PGGGQ336), but a kinked conformation reduces the number of hydrogen bonds across the interface.
The above-mentioned findings establish CTE as different from Alzheimer’s disease, even though tau inclusions of both diseases are made of all six brain isoforms. In contrast to Alzheimer’s disease, CTE is also characterised by an abundant glial tau pathology. The presence of a single CTE tau fold implies that the glial and neuronal tau inclusions are made of the same protofilament. The presence of identical CTE tau folds in the brains of a former American footballer and two ex-boxers establishes the presence of the same disease.
Conclusion
Assembled tau protein has been known to form the filamentous inclusions of a number of frontotemporal dementias since the 1980s. The finding that the same protein can be found in the inclusions of multiple diseases led some to conclude that the formation of tau inclusions is an epiphenomenon of little significance. The identification of mutations in MAPT in FTDP-17 T changed all that. To date, 65 disease-causing mutations have been identified. Most are missense mutations, but some change the ratio of 3R/4R tau. Clinicopathological studies have shown links between some mutations in MAPT and sporadic tauopathies.
Ongoing work has shown that the structures of tau filaments from sporadic PiD, CBD and CTE are different. Thus, the same protein takes on distinct structures in different diseases (Fig. 16). So far, in individuals with the same disease, be it PiD, CBD or CTE, filament structures were identical. It remains to be seen how the structures of tau filaments from the brains of individuals with MAPT mutations compare to each other and to those from sporadic diseases.
Structures of tau filament cores from human brain. (a) Protofilament from Pick’s disease (Pick fold), a 3R tauopathy; protofilament from corticobasal degeneration (CBD fold), a 4R tauopathy; protofilament from chronic traumatic encephalopathy (CTE fold), a 3R + 4R tauopathy. Red arrows point to the internal densities in CBD and CTE folds. β-Strands are marked by thick arrows (11 in the CBD fold, 9 in the Pick fold and 8 in the CTE fold). (b), Schematic depicting the microtubule-binding repeats (R1-R4) of tau and the sequence after R4, with β-strands found in the cores of tau filaments marked by thick arrows. Colours of individual β-strands are the same in (a) and (b)
References
Goedert M, Eisenberg DS, Crowther RA (2017) Propagation of tau aggregates and neurodegeneration. Annu Rev Neurosci 40:189–210
Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson DC, Goedert M (2015) Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol 41:24–46
Lee G, Newman ST, Gard DL, Band H, Panchamoorthy G (1998) Tau interacts with src-family non-receptor tyrosine kinases. J Cell Sci 111:3167–3177
Al-Bassam J, Ozer RS, Safer D, Halpain DS, Milligan RA (2002) MAP 2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol 157:1187–1196
Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, Nogales E (2018) Near-atomic model of microtubule-tau interactions. Science 360:1242–1246
Janning D, Igaev M, Sündermann F, Brühmann J, Beutel O, Heinisch JJ et al (2014) Single-molecule tracking of tau reveals fast kiss-and-hop interaction with microtubules in living neurons. Mol Biol Cell 25:3541–3551
Niewidok B, Igaev M, Sündermann F, Janning DF, Bakota L, Brandt R (2016) Presence of a carboxy-terminal pseudorepeat and disease-like pseudohyperphosphorylation critically influence tau’s interaction with microtubules in axon-like processes. Mol Biol Cell 27:3537–3549
Black MM, Slaughter T, Moshiach S, Obrocka M, Fischer I (1996) Tau is enriched on dynamic microtubules in the distal region of growing axons. J Neurosci 16:3601–3619
Qiang L, Sun X, Austin TO, Muralidharan H, Jean DC, Liu M et al (2018) Tau does not stabilize axonal microtubules but rather enables them to have long labile domains. Curr Biol 28:2181–2189
Boyka S, Qi X, Chen TH, Surewicz K, Surewicz WT (2019) Liquid-liquid phase separation of tau protein: the crucial role of electrostatic interactions. J Biol Chem 294:11054–11059
Lin Y, Fichou Y, Zeng Z, Hu NY, Han S (2020) Electrostatically driven complex coacervation and amyloid aggregation of tau are independent processes with overlapping conditions. ACS Chem Neurosci 11:615–627
Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3:519–526
Goedert M, Jakes R (1990) Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J 9:4225–4230
Yoshida H, Goedert M (2002) Molecular cloning and functional characterization of chicken brain tau: isoforms with up to five tandem repeats. Biochemistry 41:15203–15211
Tuerde D, Kimura T, Miyasaka T, Furusawa K, Shimozawa A, Hasegawa M et al (2018) Isoform-independent and –dependent phosphorylation of microtubule-associated protein tau in mouse brain during postnatal development. J Biol Chem 293:1781–1793
Brion JP, Passareiro H, Nunez J, Flament-Durand J (1985) Mise en évidence immunologique de la protéine tau au niveau des lésions de dégénérescence neurofibrillaire de la maladie d’Alzheimer. Arch Biol 95:229–235
Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A (1988) Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci U S A 85:4051–4055
Wischik CM, Novak M, Thøgersen HC, Edwards PC, Runswick MJ, Jakes R et al (1988) Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci U S A 85:4506–4510
Wischik CM, Novak M, Edwards PC, Klug A, Tichelaar W, Crowther RA (1988) Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci U S A 85:4884–4888
Berriman J, Serpell LC, Oberg KA, Fink AL, Goedert M, Crowther RA (2003) Tau filaments from human brain and from in vitro assembly of recombinant protein show cross-beta structure. Proc Natl Acad Sci U S A 100:9034–9038
Iqbal K, Liu F, Gong CX (2016) Tau and neurodegenerative disease: the story so far. Nat Rev Neurol 12:15–27
Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA (1996) Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 383:550–553
Kampers T, Friedhoff P, Biernat J, Mandelkow EM, Mandelkow E (1996) RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett 399:344–349
Pérez M, Valpuesta JM, Medina M, Montejo de Garcini E, Avila J (1996) Polymerization of tau into filaments in the presence of heparin: the minimal sequence required for tau-tau interaction. J Neurochem 67:1183–1190
Wilson DM, Binder LI (1997) Free fatty acids stimulate the polymerization of tau and amyloid beta peptides. Am J Pathol 150:2181–2195
Falcon B, Zhang W, Schweighauser M, Murzin AG, Vidal R, Garringer HJ et al (2018) Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol 136:699–708
Fichou Y, Lin Y, Rauch JN, Vigers M, Zeng Z, Srivasta M et al (2018) Cofactors are essential constituents of stable and seeding-active tau fibrils. Proc Natl Acad Sci U S A 115:13234–13239
Haj-Yahya M, Lashuel HA (2018) Protein semisynthesis provides access to tau disease-associated post-translational modifications (PTMs) and paves the way to deciphering the tau PTM code in health and diseased states. J Am Chem Soc 140:6611–6621
Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L et al (1998) Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol 43:815–825
Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H et al (1998) Association of missense and 5′-splice site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705
Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B (1998) Mutation in the tau gene in familial multiple system tauopathy with familial presenile dementia. Proc Natl Acad Sci U S A 95:7737–7741
Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J et al (2005) A common inversion under selection in Europeans. Nat Genet 37:129–137
Conrad C, Andreadis A, Trojanowski JQ, Dickson DW, Kang D, Chen X et al (1997) Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol 41:277–281
Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J et al (1999) Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 8:711–715
Di Maria E, Tabaton M, Vigo T, Abbruzzese G, Bellone E, Donati C et al (2000) Corticobasal degeneration shares a common genetic background with progressive supranuclear palsy. Ann Neurol 47:374–377
Pastor P, Ezquerra M, Munoz E, Marti MJ, Blersa R, Tolosa E et al (2000) Significant association between the tau gene A0/A0 genotype and Parkinson’s disease. Ann Neurol 47:242–245
Morris HR, Baker M, Yasojima K, Houlden H, Khan MN, Wood NW et al (2002) Analysis of tau haplotypes in Pick’s disease. Neurology 59:443–445
Zhang CC, Zhu JX, Wan Y, Tan L, Wang HF, Yu JT et al (2017) Meta-analysis of the association between variants in MAPT and neurodegenerative diseases. Oncotarget 8:4494–4507
Caffrey TM, Joachim C, Wade-Martins R (2008) Haplotype-specific expression of the N-terminal exons 2 and 3 at the human MAPT locus. Neurobiol Aging 29:1923–1929
Zhong Q, Congdon EE, Nagaraja HN, Kuret J (2012) Tau isoform composition influences rate and extent of filament formation. J Biol Chem 287:20711–20719
Lewis J, McGowan E, Rockwood J, Melrose H, Nacharaju P, van Slegtenhorst M et al (2000) Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nature Genet 25:402–405
Allen B, Ingram E, Takao M, Smith MJ, Jakes R, Virdee K et al (2002) Abundant tau filaments an nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J Neurosci 22:9340–9351
Götz J, Bodea LG, Goedert M (2018) Rodent models for Alzheimer disease. Nat Rev Neurosci 19:583–598
Macdonald JA, Bronner IF, Drynan L, Fan J, Curry A, Fraser G et al (2019) Assembly of transgenic human P301S tau is necessary for neurodegeneration in murine spinal cord. Acta Neuropathol Commun 7:44
Bussian TJ, Aziz A, Meyer CF, Swenson BL, Van Deursen JM, Baker DJ (2018) Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 562:578–582
Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A et al (2009) Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol 11:909–913
Frost B, Jacks RL, Diamond MI (2009) Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem 284:12845–12852
Jackson SJ, Kerridge C, Cooper J, Cavallini A, Falcon B, Cella CV et al (2016) Short fibrils constitute the major species of seed-competent tau in the brains of mice transgenic for human P301S tau. J Neurosci 36:762–772
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259
Kaufman SK, Del Tredici K, Thomas TL, Braak H, Diamond MI (2018) Tau seeding activity begins in transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer’s disease and PART. Acta Neuropathol 136:57–67
Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J et al (2013) Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci U S A 110:9535–9540
Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirhaba H, Li A et al (2014) Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82:1271–1288
He Z, McBride JD, Xu H, Changolkar L, Kim SJ, Zhang B et al (2020) Transmission of tauopathy strains is independent of their isoform composition. Nat Commun 11:7
Spillantini MG, Crowther RA, Kamphorst W, Heutink P, Van Swieten JC (1998) Tau pathology in two Dutch families with mutations in the microtubule-binding region of tau. Am J Pathol 153:1359–1363
Bronner IF, Ter Meulen BC, Azmani A, Severijnen LA, Willemsen R, Kamphorst W et al (2005) Hereditary Pick’s disease with the G272V mutation shows predominant three-repeat tau pathology. Brain 128:2645–2653
Van Swieten JC, Bronner IF, Azmani A, Severijnen LA, Kamphorst W, Ravid R et al (2007) The deltaK280 mutation in MAPT favors exon 10 skipping in vivo. J Neuropathol Exp Neurol 66:17–25
Murrell JR, Spillantini MG, Zolo P, Guazzelli M, Smith MJ, Hasegawa M et al (1999) Tau gene mutation G389R causes a tauopathy with abundant Pick body-like inclusions and axonal deposits. J Neuropathol Exp Neurol 58:1207–1226
Kouri N, Carlomagno Y, Baker M, Liesinger AM, Caselli RJ, Wszolek ZK et al (2014) Novel mutation in MAPT exon 13 (p.N410H) causes corticobasal degeneration. Acta Neuropathol 127:271–282
Tacik P, Sanchez-Contreras M, DeTure M, Murray ME, Rademakers R, Ross OA et al (2017) Clinicopathologic heterogeneity in frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) due to microtubule-associated protein tau (MAPT) p.P301L mutation, including a patient with globular glial tauopathy. Neuropathol Appl Neurobiol 43:200–214
Bugiani O, Murrell JR, Giaccone G, Hasegawa M, Ghigo G, Tabaton M et al (1999) Frontotemporal dementia and corticobasal degeneration in a family with a P301S mutation in Tau. J Neuropathol Exp Neurol 58:667–677
Erro ME, Zelaya MV, Mendioroz M, Larumbe R, Ortega-Cubero S, Lanciego JL et al (2019) Globular glial tauopathy caused by MAPT P301T mutation: clinical and neuropathological findings. J Neurol 266:2396–2405
Goedert M (2016) The ordered assembly of tau is the gain of toxic function that causes human tauopathies. Alzheimers Dement 12:1040–1050
Spina S, Farlow MR, Unverzagt FW, Kareken DA, Murrell JR, Fraser G et al (2008) The tauopathy associated with mutation +3 in intron 10 of Tau: characterization of the MSTD family. Brain 131:72–89
Spillantini MG, Crowther RA, Goedert M (1996) Comparison of the neurofibrillary pathology in Alzheimer’s disease and familial presenile dementia with tangles. Acta Neuropathol 92:42–48
Reed LA, Grabowski TJ, Schmidt ML, Morris JC, Goate A, Solodkin A et al (1997) Autosomal dominant dementia with widespread neurofibrillary tangles. Ann Neurol 42:564–572
Pick A (1892) Über die Beziehungen der senilen Hirnatrophie zur Aphasie. Prager Med Wochenschr 17:165–167
Alzheimer A (1911) Über eigenartige Krankheitsfälle des späteren Alters. Z ges Neurol Psychiat 22:146–148
Rasool CG, Selkoe DJ (1985) Sharing of specific antigens by degenerating neurons in Pick’s disease and Alzheimer’s disease. N Engl J Med 312:700–705
Pollock NJ, Mirra SS, Binder LI, Hansen LA, Wood JG (1986) Filamentous aggregates in Pick’s disease, progressive supranuclear palsy, and Alzheimer’s disease share antigenic determinants with microtubule-associated protein tau. Lancet 328:1211
Kertesz A, Munoz DG (eds) (1998) Pick’s disease and Pick complex. Wiley-Liss, Weinheim
Kovacs GG, Rozemuller AJM, Van Swieten JC, Gelpi E, Majtenyi K, Al-Sarraj S et al (2013) Neuropathology of the hippocampus in FTLD-tau with Pick bodies: a study of the brain net Europe consortium. Neuropathol Appl Neurobiol 39:166–178
Motoi Y, Iwamoto H, Itaya M, Kobayashi T, Hasegawa M, Yasuda M et al (2005) Four-repeat tau-positive Pick body-like inclusions are distinct from classic Pick bodies. Acta Neuropathol 110:431–433
Delacourte A, Robitaille Y, Sergeant N, Buée L, Hof PR, Wattez A et al (1996) Specific pathological tau protein variants characterize Pick’s disease. J Neuropathol Exp Neurol 55:159–168
Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R et al (2018) Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561:137–140
Probst A, Tolnay M, Langui D, Goedert M, Spillantini MG (1996) Pick’s disease: Hyperphosphorylated tau protein segregates to the somatoaxonal compartment. Acta Neuropathol 92:588–596
Delacourte A, Sergeant N, Wattez A, Gauvreau D, Robitaille Y (1998) Vulnerable neuronal subsets in Alzheimer’s and Pick’s disease are distinguished by their tau isoform distribution and phosphorylation. Ann Neurol 43:193–204
Lee SE, Rabinovici GD, Mayo MC, Wilson SM, Seeley WW, DeArmond SJ et al (2001) Clinicopathological correlations in corticobasal degeneration. Ann Neurol 70:327–340
Lhermitte J, Lévy G, Kyriaco N (1925) Les perturbations de la représentation spatiale chez les apraxiques. Rev Neurol (Paris) 2:586–600
Rebeiz JJ, Kolodny EH, Richardson EP (1968) Corticodentatonigral degeneration with neuronal achromasia. Arch Neurol 18:20–33
Gibb WRG, Luthert PJ, Marsden CD (1989) Corticobasal degeneration. Brain 112:1171–1192
Paulus W, Selim M (1990) Corticonigral degeneration with neuronal achromasia and basal neurofibrillary tangles. Acta Neuropathol 81:89–94
Feany MB, Dickson DW (1995) Widespread cytoskeletal pathology characterizes corticobasal degeneration. Am J Pathol 146:1388–1396
Kouri N, Whitwell JL, Josephs KA, Radekmakers R, Dickson DW (2011) Corticobasal degeneration: a pathologically distinct 4R tauopathy. Nat Rev Neurol 7:263–272
Sergeant N, Wattez A, Delacourte A (1999) Neurofibrillary degeneration in progressive supranuclear palsy and corticobasal degeneration: tau pathologies with exclusively “exon 10” isoforms. J Neurochem 72:1243–1249
Arai T, Ikeda K, Akiyama H, Nonaka T, Hasegawa M, Ishiguro K et al (2004) Identification of amino-terminally cleaved tau fragments that distinguish progressive supranuclear palsy from corticobasal degeneration. Ann Neurol 55:72–79
Zhang W, Tarutani A, Newell KL, Murzin AG, Matsubara T, Falcon B et al (2020) Novel tau filament fold in corticobasal degeneration. Nature 580:283–287
Ksiezak-Reding H, Tracz E, Yang LS, Dickson DW, Simon M, Wall JS (1996) Ultrastructural instability of paired helical filaments from corticobasal degeneration as examined by scanning transmission electron microscopy. Am J Pathol 149:639–651
Arakhamia T, Lee CE, Carlomagno Y, Duong DD, Kundinger SR, Wang K et al (2020) Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180:633–644
Martland HS (1928) Punch drunk. J Am Med Assoc 91:1103–1107
Millspaugh JA (1937) Dementia pugilistica. US Nav Med Bull 35:297–303
Critchley M (1949) Punch drunk syndromes: the chronic traumatic encephalopathy of boxers. In: Hommage à Clovis Vincent. Maloine, Paris, pp 131–145
McKee AC, Cantu RC, Nowinski CJ, Stern RA, Daneshvar DH, Alvarez VE et al (2013) The spectrum of disease in chronic traumatic encephalopathy. Brain 136:43–64
Corsellis JAN, Bruton CJ, Freeman-Browne D (1973) The aftermath of boxing. Psychol Med 3:270–303
Roberts GW (1988) Immunocytochemistry of neurofibrillary tangles in dementia pugilistica and Alzheimer’s disease: evidence for common genesis. Lancet 232:1456–1458
Tokuda T, Ikeda S, Yanagisawa N, Ihara Y, Glenner GG (1991) Re-examination of ex-boxers’ brains using immunohistochemistry with antibodies to amyloid β-protein and tau protein. Acta Neuropathol 82:280–285
Hof PR, Bouras C, Buée L, Delacourte A, Perl DP, Morrison JH (1995) Differential distribution of neurofibrillary tangles in the cerebral cortex of dementia pugilistica and Alzheimer’s disease cases. Acta Neuropathol 85:23–30
Schmidt ML, Zhukareva V, Newell KL, Lee VMY, Trojanowski JQ (2002) Tau isoform profile and phosphorylation state in dementia pugilistica recapitulate Alzheimer’s disease. Acta Neuropathol 101:518–524
Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R et al (2019) Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568:420–423
Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ et al (2017) Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 54:185–190
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Goedert, M. et al. (2021). Tau Protein and Frontotemporal Dementias. In: Ghetti, B., Buratti, E., Boeve, B., Rademakers, R. (eds) Frontotemporal Dementias . Advances in Experimental Medicine and Biology, vol 1281. Springer, Cham. https://doi.org/10.1007/978-3-030-51140-1_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-51140-1_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-51139-5
Online ISBN: 978-3-030-51140-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)