Abstract
Biobanks have become central structures for the provision of data. Raw data such as whole slide images (WSI) are stored in separate systems (PACS) or in biobank information systems. If segmentation or comparable information is to be exchanged in the context of the compilation of cases for the application of AI systems, the limits of proprietary systems are quickly reached. In order to provide these valuable (meta-) data for the description of biological tissue samples, WSI and analysis results, common standards for data exchange are required. Such standards are not currently available. Ongoing standardization developments, in particular the establishment of DICOM for digital pathology in the field of virtual microscopy, will help to provide some of the missing standards. DICOM alone, however, cannot close the gap due to some of the peculiarities of WSIs and corresponding analysis results. We propose a flexible, modular and expandable storage system - OBDEX (Open Block Data Exchange System), which allows the exchange of meta- and analysis data about tissue blocks, glass slides, WSI and analysis results. OBDEX is based on the FAIR data principles and uses only freely available protocols, standards and software libraries. This facilitates data exchange between institutions and working groups. It supports them in identifying suitable cases and samples for virtual studies and in exchanging relevant metadata. At the same time, OBDEX will offer the possibility to store deep learning models for exchange. A publication of source code, interface descriptions and documentation under a free software license is planned.
The work presented here was funded by the Federal Ministry of Education and Research (BMBF) as part of the “BB-IT-Boost” research project.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction and Motivation
In recent years application of artificial intelligence – especially deep learning – has made tremendous progress. In the developed world, almost everyone to some extent uses devices or services that use AI, such as photo apps on smartphones or shopping sites with product suggestions from previous purchases. The same technology is also used in many areas of ongoing medical research [1,2,3,4,5]. The possibilities are manifold and range from relatively simple tasks such as automated text indexing to more complex problems such as predicting the probability of the presence of tumors in MRI images.
Large training data sets are required for the successful training of neuronal networks. In many cases the creation of training data is only possible by a few experts. However, the compilation of larger data sets is also difficult for reasons such as data protection, data privacy or other legal reasons. If the data are collected in the current clinical context and continue to be used for the treatment of patients, the problems become even greater.
It is precisely at this interface that biobanks are increasingly prepared to make data available on a larger scale in pseudonymized or anonymized form. In the past, biobanks were mainly used for the storage of samples of different origins, but recently more and more sample-related data have come to the fore.
In this process the scope of biobanks has changed from being merely a diagnostic archive to a modern, research supporting core facility [6]. The use of whole slide images (WSI), which are managed in parallel to histological sections and blocks, offers several advantages [7]. For example, the suitability of cases for use in a study can be assessed directly on the monitor. It is not necessary to search for blocks or histological sections. Users of a biobank can search for cases independently and do not need the assistance of biobank staff if they have the right to view such cases for selection purposes. Using image analysis and pattern recognition techniques, the application examples can be extended from quality control at incoming material through quantification of biomarkers to virtual studies. Only the data from one or more biobanks are used and the analyses are performed on these data. For example, the correlation between two biomarkers and their significance for the assessment of the malignancy of a tumor could be achieved by a virtual combination of the corresponding data.
Therefore, the integration of virtual microscopy into biobank information systems has begun recently [8]. However, this integration is still limited. On the one hand, WSIs can be assigned to the respective physical slides and may be used for (remote) case selection, quality control or even viewing. On the other hand, analysis results based on image data can be stored as additional sample data using the data structures of the biobank systems, usually as key-value pairs, e.g. Ki-67 positivity. From the analysis of virtual slides, however, very high-quality data can be obtained, for example segmented cell nuclei, histoarchitectural structures (e.g. glands) or complex tissue models. An example for such high value analysis results is shown in Fig. 1. The image represents a heatmap overlay for Ki-67 positivity based on the cell nuclei classification for an entire WSI.
A biobank could increase the value of WSIs by offering such additional data to their users or even to the scientific community. In the context of machine learning (especially deep learning) where huge sets of data are needed, biobanks would be able to provide highly valuable data for training and validation of new models.
An important question at this point is whether a biobank system should actively occupy the role of data storage for the image analysis results from WSIs, or whether it should interface with systems that are more suitable for such a task. In the first case, the development of common formats and standards, due to different and incompatible solutions of the individual providers of biobank information systems, is significantly more difficult. Such fragmentation impedes the exchange of data and complicates the collaboration of different institutions and research groups. In the second case, an obvious solution would be the DICOM standard (Digital Imaging and Communications in Medicine), already well established in many clinical disciplines especially in radiology. Unlike other types of medical image data, e.g. CT or MRI, the DICOM standard in digital pathology, especially for virtual slides and supplemental data, is not yet widely used. Decisive steps - the involvement of major industrial companies, manufacturers of slide scanners and distributors of PACS (picture archiving and communication system), or the waiver of royalties for patents - have significantly advanced the DICOM standard in terms of virtual microscopy in recent years [9,10,11,12,13].
The use of DICOM would offer a valid bridge between clinical infrastructure, biobank information system and researchers. Nevertheless, there are open questions and problems that still need to be addressed, e.g. the integration of DICOM into biobank systems, the adaptation of existing PACS to fully support WSIs or the filing of analysis results in DICOM in accordance with the standard. The last point is the subject of this paper.
2 Glossary and Key Terms
- Biobank.:
-
A (core/key) facility that is responsible for storing bio samples at hospitals or research institutes, but also as an external service provider for smaller institutions. In recent years biobanks have become more important in research by providing samples and corresponding data (meta-data, data concerning the quality of samples, analytical data) to compile large collectives for e.g. clinical studies
- Digital Pathology (DP).:
-
Digital pathology includes the acquisition, management, sharing and interpretation of pathology information - including slides and data - in a digital environment. Digital slides are created when glass slides are captured with a scanning device, to provide a high-resolution image that can be viewed on a computer screen or mobile device [Royal College of Pathologist].
- Whole Slide Image (WSI, digital slide).:
-
A WSI is a digitized image of a tissue sample on a glass slide. They are digitized at a very high resolution (up to 100.000 dpi), have large dimensions (up to 1 Mio. pixel width and/or height) and are large in terms of storage space (up to several GBytes). The use of WSIs enables processing of image data of entire tissue samples and is in the focus of DP solutions.
- Virtual Microscopy.:
-
An important component of workflows in digital pathology. Virtual Microscopy refers to the work with WSIs. This includes viewing, annotating POIs/ROIs, digital image processing and the ability to collaborate directly with colleagues around the world. In the context of AI-based applications, Virtual Microscopy plays an important role in the creation of learning, validation and test sample collections.
- DICOM.:
-
Digital Imaging and Communications in Medicine (DICOM) is the standard for the communication and management of medical imaging information and related data [14]. DICOM was first presented in the 1980s (at that time under a different name) to enable the exchange of radiological images (mainly CT and MRI). After the first release more and more imaging modalities were incorporated (e.g. Ultrasound, endoscopy, microscopy). The main advantage of DICOM is that compatible devices and applications can communicate and exchange data without restriction.
- Picture Archiving and Communication System (PACS).:
-
PACS is a medical imaging technology to provide storage and access to images needed in medical facilities. The storage and transfer format used in PACS is DICOM.
- FAIR.:
-
FAIR data is a concept that describes data that meet certain conditions to facilitate the exchange of data and promote global cooperation [15]. The conditions for FAIR data are (a) findability (b) accessibility (c) interoperability and (d) reusability. In recent years, more and more researchers and institutions have endorsed the idea of FAIR data.
- Representational state transfer (REST).:
-
REST is a software architecture used to create web services. Web services that are conform to the REST architecture provide uniform and stateless operations to access and manipulate resources.
- Nginx.:
-
A free and open-source web server. Nginx supports many state-of-the-art web technologies and protocols, for example WSGI.
- Web Server Gateway Interface (WSGI).:
-
WSGI is a calling convention to forward web requests to Python services, applications or frameworks. WSGI is used to establish a connection between web servers (e.g. Nginx) and components written in Python (e.g. a Flask web service).
- FLASK.:
-
A lightweight micro web framework written in Python. Flask incorporates the REST architecture and is WSGI compliant. The main advantage of Flask is the ease of use, low system requirements, fast development and extensibility.
- Relational Database Management System (RDBMS).:
-
A database management system based on the relational model to represent data as tuples, grouped into relations. A RDBMS can be queried in a declarative manner.
- Object-relational mapping (ORM).:
-
ORM is a technique to convert data between incompatible type systems using object-oriented programming languages. A common use case for ORM is to map objects used in program code onto tables in a relational database. All access to or changes in the state of objects are automatically translated into commands understandable by the database.
3 State-Of-The-Art
The possibilities of machine learning in general and recent developments in artificial neural networks (especially convolutional neural networks – CNNs) are applied in more use cases every year. These applications of artificial intelligence (AI) to process huge amounts of data are the basis for many products and services presented in the last few years. For many medical disciplines, including pathology, AI-based applications have been the subject of extensive research in recent years. In the following section we will present and discuss some of the up-to-date AI-focused research in digital pathology. This overview is very limited, but gives a general impression of the current development.
3.1 Current Developments of AI in Digital Pathology
In line with the objective of OBDEX, we concentrate on AI-based applications for the processing of histological sections. AI is already successfully used for many technical approaches for the evaluation of histological sections, e.g. image analysis, 3D reconstruction, multi-spectral imaging and augmented reality [16]. It provides both commercial and open source tools to assist pathologists with specific tasks (e.g. quantifying biomarkers or registering consecutive slides). The authors of the cited survey come to the conclusion that the AI will continue to expand its influence in the coming years.
A similar prediction of developments in digital pathology is outlined in [17]. This overview focuses on the integrative approach to combine different data sources, such as histological image data, medical history or omics data, to make predictions to assist pathologists. Such a combination of heterogeneous data into a Machine Aided Pathology (also [17]) would benefit development towards personalized medicine.
Another important aspect when using the assistance of computer generated/analyzed data to make a decision (for diagnosis as well as in research) is the ability to understand and explain why this decision was made. There are researchers who are working on approaches fostering transparency, trust, acceptance and the ability to explain step-by-step the decision-making process of AI-based applications [17, 18]. First steps to understand and evaluate the predictions of trained AI models for image data have been taken [19]. These methods were used to evaluate the automated detection of tumor-infiltrating lymphocytes in histological images [4]. Building trust in predictions from AI-based applications is of utmost importance. A pathologist who does not trust the results of a software will not use it.
Trust, understanding and explainability is not only important regarding the acceptance of AI among pathologists but also when legal or regulatory approval is concerned. In [20] a roadmap is set out to help academia, industry and clinicians develop new software tools to the point of approved clinical use.
3.2 Conclusion
Many researchers predict an increasing importance of AI-based applications in digital pathology for the next years. In addition to the ongoing development of new methodologies and the transfer to new use cases other aspects are equally important. Integration data from different sources, building trust among users and satisfying legal and regulatory conditions must not be neglected.
In the scope of this work the standardization of data formats, communication and transfer protocols and interfaces is of particular interest. The impact of AI in digital pathology could be increased further, if (meta-)data of biological samples, analysis results, (labeled) data sets for training, validation and testing and/or partially or fully trained prediction models would be shared within the medical and scientific community. To realize this, common standards are needed as soon as possible to counteract the development of many individual solutions that are not compatible.
3D reconstruction of tissue (example reconstruction of mouse brain) [21].
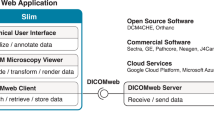
4 OBDEX – Open Block Data Exchange System
We propose an extensible and flexible storage system for analysis data of virtual slides to address the issues described above. The scope of this system is to provide access to and to allow the exchange of detailed results based on the analysis of WSIs. On one hand it will be possible to access a complex step-by-step evaluation (e.g. model of histoarchitecture) on already existing results (e.g. classified cell nuclei) and on the other hand to compare the results between different algorithmic solutions on several levels.
The general structure of meta and analytical data for tissue slides (or more generally whole tissue blocks) is more or less hierarchical. The topmost (first) level contains data describing the physical tissue block. The type of material (biopsy, OP material) and the spatial location within the organ are examples for such data. The first level is followed by the second level, with all data concerning the glass slides generated from the tissue samples and their corresponding WSIs. It should be noted here that no primary image data (raw data) is stored in OBDEX, but only data newly generated by analysis (e.g. contours of cell nuclei) or transformation (e.g. registration) is stored. Image data must be stored in other information systems designed for such a task, ideally a fully DICOM conform PACS.
Between the first (tissue block) and second level (individual slides/WSIs) there is an intermediate level to address data that combines information from more than one single slide. An example for such multi-slide data is the registration of consecutive WSI of the same block. Such registration is necessary if WSI with different staining are to be viewed in parallel. Reconstructing 3D models of complex anatomical structures based on large stacks of consecutive slides is another use case where registration data is needed (see Fig. 2).
On the third level (and all levels following thereafter) analytical results based on individual slides are arranged. The data on these levels can be very diverse and can depend on each other (e.g. a tissue classification depends on a tissue segmentation). Figure 3 illustrates the basic structure of the hierarchical data arrangement and possible dependencies.
Taking into account the increasing adaptation of DICOM to the requirements of digital pathology, the system we propose can be used in conjunction with an appropriate PACS, perhaps as a DICOM addition in the future. Beside the image data DICOM supports handling of other, even sensible data in a save way. So (meta-)data concerning the slide (e.g. image acquisition parameters, modality data) are stored in the DICOM series. Complex analysis results (e.g. graph-based topological/object representations as discussed in [22]) that cannot be represented on the basis of DICOM are stored in OBDEX. In some cases (e.g. segmentation) a redundant storage in both systems can be desirable to address different use cases more comfortably. A DICOM viewer expects segmentation data according to the DICOM standard. A data scientist performing further data analysis or composing a data set to train artificial neural networks based on segmentations may use different data formats (e.g. XML or CSV files). From their point of view, it is neither necessary nor useful to work directly with the DICOM format. In this case, parallel storage in both systems makes sense, provided that the data is synchronized.
4.1 OBDEX Implementation
The technical infrastructure of hospitals and research facilities can be very versatile. An important feature of a storage system is its flexibility to be used in as many contexts as possible. In order to achieve this necessary flexibility, OBDEX is divided into the two components storage engine and middleware.
Storage Engine. The purpose of the storage engine is to store the analysis data. The actual implementation of the Storage Engine is interchangeable to be as flexible as possible so that it can be adapted to the needs or requirements of a particular infrastructure, provided a common interface is used with the middleware. This offers the possibility to use different database systems (e.g. PostgreSQL, MySQL) or file-based formats (HDF5 - hierarchical data format) as storage engines.
The middleware component is used as gateway between the data stored by the storage engine and the user. Middleware and storage engine are connected by the interface as described in Sect. 2. The middleware is able to access as many storage engines as necessary. A second interface serves to interact with the users by providing all necessary endpoints for creating, reading, updating and deleting data from connected storage engines.
Middleware. From a technical perspective, the middleware is a RESTful web application implemented in Python using the web micro-framework Flask [23] and is served by a Nginx [24] proxy server and uWSGI [25] to be accessible for multiple users simultaneously.
If the middleware is used in combination with database systems as storage engines, the ORM toolkit SQLAlchemy [26] is used to handle all accesses to the databases. Additionally, if a supported relational database management system is used, all database tables can be created automatically as well.
Extensibility. In its base form the user-interface of the middleware can handle standard analysis results, for example annotations, segmentations and classifications, in a predefined JSON format. Both data format and supported analysis result types can be extended. To add a new data format a parser for this format must be provided. A new parser can include only data input or output or both and must implement methods to access (input only), validate and write (output only) the data in the new format. If needed the validation can be done by using an additionally provided XML or JSON schema.
To add a new analysis type the provided data model must be extended. This can be done by using all existing models (e.g. deriving a new model or aggregating existing models) or by defining completely new models. All user provided data formats and analysis results will be loaded and integrated beside the standard format and data models on server start.
System Overview. Based on the components introduced the storage system is a flexible composition of middleware and one or more storage engines. Figure 4 shows an example of the general structure of the storage system.
The FAIR data principles [15] for research data as a basis for international scientific cooperation are a crucial reason for the specific form of the proposed storage system. In accordance with these principles all used protocols, software libraries and data structures are freely available, open source and documented.
4.2 Use Cases
To demonstrate the flexibility and ease of use of the OBDEX system, we will consider the following two use cases: (1) use of intermediate results, (2) composition of ground truth data for AI-based analysis.
Ki-67 Positivity, Original Data Access Versus Heatmap Exchange. A tile-based Ki-67 positivity analysis should be performed for a WSI to generate a heatmap overlay (as shown above). The planned analysis is composed of four steps for each image tile of the WSI: cell nuclei segmentation, nuclei feature calculation, nuclei classification and Ki-67 positivity score calculation (as described in [27]). It would be sufficient to store only the final results for each tile (the positivity score) to generate the heatmap. But in terms of reusability and data exchange the storage of all intermediate results would be much better. The OBDEX system can provide that possibility.
If intermediate analysis results are stored and arranged in an appropriate structure, researchers can retrieve them at any stage in the analysis process and use them for their own research purposes. In the case of a Ki-67 analysis with the results of all stored process steps, it would be possible to retrieve only the classification of the nuclei (together with the segmentation result and the calculated features) without considering positivity. The obtained results could be used for a different kind of analysis, e.g. to determine morphological characteristics based on nuclei of a certain type or to generate basic truth data for artificial neural networks. In addition, it would allow users to retrieve only subsets of result data that meet a number of requirements, such as nuclei of a particular class that are larger than a minimum area. Figure 5 shows an intermediate classification result of a Ki-67 positivity analysis.
Composition of Ground Truth Data for AI Based Analysis. The aim is to train an AI model to recognize certain morphological structures in histological images. In order for a data scientist to be able to fulfill this task, corresponding annotated image data are required as basic truth. If the existing infrastructure consists for example of a biobank information system for samples and clinical data, an image archive (PACS) for WSIs and OBDEX for the storage of analysis results, such a basic truth could be put together. Figure 6 shows the information flow and interaction between the systems involved in such a use case.
4.3 First Reference Implementation
A first implementation of the OBDEX system is used in conjunction with the ePathology platform CoPaW (Collective Pathology Wisdom) [28]. In this project it is possible to exchange cell nuclei segmentations for entire WSIs or a specific field of view including the possibility of live viewing of all segments. The same mechanism is used for the exchange of annotations within an open discussion of cases between groups of pathologists. Figure 7 show a screenshot of the CoPaW platform.
This reference implementation of OBDEX uses the middleware as described in Sect. 4.1. The REST-interface provides endpoints to create, access, update or delete annotations for each WSI shared on the CoPaW platform. When accessing annotations, additional parameters can specify a certain field of view to filter the returned annotations by means of visibility. Similar endpoints are provided to handle segmentation results. All endpoints enable the user to send an optional format parameter to choose between different data formats (in the current implementation state predefined XML or JSON). As storage engine a PostgreSQL database is used.
5 Open Problems
Due to the fact that OBDEX is designed as a supplement to DICOM, the ongoing development of the DICOM standard and the integration of digital pathology and WSIs into it, is of particular importance.
5.1 OBDEX and the DICOM Standard
The first step towards a common standard for the exchange of medical images took place at the beginning of the 1980s, when radiologists demanded the possibility of using images from computer tomography (CT) and magnetic resonance imaging (MRI) in therapy planning (e.g. dose planning for radiation therapy). In 1985 the American College of Radiology (ACR) and National Electrical Manufacturers Association (NEMA) introduced the ACR/NEMA 300 standard. After two major revisions the third version of the standard was released in 1993 and renamed to DICOM. Currently DICOM is used in all imaging systems in radiology and nuclear medicine to store, visualize and exchange image and corresponding supplemental data. DICOM has been extended to most other medical specialties that use images, videos or multidimensional image data, such as sonography, endoscopy or dentistry.
The use of DICOM in digital pathology was somewhat hesitant. A major problem is the huge size of WSIs, so specific parts of the DICOM standard could not be adapted one-to-one. This can be seen, for example, in the use of annotations and segmentations. Of course, these can be managed in DICOM, but not for millions of segmented objects, neither in number nor in speed. We discussed the possibilities of DICOM for digital pathology and WSIs with David ClunieFootnote 1 in an email conversation in 2018. His answer concerning the huge number of segments or annotations in a single WSI was:
Without extending the DICOM standard, the two ways to store WSI annotations that are currently defined for general use are: On a pixel by pixel basis, as a segmentation image, where each pixel is represented as a bit plane \(\ldots \); there may be multiple segments (e.g. nuclei, membranes), but this approach does not anticipate identifying individual nuclei \(\ldots \) As contours (2D polylines) outlining objects, stored as either DICOM presentation states (not ideal, no semantics) or DICOM structured reports (SR) (each graphic reference may have attached codes and measurements and unique identifiers (UIDs). The biggest problem with the coordinate (SR) representation is that it is not compact (though it compresses pretty well), so while it may be suitable for identifying objects in small numbers of fields (tiles, frames) it probably doesn’t scale well to all the nuclei on an entire slide.
His comment on dealing with graph based analysis results (e.g. Delaunay triangulation with different cell types) was:
However, there is no precedent for encoding the semantics of graph-based methods as you discuss.
Our conclusion from these statements is that DICOM is currently not suitable to cover all of the use cases described in the above sections. Most are more or less feasible (in a cumbersome way), some are not.
One of the main challenges will be to find out where the limits of DICOM as the used standard are and where OBDEX can be used additionally to enable applications beyond the limits of DICOM.
5.2 Integration into Existing Clinical Infrastructures
As discussed in the previous sections OBDEX is focused to handle all relevant data concerning tissue samples. This includes data for the entire block but also for a single slide/WSI. When OBDEX is used as a supplement to other systems (e.g. biobank system or laboratory information management system) collaboration between the systems is needed to link the data for individual samples. It is to be expected, that such links could require an adaptation of other systems or the need to provide a joint search interface to fully integrate all available data of a sample into a single query. Another aspect of integration that needs to be taken into account is the synchronization of data that is stored redundantly in both systems for convenience and ease of use.
Some clinical use cases require that different samples (blocks) of a patient must be considered together, for example a metastatic tumor. In the case of two tumors, for example, the question may arise as to whether one tumor is a metastasis of the other or whether it is an independent tumor. All samples and their associated data are needed to make the final diagnosis. In laboratory information systems, this information is linked on a case-by-case basis. In its current design state OBDEX is not able to link samples from different tissue blocks. If such links are needed, they have to be established in other systems. From a technical perspective OBDEX could be extended to be able to realize such links by itself. However, it would be preferable if links between different blocks could be established by other systems, e.g. biobank systems. In a biobank system, at best all samples of a patient are semantically connected. Adding the functionality to link samples in OBDEX would be comparatively easy. However, bi-directional interfaces to LIS or LIMS would have to be established and data protection aspects would have to be taken into account.
6 Conclusion, Further Developments and Future Outlook
The OBDEX system for meta and analytical data for tissue blocks, glass slides and WSI is flexible, modular and extensible. By using open and extensible interfaces and complying with FAIR data principles, it offers the possibility to facilitate data exchange between institutions and working groups. By linking to biobank, pathology and other medical information systems, relevant data can be provided to support AI applications.
The hierarchical structure of the interconnected measurement results and data opens a channel for the exchange of intermediate results at different levels of a complex algorithm.
In the future, DICOM will be an important standard in both digital pathology and biobanking. Regardless of the specific features of DICOM discussed in this paper, dimensions such as image acquisition, image management, patient planning information, image quality, media storage, security, etc. form the basis for future use of the standard in pathology and biobanking.
The OBDEX system can help to remedy the current lack of common standards until DICOM becomes more widely used, covering applications that DICOM cannot or cannot comfortably handle.
6.1 Further Developments
The further development of OBDEX will concentrate on four essential aspects: Data protection, support of additional data formats, simplification of use and integration with other information systems (Biobank IS, Pathology IS).
In the current implementation the system does not contain any ID data and does not log any data to identify individual users by their IP addresses. Within specific research context or clinical studies it may be necessary to protect data from unauthorized access. Therefore, an authentication and authorization concept is required. A sensible solution is to control access to the data via another platform on which the users are already authenticated. If, for example, analysis data is to be accessed for visualization with a viewer, the user’s login information is transferred to the query at OBDEX in order to check whether he has access to the queried data. When implementing such scenarios, the GDPR (General Data Protection Regulation) must of course be taken into account.
To increase the number of input and/or output data formats supported by OBDEX, additional format parser must be provided. The focus is on common formats of widely used analysis tools or platforms. Import and export from or to DICOM should be realized as soon as possible.
Analysis results derived from deep learning applications should be sufficiently complemented by metadata describing important attributes of the model used and the underlying ground truth data. Ideally, the results could be complemented by the model from which the data was generated. Then other users who do not have the ability to create a sufficient model themselves can reuse it for their own analysis. This requires a common standard for the exchange of these models. For example, ONNX (Open Neural Network Exchange) [29] could be used. However, it should be examined whether this can be brought into line with the FAIR principles.
Easy handling is also an important goal. Therefore, the storage system should be as easy to install and operate as possible. A direct way to achieve this would be to use preconfigured docker images that can be customized with only a few configuration files. In a standard installation (default settings for ports of the web server, number of processes and threads of the Flask application) without self-created format or analysis extensions, only the external IP address would have to be configured. In order to use OBDEX with such default settings, only two Docker Container (middleware and storage engine) have to be started. New types of storage engines could also be easily distributed as docker images. Docker would also make it easier to connect several separate middleware instances to shared storage engines and could form the basis of a decantralized research infrastructure. The first steps towards a docker-based distribution have already been taken.
OBDEX and all necessary supplemental documentation will be released under a suitable open source software license in the near future.
Notes
- 1.
David Clunie is a member of the DICOM committee.
References
Geert, L., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Song, Y., et al.: A deep learning based framework for accurate segmentation of cervical cytoplasm and nuclei. In: 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 2903–2906, IEEE (2014)
Henning, H., et al.: Deep learning nuclei detection: a simple approach can deliver state-of-the-art results. Comput. Med. Imaging Graph. 70, 43–52 (2018)
Klauschen, F., et al.: Scoring of tumor-infiltrating lymphocytes: from visual estimation to machine learning. Semin. Cancer Biol. 52(2), 151–157 (2018)
Sharma, H., et al.: Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput. Med. Imaging Graph. 61, 2–13 (2017)
Hufnagl, P., et al.: Virtual microscopy in modern tissue-biobanks - the ZeBanC example. In: 27th European Congress of Pathology, Extended Abstracts, pp. 41–45. Springer (2015)
Weinstein, R.S., et al.: Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Human Pathol. 40(8), 1057–1069 (2009)
Kairos - Biobanking 3.0. https://www.kairos.de/referenzen/konsortium/biobanking-3-0. Accessed 29 Nov 2019
Jodogne, S.: The orthanc ecosystem for medical imaging. J. Digit. Imaging 31(3), 341–352 (2018)
Clunie, D., et al.: Digital imaging and communications in medicine whole slide imaging connectathon at digital pathology association pathology visions 2017. J. Pathol. Inform. 9 (2018)
Herrmann, M., D. et al.: Implementing the DICOM standard for digital pathology. J. Pathol. Inform. 9 (2018)
DICOM Working Group 26. https://www.dicomstandard.org/wgs/wg-26/. Accessed 29 Nov 2019
Godinho, T.M., et al.: An efficient architecture to support digital pathology in standard medical imaging repositories. J. Biomed. Informat. 71, 190–197 (2017)
DICOM PS3.1 2019d - Introduction and Overview - 1 Scope and Field of Application. dicom.nema.org/medical/dicom/current/output/chtml/part01/chapter1.html. Accessed 29 Nov 2019
Wilkinson, M.D., et al.: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3 (2016)
Janardhan, K.S., et al.: Looking forward: cutting-edge technologies and skills for pathologists in the future. Toxicologic pathology (2019). 0192623319873855
Holzinger, A., et al.: Machine learning and knowledge extraction in digital pathology needs an integrative approach. In: Holzinger, A., Goebel, R., Ferri, M., Palade, V. (eds.) Towards Integrative Machine Learning and Knowledge Extraction. LNCS (LNAI), vol. 10344, pp. 13–50. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-69775-8_2
Pohn, B., et al.: Visualization of histopathological decision making using a roadbook metaphor. In: 23rd International Conference Information Visualisation (IV). IEEE (2019)
Bach, S., et al.: On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 10, 7 (2015)
Colling, R., et al.: Artificial intelligence in digital pathology: a roadmap to routine use in clinical practice. J. Pathol. 249(2), 143–150 (2019)
Norman, Z., et al.: Creation and exploration of augmented whole slide images with application to mouse stroke models. In: Modern Pathology, vol. 31 Supplement 2, p. 602 (2018)
Sharma, H., et al.: A comparative study of cell nuclei attributed relational graphs for knowledge description and categorization in histopathological gastric cancer whole slide images. In: 2017 IEEE 30th International Symposium on Computer-Based Medical Systems (CBMS), pp. 61–66. IEEE (2017)
Flask (A Python Microframework). http://flask.pocoo.org/. Accessed 29 Nov 2019
NGINX. https://www.nginx.com/. Accessed 29 Nov 2019
The uWSGI project. https://uwsgi-docs.readthedocs.io/en/latest/. Accessed 29 Nov 2019
SQLAlchemy - The Database Toolkit for Python. https://www.sqlalchemy.org/. Accessed 29 Nov 2019
Klauschen, F., et al.: Standardized Ki67 diagnostics using automated scoring–clinical validation in the GeparTrio breast cancer study. Clin. Cancer Res. 21(16), 3651–3657 (2015)
CoPaW - Collective Pathology Wisdom, A Platform for Collaborative Whole Slide Image based Case Discussions and Second Opinion. http://digitalpathology.charite.de/CoPaW. Accessed 29 Nov 2019
ONNX: Open Neural Network Exchange Format. https://onnx.ai/. Accessed 29 Nov 2019
Acknowledgements
We thank David Clunie for his feedback on the current state of DICOM for digital pathology and his comments on OBDEX ideas. We also thank Tobias Dumke, Pascal Helbig and Nick Zimmermann (HTW University of Applied Sciences Berlin) for implementing the CoPaW ePathology platform and collaborating on the integration of the OBDEX system. Finally, we would like to thank Erkan Colakoglu for his work on adaptations for distributing OBDEX via Docker.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Lindequist, B., Zerbe, N., Hufnagl, P. (2020). OBDEX – Open Block Data Exchange System. In: Holzinger, A., Goebel, R., Mengel, M., Müller, H. (eds) Artificial Intelligence and Machine Learning for Digital Pathology. Lecture Notes in Computer Science(), vol 12090. Springer, Cham. https://doi.org/10.1007/978-3-030-50402-1_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-50402-1_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-50401-4
Online ISBN: 978-3-030-50402-1
eBook Packages: Computer ScienceComputer Science (R0)