Abstract
Protein phosphatases especially type 2C (PP2C) are unique and highly conserved and play pivotal roles in stress signaling in plants by counter affecting kinases. During potassium (K+) deficiency in the soil as well as salt stress conditions, protein phosphatases have been recognized to fine-tune and help the plant systems in the uptake of K+ ions. K+ is an essential macronutrient and performs vital functions in plants such as osmoregulation, regulation of membrane potential, cotransport of sugars, growth, stress adaptation, and others. Diverse transporters exist for the transport of K+, but their fine regulation under fluctuating (high and low) K+ concentrations in the external medium remains largely unclear. Studies have identified major calcium (Ca2+)-signaling pathway in its regulation. Activation of voltage-gated K+ channel, Arabidopsis K+ transporter (AKT1), is dependent on calcineurin B-like Ca2+ sensor1/9 (CBL1/9) and a target kinase 23 (CBL-interacting protein kinase 23 or CIPK23). This Ca2+ sensor-kinase complex phosphorylates AKT1 for its activation to acquire K+ in roots as a response to low K+ availability. Protein phosphatases have also been recognized as important regulators of the activity of different K+ channels through a reversal of kinase activity by the process of dephosphorylation. Several findings provide insights into the signaling networks consisting of Ca2+ responder module CBL-CIPK and PP2C interactions in the activation-deactivation of channels for K+ acquisition. Existing information is collated in this chapter on the K+ transporters that are triggered by low K+ levels under K+ deprivation and high saline conditions. Taken together, it seems an intricate phosphorylation-dephosphorylation network mediated by Ca2+ signaling is regulating the K+ acquisition and signaling in plants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Abiotic stress
- Calcium signaling
- CBL
- CIPK
- AKT1
- HAK5
- Protein phosphatases
- Potassium uptake
- Potassium deficiency
- Potassium homeostasis
11.1 Introduction
Plants experience different types of biotic and abiotic stresses and nutrient deficiency during their life cycle. In a given environment, multiple stress factors are exerting their effect on plant growth and development simultaneously. Enhancing crop resilience in response to these abiotic stresses or mineral deprivation is a challenge in the endeavor to improve crop productivity (Mittler and Blumwald 2010). Abiotic stresses, in general, hamper plant growth and productivity by affecting several physiological, biochemical, and molecular responses (Zhu 2016). The role of potassium (K+) during plant stress tolerance especially salt and drought as well as in plant growth and development is well known. In comparison, it has been reported that K+ retains the photosynthetic electron transport activity by decreasing the activity of nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (Waraich et al. 2012). This phenomenon, in turn, decreases the production of ROS under stress conditions. On the other hand, deficiencies in K+ can reduce the dark CO2 fixation and productivity (Waraich et al. 2012). K+ reduces the effects of drought, cold, and high light intensities (Waraich et al. 2012). Its accumulation is triggered under a combination of stresses like drought and high temperature (Halford 2009). K+ homeostasis in the cytoplasm and the ability of plant tissues to retain K+ under stress conditions is an important mechanism of salinity stress tolerance (Shabala et al. 2016). A clear positive correlation between tissue retention of K+ and salt stress tolerance was noticed in cotton, pepper, and other crops (Shabala and Cuin 2008). Further, it appears that the differential K+ retention ability imparts diverse salinity stress tolerance between halophytes and glycophytes (Assaha et al. 2017). Thus, K+ appears to play crucial roles during abiotic stress tolerance. It is not clear how K+ transporters are regulated by the phytohormonal network, reactive oxygen species (ROS) signaling, and other factors especially under K+-deficient or high salt/drought stress conditions. Fifty-six K+ transporters have been identified in higher plants like Pyrus bretschneideri, Fragaria vesca, Vitis vinifera, and wheat (Cheng et al. 2018; Li et al. 2018), but only few of them (e.g., Arabidopsis K+ transporter 1 (AKT1), high-affinity K+ transporter 5 (HAK5), K+ channel in Arabidopsis thaliana (KAT), gated outwardly rectifying K+ channel (GORK), K+ efflux antiporter (KEA), tandem-pore K+ channel (TPK/KCO)) have been characterized functionally under K+ nutrition/transport (Dreyer and Uozumi 2011; Hedrich 2012; Ragel et al. 2019). These transporters play crucial roles in K+ deprivation conditions and help the plants to survive under harsh environments.
Kinases and protein phosphatases (PPs) are the two enzymes involved in protein phosphorylation and dephosphorylation, respectively (Luan 2003). PP is an enzyme, which removes the phosphate group from the phosphorylated amino acid residue by counteracting the action of the protein kinases and provides modulation and reversibility of the phosphoregulatory mechanism (Luan 2003). In plant and animal, phosphorylation occurs on nine different amino acids, but the most common are Ser, Thr, and Tyr (Moorhead et al. 2009). Phosphorylation of His has been reported in fungi (Ota and Varshavsky 1993) and higher plant (Chang et al. 1993; Nongpiur et al. 2012). PPs occur in chloroplast, cytosol, mitochondria, and nuclei (MacKintosh et al. 1991; Huber et al. 1994). Based on the function, sequence, and structure, plant and animal PPs are evolutionarily conserved and are classified into four groups: (1) the phosphoprotein phosphatases (PPP), (2) the Mg2+/Mn2+-dependent enzymes (PP2C/PPM), (3) the protein tyrosine phosphatases (PTP), and (4) the Asp-based enzymes (Uhrig et al. 2013b). The PPPs are responsible for ∼80–90% protein dephosphorylation in animal. In a plant cell, the PP2C/PPMs have a higher number of PP2Cs (compared to animal), and in the plant, the above ratio (of PPPs performing 80–90% dephosphorylation) may be altered (Uhrig et al. 2013a). Many studies point out that protein phosphatases participate in signaling cascades including abscisic acid (ABA) (Luan 1998), auxin (Xu and Zhang 2015), brassinosteroid (Tang et al. 2011), and biotic stress responses (Durian et al. 2016). Phosphatases play an important role in plant growth and development (Luan 2002) and are also involved in the regulation of mineral deprivation conditions (Pandey et al. 2014). This chapter summarizes some of the recent advances made on the regulation and functional aspects of phosphatases and their role in K+ homeostasis under low K+ availability and abiotic stress. Elucidation of such a mechanism(s) would help in designing crop plants that can cope with high salinity stress conditions and yet produce better yields.
11.2 Protein Phosphatases (PPs): Their Classes and Structure
Post-transcriptional regulation of proteins by reversible activities of kinases (phosphorylation) and phosphatases (dephosphorylation) is an important aspect in the life cycle of a plant and involves in diverse functions such as metabolism, ion transport, developmental control, and stress responses (Luan 2003). Attachment of a phosphate group to a protein regulates the enzyme activities by the initiation of allosteric conformational changes. Such an attachment blocks the access to the active site of an enzyme (Johnson et al. 1993; Johnson and O’Reilly 1996) and also the interaction among protein partners for proper functioning at the destination sites (Pawson 1995). Therefore, it is interesting to study the key roles being played by both phosphatases and kinases during plant development. Both diversity and functions of plant protein phosphatases and kinases have been well recognized.
In the PPP group, the catalytic subunit is the core enzyme and is more specific for Ser/Thr residues than Tyr (Moorhead et al. 2009). The PPP can be divided into seven members, protein phosphatase PP1, PP2A, PP2B (aka PP3), PP4, PP5, PP6, and PP7 (Moorhead et al. 2009). All the four groups are conserved in plants except PP2B/PP3 (aka calcineurin), which is absent in plants (Singh et al. 2015). Probably as compensation, plants have Kelch-repeat domain-containing protein phosphatases (associated with brassinosteroid signaling) (Mora-Garcia et al. 2004) and Shewanella-like phosphatases (SLPs) (Andreeva and Kutuzov 2004). These two also fall under the PPP and are unique to plants although they may also show Tyr specificity (Uhrig et al. 2013b). The PP2Cs in Arabidopsis and rice are divided into 11 subclades (A-K) and, like PPPs, are Ser/Thr dephosphorylating enzymes (Singh et al. 2015). The PTPs and Asp-based enzymes can be phosphatases dephosphorylating Tyr, Ser/Thr, and both (aka dual-specificity phosphatase) (Uhrig et al. 2013b). The Like-SEX4-1 and Like-SEX4-2 (LSF1 and LSF2) are the best-characterized examples of PTP (Silver et al. 2014; Gentry et al. 2016). Both PPP and PPM members share similar catalytic mechanisms but differ in many ways (Uhrig and Moorhead 2011). PPMs are Mg2+/Mn2+ dependent, without any associated regulatory subunits, and are inhibited by okadaic acid and microcystin (Shi 2009). On the other hand, members of the PPP family lack accessory domains in their catalytic subunits but are associated with regulatory subunits (Moorhead et al. 2009). Members of the PPP family have a catalytic domain of nearly 280 amino acids, and the sequence is mostly conserved. On the other hand, members of the PP2C family are similar to animal homologs but diversified (Luan 2003). Though the primary sequence of PP2C and other members of PPM family do not share homology with PPP enzymes, the structural folds are mostly similar (Das et al. 1996). The N-terminal catalytic region forms a central β-sandwich that binds to Mn2+ surrounded by α-helices, and the C-terminal region is an antiparallel helix structure attached to the catalytic domain (Das et al. 1996). In Arabidopsis PPP and PPM families comprise 102 of the nearly 150 protein phosphatases, while there are only 31 members (PPP and PPM proteins) out of 148 protein phosphatases in humans (Kerk et al. 2008; Uhrig and Moorhead 2011). In Arabidopsis, 80 PP2C members have been annotated so far (Xue et al. 2008; Singh et al. 2015). Identification of such a large number of PP2C members indicates that they may have diverse functions to perform during the growth and development of a plant.
The PTPs and Asp-based enzymes are both Tyr phosphatases, the difference between them arising from the residue (Cys in case of PTP and Asp in the latter case) that mediates dephosphorylation (Hobiger and Friedrich 2015). PTPs can be divided into three classes (I to III). The Class I comprises classical and dual-specificity phosphatases (DSPs). The classical PTPs show specificity toward Tyr, but the DSPs toward both Ser/Thr and Tyr. The DSPs can also choose non-protein substrates (mRNA, phosphoinositol, myotubularins, inositol-4-phosphatases, and carbohydrate). The Class II phosphatases often work at low pH values (≤6) and hence are also called acid phosphatases. These are smaller than Class I substrates (and hence also termed as low molecular weight phosphatases) and are also Tyr specific. The Class III phosphatases are involved in cell division and are also known as cell division control (CDC) phosphatases. Some of the members of the CDC can target Thr as well as Tyr (Hobiger and Friedrich 2015). Plants lack typical Tyr kinase and consequently typical classical PTPs. Class II and Class III PTPs are absent in plants (Moorhead et al. 2009; Hobiger and Friedrich 2015). The Asp-based enzymes consist of the FCP/SCP, eyes absent phosphatases and haloacid dehalogenase (HAD) family enzymes (Moorhead et al. 2009; Shankar et al. 2015). The PTPs and DSPs share a common catalytic domain of around 250 amino acids. At the N-terminal extremity of the catalytic domain has a conserved Lys residue for ATP binding. The catalytic signature of the PTP family is CX5R and the Asp-based enzyme is DXDXT/V (D is Asp) (Moorhead et al. 2009; Shankar et al. 2015). The 24 members of PTP identified in Arabidopsis are one Tyr-specific PTP, 22 DSPs, and a low molecular weight PTP (probably a homolog of Class II PTPs) (Shankar et al. 2015). Rice genome sequence revealed 23 DSPs, one PTP, and one low molecular weight PTP (Shankar et al. 2015). Till date, no Asp-based phosphatase has been identified in plants. All information on the classification of phosphatases has been summarized in Fig. 11.1.
Classification of protein phosphatases. The classification of protein phosphatases present in animal and plant is presented. The PPP and the PP2C/PPM group are the Ser/Thr phosphatases. Except for PP2B/PP3, all others are present in the plant. The plant additionally has some more phosphatases of this class. The PTP- and ASP-based enzymes are the Tyr phosphatases. Till date, more DSPs have been identified in the plant, which serves as the plant PTPs
11.3 The CBL-CIPK Family, Plants Modified Version of PP2B Family Phosphatases
As mentioned in the previous paragraphs, the PP2B family members are absent in plants. But animals have calcineurin, which consists of two subunits, i.e., CNA (PP2B phosphatase) and CNB (Ca2+ sensor), which is one of the important signaling molecules involved in Ca2+-mediated signal transduction in animal and fungi. Search for a similar molecule in plant led to the identification of calcineurin B-like proteins (CBLs), a reminiscent of CNB (Luan 2009). CBL interacts and targets a specific group of kinases called CBL-interacting protein kinases (CIPKs) and mediates the Ca2+ signaling in plants (Sanyal et al. 2015). Studies on the CBL-CIPK module have led to the identification of multiple CBLs and CIPKs in diverse plant systems. A total of ten CBLs and CIPKs (26 and 33) have been identified in Arabidopsis and rice, respectively (Kolukisaoglu et al. 2004; Kanwar et al. 2014). Over the years, the CBL-CIPK module has been identified in several plant species (Pandey et al. 2014). But lower plants like Selaginella and charophytes have lesser number of CBLs and CIPKs (Weinl and Kudla 2009; Kleist et al. 2014).
The CBL proteins contain calcium-binding four EF-hand (helix-loop-helix) domains (Sanyal et al. 2015). The N-terminal sequence of CBL protein contains the typical signal sequence that leads to the subcellular localization of a CBL and its interacting CIPK. Majorly, the myristoylation and acetylation sites (type I), the tonoplast targeting sequence (type II), and the transmembrane helix (type III) are present in the CBLs (Kleist et al. 2014). There is also a C-terminal phosphorylation motif present, phosphorylated by an interacting CIPK to enhance the activity of the module (Sanyal et al. 2016). On the other hand, CIPKs contain an N-terminal catalytic and C-terminal regulatory domains [having the NAF (CBL binding) and PPI (protein phosphatase binding domain)] (Sanyal et al. 2015). Similar to the SNF1 protein kinase, the CIPKs are Ser/Thr kinases with ATP binding site and a catalytic domain with an activation loop (Sanyal et al. 2015). CBLs are the Ca2+ sensors that bind to Ca2+ and change their conformation thus allowing CIPKs to bind to them through NAF domain. This binding relieves the autoinhibition of CIPK making the kinase active and ready to phosphorylate the targets (Sanyal et al. 2015). It has also been reported that CBL can interact with other proteins (than CIPK) indicating that the sensor module might also act independently (Sanyal et al. 2015).
11.4 Phosphatases and Signal Transduction
In eukaryotes, PP regulates all signaling pathways at different levels such as regulation of gene expression and phosphoregulation of a substrate(s). Here, we discuss the role of phosphatases in some well-known signal transduction pathways.
11.4.1 Phosphatases and Abscisic Acid (ABA) Signaling
PPs have an elaborate role in ABA signaling. ABA is a key plant hormone involved in abiotic stress, signal transduction, as well as during development (Zhu 2016). Accumulation of ABA under varied environmental stress conditions is common. It’s signaling modulates many responses to the stress conditions. ABA regulates several of the downstream genes associated with stress (Verslues and Zhu 2005). Five components such as ABA receptors, positive regulators, negative regulators, ABA-responsive genes, and ABA-responsive transcription factors are associated with its signaling (Yang et al. 2017). Members of ABA receptors like pyrabactin resistance 1 (PYR)/PYR1-like (PYLs)/regulatory components of ABA receptors (RCAR), type 2C protein phosphatases (negative regulators), SnRK2 protein kinases (positive regulators), transcription factors, and ion channels play pivotal roles in ABA signaling (Wang et al. 2013). Dephosphorylation by PP2Cs and phosphorylation by SnRK2s are essential components in ABA signaling events (Fujii et al. 2009). The interaction of PYL and ABA is vital for the PP2C activity and downstream components like sucrose non-fermenting 1 related kinase group 2 (SnRK2s) to be phosphorylated (Fujii et al. 2009). Under normal condition, PP2Cs bind to SnRKs and keep them dephosphorylated. Stress-induced ABA binds to PYR/PYL; it interacts with PP2Cs repressing the phosphatase activity. Thus, the inhibition of SnRK2s is relieved by the above complex components (Fujii et al. 2009). SnRK2s are then autophosphorylated, and activated SnRKs phosphorylate the downstream targets like transcription factors and other targets (Wang et al. 2013). Such phosphorylations of SnRK2s bring about ABA-associated responses including stomatal closure (Wang et al. 2013). Both protein phosphorylation and redox modification play key roles in stomatal regulations.
Out of nine clade A protein phosphatase 2Cs in A. thaliana, functions are known for six of them [ABA Insensitive1 (ABI1), ABI2, ABA Hypersensitive Germination1 (AHG1), AHG3/AtPP2CA, Hypersensitive to ABA1 (HAB1), and HAB2] as negative regulators of ABA signaling (Sheen 1998; Yoshida et al. 2006; Nishimura et al. 2007; Xue et al. 2008). It has been demonstrated that PP2Cs have overlapping and redundant functions in ABA signaling (Rubio et al. 2009). It has been shown that ABA receptors such as ABI1, ABI2, HAB1, and AHG3 interact with RCAR/PYR/PYL (Singh et al. 2015).
11.4.2 Protein Phosphatases in Defense Signaling and the Regulation of Primary and Secondary Metabolism and the Regulation of Mitogen-Activated Protein Kinases (MAPKs)
It is vital to understand how pathogen infection produces signals that can succor the plants to prepare for combat mechanisms. We also need to find out how plants deploy converging signaling pathways to decipher stimuli generated at the membrane (in response to pathogen attack) and get it translated into varied physiological and biochemical changes to bring about disease resistance in the plants in addition to other stresses. Dephosphorylation of proteins is slowly becoming prominent as an important regulatory mechanism. Activities of both protein kinases and phosphatases appear to be involved in such combat mechanisms. Receptor-like kinases (RLKs) or proteins (RLPs) sense the presence of pathogen-associated molecular patterns (PAMPs) and elicit the downstream response in plants (Gomez-Gomez and Boller 2000; Gust and Felix 2014; Prince et al. 2014). PP2A is a crucial component and performs many functions like the regulation of signaling (receptor and organellar), metabolic pathways, and gene expression during such biotic stresses or plant immunity (Durian et al. 2016). PP2A is a trimeric protein with catalytic subunit C, scaffold subunit A, and regulatory subunit B (Trotta et al. 2011). Subunit B is generally referred to as “specificity unit” since it determines the target specificity of PP2A enzyme (Rahikainen et al. 2016). They are highly conserved both in animal and plant and modulate stress signaling (Rahikainen et al. 2016). It appears that different subunits of PP2A conciliate posttranslational regulation of metabolic enzymes besides signaling events (Durian et al. 2016). It has also been noticed that transcript levels of some of the catalytic subunits of PP2A like PP2A-C5 and PP2A-C2 enhance in response to plant pathogen attack (Durian et al. 2016). Similarly, transcript levels of subunits of PP2A-B (regulatory subunit) like B′ζ, B′ƞ, and B′θ are also upregulated under biotic stress conditions (Durian et al. 2016). This infers that both the catalytic and regulatory subunits are involved in perceiving biotic cues. The information accumulated till date reveals that PP2A is functionally controlled at multiple levels perhaps to safeguard specificity in cellular signaling cascades with different heterotrimeric compositions (Bheri and Pandey 2019a, b).
PP2A also acts as a modulator of cell death (Durian et al. 2016). PP2A is negatively regulated by bacterial type III effectors (AvrEs) (Durian et al. 2016). They bind to PP2A and inhibit molecular pattern-triggered immunity (Degrave et al. 2015). Further, it has been reported that a mutant deficient in B′θ of PP2A exhibits resistance to pathogens (Kataya et al. 2015a). The B′θ subunit of PP2A co-localizes with PP2A-C2, PP2A-C5, and PP2A-A2 subunits in peroxisomes and influences β-oxidation of fatty acids (Kataya et al. 2015b). Overall it is inferred that PP2A is involved in complex and tightly interconnected molecular events in plant biotic stress such as perception, signaling, and response (Durian et al. 2016). The appearance of several primary metabolites like carbohydrates and amino acids is central to the plant immunity (Bolton 2009). In an interesting discovery, it was found that PP2A-B′γ subunit regulates the phosphorylation level of the cytosolic ACONITASE 3 (Konert et al. 2015). Metabolic reprogramming in plants appears partially mediated by PAMP-induced genes involved in the biosynthesis of carbohydrates, amino acids, lignins, flavonols, and phytoalexins (Truman et al. 2007). PP2A is also involved in the control of metabolic responses (Durian et al. 2016). Several proteins like MPK3 and MPK6 are constitutively active and trigger secondary metabolic activities and biosynthesis of camalexin and agmantine derivatives (both are defense-related metabolites) (Lassowskat et al. 2014). PP2A also appears to negatively regulate ethylene biosynthesis under such stress conditions (Durian et al. 2016). Overall PP2A is well connected with primary and secondary metabolic pathways, metabolic adjustments, and cellular signaling cascades indicating that PPs are key central components in plant immunity.
A chloroplast-localized DSP has been discovered which contains an ancient carbohydrate-binding domain and binds the starch (Kerk et al. 2006). The data suggest that DSPs in plant systems could be part of a protein assemblage at the starch granule. Thus, some DSPs bind to starch and modulate starch metabolism through reversible phosphorylations (Shankar et al. 2015). Dual-specificity plant tyrosine-specific phosphatases (PTPases) can modulate the mitogen-activated protein kinases (MAPK) (He et al. 2012). The Arabidopsis dual-specificity PTPase (AtDsPTp1) can inactivate MAPK by dephosphorylation (Gupta et al. 1998).
11.4.3 Role of Auxin and Brassinosteroid and Protein Phosphatases
Both auxin and brassinosteroid are two important phytohormones implicated in plant growth and development. They act synergistically to usher in multiple physiological and biochemical changes like hypocotyl elongation, vascular bundle and root development, and abiotic stress tolerance (Tian et al. 2017). They share the same target genes and regulate multiple processes on multiple levels. Auxin perception and degradation of AUX/IAA proteins and consequent release of auxin response factors (ARFs) in higher plants mediate auxin response (Tian et al. 2017). The PP2A protein is vital for the maintenance of auxin transport streams and hence plant growth and responses to salt stress (Han et al. 2017). Further, PP2A can bind to phosphatidic acid (a product of phospholipase D) to regulate auxin transport (Gao et al. 2013). Decreased PP2A activity enhances the level of phosphorylation of PIN1, PIN2, and PIN4 (Gao et al. 2013). The phosphatases thus play a major role in plant signaling by cross talking with hormone (ABA, brassinosteroid, auxin) and in biotic stress pathway.
11.5 Conditions that Necessitate K+ Uptake Systems (AKT1, HAK5, KUP7) in Plants and K+ Deficiency Sensing
Conditions such as low soil K+ concentrations and salt and drought stresses can necessitate K+ uptake systems in higher plants. It was reported that plant could sense the change of external K+ levels and initiate many physiological responses (Schachtman and Shin 2007). The root epidermal cells and root hairs help the plant systems to absorb soil K+ (Aleman et al. 2011). But, what is vital for its transport is the membrane potential of plant cells that depend mostly on the level of externally available K+ (Cheeseman and Hanson 1979; Maathuis and Sanders 1994). It appears that plant cells behave similarly to K+-specific electrodes with respect to external K+ concentrations (Wang and Wu 2010). Hyperpolarization of root cell membrane potential is the first symptom that appears due to K+ deficiency in plants (Maathuis and Sanders 1993), but depolarization occurs if the external K+ concentration increases (Spalding et al. 1999). Uptake of K+ is coupled to H+ extrusion (Behl and Raschke 1987). The ATPases also help in K+ uptake from the soil by establishing an electrical gradient and/or proton motive force (Dreyer and Uozumi 2011). Importantly, the activities of these channels are increased by protons (Fuchs et al. 2005). Thus, low external K+-induced membrane hyperpolarization, coupled with extracellular acidification, is responsible to absorb K+ under conditions of K+ deprivation/salt stress (Wang and Wu 2010). The K+ channels actually responsible for K+ uptake from soil are HAK5 (at external K+ concentrations below 0.01 mM), HAK5 and AKT1 (at external K+ concentrations between 0.01 mM and 0.05 mM), and AKT1 and other unknown low-affinity K+ uptake systems (at further higher K+ concentration) (Sharma et al. 2013). Thus, the uptake of K+ from the soil is carried out by transport proteins in an organized and well-coordinated fashion (Aleman et al. 2011). In addition to the AKT1 channel and HAK5 transporter proteins, KUP7 (a member of the HAK family) helps in K+ uptake in the roots (Han et al. 2016). KUP7 mediates K+ transport into the xylem as well as into shoots (Han et al. 2016). AKT1, HAK5, and KUP7 are localized to the plasma membrane of the Arabidopsis roots (Lagarde et al. 1996; Qi et al. 2008; Han et al. 2016). Once K+ is absorbed by peripheral root cells, it is transported into root stelar tissues and then to shoots by gated outwardly rectifying K+ channel (GORK) and stelar K+ outward rectifier (SKOR) (Han et al. 2016). While GORK regulates K+ efflux from roots (Ivashikina et al. 2001), SKOR is expressed in stelar tissues and releases K+ into xylem sap (Gaymard et al. 1998).
AKT1 is a member of the plant shaker family of voltage-gated K+ channels. In Arabidopsis, the family has nine members divided into four subfamilies depending on their response to the membrane voltage (Lebaudy et al. 2007; Dreyer and Blatt 2009). Five members (including AKT1, AKT5, AKT6/SPIK, KAT1, and KAT2) get activated upon membrane hyperpolarization and are closed when the driving force for K+ is outwardly directed (and elicit only inward K+ currents (Kin)) (Lebaudy et al. 2007). GORK and SKOR get activated upon membrane depolarization and function opposite to Kin (i.e., they are closed when the driving force for K+ is inwardly directed and elicit only outward K+ currents (Kout)) (Lebaudy et al. 2007; Sharma et al. 2013). Both of them regulate the release of K+ (leakage pathways) from the cells (Demidchik et al. 2014). It was demonstrated that SKOR-encoded channels catalyze the K+ leakage from stelar cells of roots into the xylem (Gaymard et al. 1998). They are activated at voltages more positive than resting potential (Demidchik et al. 2014). The GORK-encoded channels are expressed in the root atrichoblasts and trichoblasts (epidermal cells) and guard cells (Demidchik et al. 2014). GORK has the functions of ABA and ethylene-controlled K+ efflux (Demidchik et al. 2014). Once K+ is released through GORK, stomata are closed, which is regulated by Ca2+ and phytohormones (Demidchik et al. 2014). AKT2 exhibits weak voltage dependence and can mediate both, K+ efflux and K+ influx (Kweak) (Dreyer and Uozumi 2011; Sharma et al. 2013). The KC1 (aka KAT3) can modify the property of some Kin channels and is classified in Ksilent category (Dreyer and Uozumi 2011). However, Kout channels are not regulated by the KAT3 protein (Ragel et al. 2019). But the interaction of KAT3 with Kin channels is important for negative regulation of the Kin channels (Ragel et al. 2019). Also, heteromerization of different subunits of Kin channels plays a crucial role in the functional diversity (Ragel et al. 2019). The initial identification of the HAK family of transporters in different systems by different groups led to the HAK, KUP, or KT nomenclature, but presently they are recognized as one group KT/HAK/KUP (Grabov 2007). The phylogenetic tree prepared for the plant KT/HAK/KUP transporters revealed that it contains four clades (Rubio et al. 2009). All plants have transporters homologous to members of clades I or clades II (Grabov 2007). Clade III members are majorly found in Arabidopsis and rice (Grabov 2007). The smallest is cluster IV, comprising only four rice genes (Grabov 2007). Recent work has increased this classification to six clusters, with the sixth cluster only present in bryophytes (Nieves-Cordones et al. 2016; Santa-Maria et al. 2018). The current information states that the family has members in Arabidopsis, peach, grapevine, Medicago, Cassava, rice, maize, Brachypodium, and Panicum (Ragel et al. 2019). Important members from Arabidopsis fall in clade Ia (HAK5), clade IIa (KUP3, KUP4/TRH1), clade IIb (KUP1), clade IIc (KUP2, KUP6, KUP8), clade III (KUP9, KUP10, KUP11, KUP12), and clade V (KUP7) (Nieves-Cordones et al. 2016; Ragel et al. 2019). In general terms, transporters involved in root high-affinity K+ uptake fall into clade Ia, and transporters associated with developmental processes, especially those which demand turgor-driven cell expansion, fall into clade II (in Arabidopsis only) (Nieves-Cordones et al. 2016). However, some members of the clade I show different functions. For example, the CqHAK5-like transporters from quinoa are involved in K+ influx into the cells of the leaf salt bladders to contribute to the osmotic balance of the cytosol (Bohm et al. 2018). The other clade members have not been characterized sufficiently to affix a general function with these clades.
11.5.1 Plant Non-voltage-Gated K+ Channels (TPK /KCO) and KEA K+ Transporters
Other than the voltage-gated class, plants also possess the non-voltage-gated class of K+ transporters. In Arabidopsis, five tandem-pore channels (TPK1–TPK5) (aka KCO) have been reported along with a sixth duplicated (and partially deleted) KCO3 (Dreyer and Uozumi 2011). A functional TPK channel would exist as a dimer and is more relaxed in their cellular localization (they can be found in the tonoplast, or other organelle membranes, and the plasma membrane) (Dreyer and Uozumi 2011). The KEA group of efflux proteins are the final group of K+ transporters in plants having homology to bacterial K+/H+ transporters (Grabov 2007). Arabidopsis has six KEA members in its genome (Tsujii et al. 2019). KEAs belong to the monovalent cation/proton antiporter (CPA) superfamily. The CPA family consists of two subfamilies, CPA1 (Na+-H+ exchangers (NHX) are members) and CPA2 (KEA are members) (Tsujii et al. 2019). The KEAs themselves can be divided into two clades, KEAI (having KEA1-KEA3 as members) and KEAII (having KEA4-KEA6 as members). The KEAI group can be further divided into two groups, KEA-Ia (with KEA1 and KEA2) having a longer N-terminal sequence and KEA-Ib (with KEA3) (Tsujii et al. 2019). KEAI members possess a putative chloroplast transit peptide in their N-terminal regions. KEA1 and KEA2 are localized in the inner envelope of the chloroplast, and that KEA3 is present in the thylakoid membrane (Tsujii et al. 2019). The role of KEA proteins has been implicated in photosynthetic regulation, chloroplast development, and stress tolerance (Tsujii et al. 2019). Using a bacterial system, it has been demonstrated that the Arabidopsis KEAs show K+ transport activity (Tsujii et al. 2019).
11.5.2 K+ Deprivation and Calcium Signaling
Other intracellular events help in the activation of the K+ channels once low K+ is sensed by the plant. The decrease in external K+ concentration elevates cytoplasmic Ca2+ in root cells, which is mediated by hyperpolarization and ROS-activated Ca2+ channels (Shin et al. 2005; Demidchik and Maathuis 2007). These Ca2+ signatures generated in the cytoplasm due to K+ concentration are decoded by members of several Ca2+ sensors such as CDPKs and CBL-CIPK module (Wang and Wu 2017). Further, it has been observed that K+ deficiency causes the accumulation of ROS and activation of low K+-responsive genes like HAK5 (Wang and Wu 2010). So in the following sections, we discuss our current understanding of the K+ sensing mechanism of plants and the role of phosphatases in it.
11.5.3 Regulation of Arabidopsis K+ Transporter 1 (AKT1) K+ Selective Channel
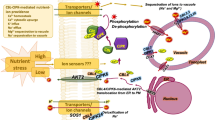
As already mentioned, several of the Shaker-like K+ channels function in K+ nutrition in plants (Very and Sentenac 2003). AKT1 was the first K+ transporter identified in plants (Hirsch et al. 1998). As already mentioned, AKT1 majorly contributes to K+ uptake along with HAK5 in plants. Therefore, AKT1 is one of the main targets of the regulatory network in higher plants. The search for this regulatory network resulted in the identification of the CBL-CIPK module that could modulate AKT1 activity (Li et al. 2006; Xu et al. 2006). CIPK23 phosphorylates AKT1 and increases the uptake of K+ under conditions of low-K+ stress (Li et al. 2006; Xu et al. 2006). On further probing, they identified that CBL1 and CBL9 are necessary to activate AKT1 and also act as the upstream regulators of CIPK23 (Li et al. 2006; Xu et al. 2006). On sensing the Ca2+ signature generated due to low K+, the CBLs bind to Ca2+ and get activated (Behera et al. 2017). They then bind to the CIPK23 and direct its localization to the plasma membrane (where AKT1 is located). Here the module CBL1/CBL9-CIPK23 phosphorylates AKT1 and activates it (Li et al. 2006; Xu et al. 2006). This paradigm that the CBL-CIPK module leads to phosphorylation of AKT1 for its modulation has been conclusively proved in the oocytes of Xenopus laevis (Lee et al. 2007). However, a regulatory network is not complete until the antagonist can be identified that can reverse the action of the agonist. So, this search resulted in the identification of a protein phosphatase2C (AIP1) that binds to the ankyrin domain of AKT1 and keeps it deactivated by dephosphorylating it (Lee et al. 2007) (Fig. 11.2). Further investigation on the CBL-CIPK module and the PP2C for their role in AKT1 regulation has proved that the PP2C can also bind directly to the CIPK (CIPK6) (that is already bound to ankyrin domain of AKT1). The bound PP2C can inhibit AKT1 activation by probably inhibiting the kinase activity of the CIPK6 (Lan et al. 2011). This is supposed to be another mechanism that allows the regulation of CIPK kinase activity (besides the CBL binding and relieving autoinhibition of CIPK) (Lan et al. 2011). Another CBL (one different from the CBL present in the module) can interact with the PP2C and relieves its inhibitory effect on CIPK6 (Lan et al. 2011). The entire mechanism points to very complex control of the AKT1 by the CBL-CIPK module and the PP2Cs (Lan et al. 2011). In the same work, it was also shown that CIPKs could directly interact with PP2Cs belonging to ABA signaling pathway in addition to the ones involved in K+ signaling pathway (Lan et al. 2011). So this clearly shows that PP2Cs and CIPKs form major phosphorylation/dephosphorylation pair controlling important physiological processes (Lan et al. 2011).
The role of phosphatases in kinases in plant K+ uptake. HAK5 transporters are modulated by the CBL1/9-CIPK23 module. The CBL1/9 can take the CIPK23 to the plasma membrane where it phosphorylates and modulates HAK5 for K+ uptake. At further higher K+ concentrations, the AKT1 channel is activated. It is under the control of AIP1 and CBL10 (and others) that are its negative regulators. CIPK23 and KC1 help in modulating AKT1 positively. The CBL4-CIPK6 module helps in targeting of AKT2 channel from endoplasmic reticulum (ER) to plasma membrane. The CBL2/3-CIPK9 module is targeted to vacuole where it can regulate a yet unknown transporter/channel for K+ movement to or from the vacuole. AP2C1 can directly interact with CIPK9 and can directly inhibit its activity, acting as a dephosphorylation switch
The importance of CIPK23 and CBLs in the regulation of AKT1 is unambiguous. As already mentioned, there is another very robust mechanism that plants further employ to regulate the K+ uptake through AKT1. The KC1 inhibits the AKT1 channel and is thought to control any leakage of K+ from AKT1 (Reintanz et al. 2002; Duby et al. 2008). The effects of KC1 on inward and outward currents of AKT1 and also the K+-dependent stability of the pore alteration in AKT1-KC1 heteromers under varying K+ concentrations were reported (Geiger et al. 2009). The heteromerization of AKT1/KC1 makes the channel highly efficient at blocking K+ permeation in the outward direction (Geiger et al. 2009). The CIPK23 (along with CBLs) and KC1 are thought to work synergistically to control plant K+ uptake (Wang et al. 2016). The CBL(1/9)-CIPK23 complex would then activate AKT1 to take up more K+, and KC1 should lock it preventing K+ leakage (Wang et al. 2016). Exploration for other controllers of AKT1 has shown that there are several posttranslational mechanisms for AKT1 control. One interesting report states that CIPK and PP2Cs are not required at all for the regulation of AKT1 (Ren et al. 2013). CBL10 can directly bind to AKT1 and reduces its activity (Ren et al. 2013). This is summarized in Fig. 11.2. This activity is interestingly concentration-dependent, but CIPK-independent (Ren et al. 2013). The CBL(s)-CIPK23-mediated modulation of AKT1 is present in orthologous plant species as well. AKT1-like channels from HvAKT1 (Hordeum vulgare) and VvK1.1 (Vitis vinifera) are activated by CBL1 and CIPK23 (Boscari et al. 2009; Cuellar et al. 2010). Further, voltage-gated inward K+ channel from grapevine was detected and named as VvK1.2 (Cuellar et al. 2013).
11.5.4 Regulation of Arabidopsis K+ Transporter 2 (AKT2) K+ Selective Channels
Interaction of PP2CA with the plasma membrane K+ transporters like Arabidopsis K+ transporter 2 (AKT2) and Slowly Activating Anion Channel 1 (SLAC1) has been reported (Chérel et al. 2002; Lee et al. 2009). The coexpression studies of the AKT2 and AtPP2CA genes were performed in mesophyll cells and observed that AtPP2CA gene expression levels were overlapping with that of AKT2 and were upregulated by ABA in root and shoot tissues (Chérel et al. 2002). Dephosphorylation events mediate this protein-protein interaction because the changes in AKT2 activity are inhibited by vanadate (Chérel et al. 2002). Further, it is also suppressed by a point mutation in the catalytic site of the AtPP2CA protein, resulting in the loss of its phosphatase activity (Chérel et al. 2002). The Ca2+ sensor CBL4 interacts with CIPK6 and mediates the translocation of AKT2 to the plasma membrane in Arabidopsis (Held et al. 2011). The Ca2+ sensor CBL4 interacts with CIPK6 and forms CBL4-CIPK6 module, which enables translocation of AKT2 K+ channel to the plasma membrane and hence regulates its activity in phosphorylation-independent manner (Held et al. 2011). This is described in Fig. 11.2.
11.5.5 Role of Other CBL-CIPK Modules in Ion Homeostasis Pathways
Ca2+ sensors such as CBL2 and CBL3 are located in the vacuolar membrane (Tang et al. 2012). Therefore, they can interact with tonoplast-bound H+-ATPase and modulate intracellular ion homeostasis (Tang et al. 2012). It has been shown that cbl2cbl3 double mutants are sensitive to several metal ions like Ca2+, Cu2+, K+, Zn2+, and Mg2+, but not to Na+ (Tang et al. 2012). Later, it was demonstrated that CBL2 and CBL3 function in the sequestration of vacuolar Mg2+ along with a quartet of CIPKs (CIPK3/9/23/26) (Tang et al. 2015). Another CBL-CIPK module, CBL3, and CIPK9 are involved in the transport of K+ (Liu et al. 2013). CBL2 and CBL3 share 92% similarity at the level of amino acid sequence (Liu et al. 2013). Accordingly, overexpression of CBL2 and CBL3 displayed phenotypes that are sensitive to low K+ conditions (Liu et al. 2013). Therefore, it is inferred that CBL2 and CBL3 are paralogous gene pair that mostly have similar physiological functions. The role of CIPK23 has already been discussed in K+ homeostasis. Besides K+, CIPK23 also plays a role in nitrate and iron homeostasis (Ho et al. 2009; Dubeaux et al. 2018). Two other CBL-CIPK modules involved in Na+ homeostasis are CBL4-CIPK24 (SOS pathway members) and CBL10-CIPK24 (Kim et al. 2007; Zhu 2016). Further, it has been shown that CBL10 functions in the shoots and increases the tonoplast-bound Na+/H+ exchanger (NHX) activity by interacting with CIPK24 protein and brings about ion homeostasis to impart salt stress tolerance (Kim et al. 2007).
11.5.6 The GORK and SKOR Channels in K+ Homeostasis and Other Functions in Plant Cell
It is not only the uptake of K+ that is pivotal, but also its internal distribution equally within the plant and its homeostasis under abiotic stress conditions. The gating of GORK depends on the extracellular concentrations of K+, and GORK can sense the changes in K+ levels and enable the roots to react accordingly (Sharma et al. 2013). The transport of K+ from roots to the leaf is vital and takes place via the xylem tissues (Gaymard et al. 1998). The SKOR channel has been found to express strongly in root tissues like pericycle and the xylem parenchyma (Gaymard et al. 1998). SKOR can be modulated by external K+ concentrations which increases the voltage required by the channel to open to a higher threshold (Johansson et al. 2006). This is achieved by a complex interplay between the pore region of SKOR and last transmembrane domains of the channel responsible for opening and closing (Johansson et al. 2006). At high external K+ concentration, the pore region strongly interacts with the last transmembrane domain of the channel and keeps the channel in a closed state (Johansson et al. 2006). At low external K+ concentration, the pore region has fewer K+, and as a result, the pore region of SKOR does not interact with the transmembrane domain (Johansson et al. 2006). This brings down the channel opening voltages at a lower threshold. Following the molecular rearrangement during low-sK+ state, the transmembrane domain arranges itself in such a way that a K+ outward current is allowed so that the SKOR can now allow K+ to the stellar apoplast for K+ transport within the cell (Johansson et al. 2006; Sharma et al. 2013).
It has been shown that the expression of SKOR is negatively regulated by ABA (Gaymard et al. 1998). Further, acidification (pH) also inhibits the SKOR currents, thus preventing the loss of K+ from roots to shoots (Lacombe et al. 2000). Since reactive oxygen species (ROS) (of which H2O2 is a part) acts as a signal and impacts plant growth and development under abiotic stress conditions (Gapper and Dolan 2006), exogenous application of H2O2 leads in the enhancement of SKOR outward currents (Garcia-Mata et al. 2010). Thus, K+ uptake and its redistribution/partitioning within the root and shoot systems appear to be part of the complex stress response preeminently played by the ROS. Investigations have shown that the regulation of the GORK channel, which mediates K+ efflux, is carried out by PP2CA in A. thaliana (Lefoulon et al. 2016). It has also been recently shown that Ca2+-dependent protein kinase 21 (CPK21) activates the GORK channel (van Kleeff et al. 2018). So like the AKT1-PP2C and CBL-CIPK functioning, we can imagine a similar fine-tuning of GORK activity by the kinase-phosphatase pair.
11.5.7 The Regulation of AtHAK5
Since the availability of soil K+ varies, plants utilize multiple transporters and mechanisms for acquiring it (Maathuis 2009). Members of the family KT/HAK/KUP are ubiquitous, distributed in different subcellular compartments with variable numbers and involved in high-affinity K+ uptake from the soil across membranes and K+ supply, besides K+ translocation, control of water movement at the plant level, and salt and drought tolerance also (Li et al. 2018; Santa-Maria et al. 2018). As already mentioned, members of the clade Ia of KT/HAK/KUP transporters are associated with K+ uptake from the soil (e.g., AtHAK5, OsHAK1, and OsHAK5) (Ragel et al. 2019). The HAK5 (and other HAK-like transporters) is generally upregulated under K+ deprivation and downregulated when K+ is resupplied (Ragel et al. 2019). It is generally accepted that under any stress conditions that affect K+ acquisition (including salinity), high-affinity K+ uptake systems should be transcriptionally (or posttranslationally) activated to maintain the K+ supply and K+/Na+ homeostasis (Ragel et al. 2019). It must also be mentioned that most of clade II to V members of KT/HAK/KUP do not exhibit transcriptional regulation in response to K+ deficiency (Ragel et al. 2019).
As HAK5 is the most investigated protein of the KT/HAK/KUP class, we focus more on it to understand the regulation of this class. Both K+ deficiency and salt stress regulate HAK5 in Arabidopsis and tomato and are associated with hyperpolarization of the plasma membrane of root cells (Nieves-Cordones et al. 2008; Rubio et al. 2014). Roots deprived of K+ induce the expression of ethylene (biosynthesis and signaling) and ROS metabolism genes promoting higher levels of ethylene and the increase in hydrogen peroxide (H2O2) (Ragel et al. 2019). Both, as a result, enhance transcription of HAK5 (Ragel et al. 2019). Salt stress also results in modifications of AtHAK5 expression or the low-K+ response. As only high salt stress induces AtHAK5 expression, it may be inferred that plants may recognize high Na+ levels as K+ under K+ deprivation (Ragel et al. 2019). It appears that the expression of HAK5 gene is also dependent on several other factors like the hormones like auxin, cytokinin, jasmonic acid, ABA (K+ deficiency also induces ethylene biosynthesis), and many transcription factors in plants (Ragel et al. 2019). Taking a cue from the above findings, if we look into the transcription factors (TFs) that have been identified in HAK5, we find that HAK5 is under a strict regulation depending on the availability of external K+. Under normal condition (external K+ sufficient), ARF2 (Auxin Response Factor 2) binds to the auxin-responsive elements (AuxREs) within the AtHAK5 promoter and represses transcription, so that the K+ uptake is only through channels and not through more energy requiring HAK5 (which is a symport system) (Zhao et al. 2016). Under low external K+ condition, ARF2 is phosphorylated by a kinase (yet unknown) and loses DNA binding activity and is removed from the AtHAK5 promoter, which relieves the repression on AtHAK5 transcription (Zhao et al. 2016; Ragel et al. 2019). Now another set of TFs (including RAP2.11, DDF2, JLO, bHLH121, and TFII_A) binds to the promoter of HAK5 to initiate its transcription (Kim et al. 2012; Hong et al. 2013). The RAP2.11 binds to the ethylene-responsive element (ERE) and the GCC-box of the AtHAK5 promoter (Kim et al. 2012). The information on the binding sites of others is still unavailable (Ragel et al. 2019).
Moving from transcriptional regulation of HAK5 to posttranslational regulation, it has been reported that the CBL1/9-CIPK23 module can also regulate the HAK5 protein (through phosphorylation) (Ragel et al. 2015; Scherzer et al. 2015; Bohm et al. 2018). The phosphorylation of the cytosolic N-terminal of HAK5 by CBL-CIPK23 complex is similar to the AKT1 activation by the same module (Li et al. 2006; Xu et al. 2006). The regulation could also be like CBL4-CIPK6 and AKT2, i.e., the CBL-CIPK23 module could traffic HAK5 to the plasma membrane (as under K+ starvation HAK5 enriches in the plasma membrane) (Qi et al. 2008; Ragel et al. 2019). HAK5 can also be regulated by raf-like integrin-linked kinase1 (ILK1) (probably undergoing phosphorylation). The calmodulin-like protein 9 (CML9) and ILK1 are together required for regulation of HAK5 (Brauer et al. 2016). However, it is unknown at this point of time if this happens in coordination with CBL1/9-CIPK23 pathway or independent of it. The direct phosphatase modulator of HAK5 is yet to be identified. ABI2 has been identified as direct interactors of CBL1 and CIPK23 (although during nitrate sensing) (Leran et al. 2015). The same phosphatase may modulate the HAK5 activity by controlling the CBL1 and CIPK23.
11.5.8 K+ Uptake Regulation by a Novel CIPK and PP2C Pair
Pandey and colleagues first identified CIPK9, a calcium sensor-interacting protein kinase which is required for low K+ tolerance in A. thaliana (Pandey et al. 2007). Its transcript levels both in roots and shoots are upregulated by K+ deprivation conditions (Pandey et al. 2007). Based on the T-DNA inserted loss-of-function mutant analysis, they have suggested the functional role of CIPK9 in K+ utilization or sensing mechanism (Pandey et al. 2007). Another Ser/Thr phosphatase type 2C1 (AP2C1), a stress signal regulator in A. thaliana, negatively regulates both MAPK4 and MAPK6 (Schweighofer et al. 2007). It also modulates innate immunity, jasmonic acid, and ethylene levels in A. thaliana. The authors further showed that ap2c1 mutant plants produce higher amounts of jasmonate upon wounding. Their experiments revealed an important role for AP2C1 phosphatase in moderating defense responses and activities of MAPK. While searching for up- and downstream components of CIPK9-mediated K+ deficiency signaling, AP2C1 was identified as the interactor of CIPK9 in a yeast two-hybrid screen (Singh et al. 2018). AP2C1 physically interacts with CIPK9 in vitro and in planta (Singh et al. 2018). Just like CIPK23, CIPK9 is known to be associated with the modulation of K+ signaling in Arabidopsis (Pandey et al. 2007). As mentioned above, AP2C1 was recognized earlier as a MAPK phosphatase, but the discovery that it interacts with CIPK9 was a new finding. AP2C1 was characterized as a negative regulator of K+ signaling under low K+ availability (Singh et al. 2018). Genetic analysis of null mutants of AP2C1 and CIPK9 and AP2C1-overexpressing transgenic A. thaliana lines revealed that they indeed modulate K+ deprivation conditions (Singh et al. 2018). Further, AP2C1 has KIM domain (a conserved structural feature) necessary for interaction with different kinases including CIPK9 (Singh et al. 2018). Though CIPK proteins are known to be cytoplasmic, their cellular location and action depend on the interaction with CBLs (Batistic et al. 2010). CIPK9 also interacts with a Ca2+ pump, ACA8, in Arabidopsis and brings about changes in cytosolic Ca2+ levels (Costa et al. 2017). Although the target/substrate of CIPK9 has not been identified, authors speculate that CIPK9 might be regulating some of the transporters/channels present on the tonoplast under K+ deficiency condition (Singh et al. 2018). Authors have shown that CIPK9 and AP2C1 act as an important phosphorylation-dephosphorylation switch where CIPK9 might act as a positive regulator while AP2C1 might be acting as a negative regulator of K+ deficiency response (explained in Fig. 11.2). The phosphatases thus play a very crucial and critical role in plant K+ homeostasis.
11.6 Ser/Thr Protein Phosphatases in Stress Adaptation
Protein phosphatase 2Cs are the negative modulators of protein kinase pathways and are associated with many environmental stress responses (Xue et al. 2008). Since diverse gene family members are present, the tissue-specific expressions under diverse environmental stresses are critical for the functional understanding of the genes. So in the following section, we look into the role of protein phosphatases in abiotic stress.
11.6.1 Protein Phosphatase Expression Profile Under Stress Conditions
Several PP2C members in the subfamily A displayed their expressions under stress conditions and also during ABA responses (Yang et al. 2018). Expression analysis in Arabidopsis, rice, maize, and tomato has revealed that group-A PP2C genes are highly inducible in response to different abiotic stresses (Singh et al. 2015). The PP2C genes, both from Arabidopsis and rice, exhibit specific as well as an overlapping expression during drought and high salinity (Singh et al. 2015). One hundred and thirty-two PP2Cs were identified in the rice genome (Singh et al. 2010). Of the 132 identified, 128 genes were differentially expressed under environmental stresses such as salinity, cold, and drought and 11 under reproductive developmental stages (Singh et al. 2010). The catalytic subunit genes of OsPp2A-1-5 are upregulated in leaves under salt stress conditions (Yu et al. 2003, 2005). The transgenic tobacco plants overexpressing TaPP2Ac-1 (a catalytic subunit) could tolerate salt and drought stress conditions (Xu et al. 2007). In potato, during salinity stress, high transcript levels were recorded for StPp2Ac1, StPP2Ac2b, and StPP2Ac3 (Pais et al. 2009). In Medicago truncatula, the expression profiles revealed differential expression patterns under cold, drought, and ABA stress conditions (Yang et al. 2018). In wheat, the TaPP2C genes regulate developmental processes as well as stress responses (Yu et al. 2019). Taken together, it appears that PP2Cs are implicated in regulating both stresses and in developmental conditions in plants.
11.6.2 Phosphatases Are Involved in Modulating Kinases During Salt Stress
As already mentioned some of the clade A PP2C members are involved in ABA signaling. The remaining three members of Highly ABA-Induced1 (HAI1), AKT1-Interacting PP2C1/HAI2, and HAI3 (collectively HAI) exhibited a more ABA-independent role in plants (Bhaskara et al. 2012). It was speculated that this HAI group may cross talk with the ABA-dependent and ABA-independent pathway during abiotic stress (Bhaskara et al. 2012). Clade A PP2Cs (ABI1 and PP2CA) inhibit ABA-activated SnRK2s during salt stress (Krzywinska et al. 2016; Krzywińska et al. 2016). Thus, PP2CA along with ABI1 inhibits the activity of SnRK2.4 and regulates root growth and salt stress tolerance (Krzywinska et al. 2016; Krzywińska et al. 2016). Further, they showed that salt-induced SnRK2.4/SnRK2.10 activity is better in the double-mutant abi1-2 pp2ca-1 in comparison with controls and also single-mutant plants (abi1 or pp2ca) (Krzywinska et al. 2016; Krzywińska et al. 2016). This points out that these phosphatases are inhibitors of SnRK2.4 activity in plants under stress conditions. Another member of PP2C-type protein phosphatase ABI2 was identified as a CIPK24 [aka SOS2 (salt overly sensitive)]-interacting phosphatase (Ohta et al. 2003). ABI2 plays a critical role in dephosphorylating CIPK24. The abi2 mutant plants are salt-tolerant unlike the sos2 mutant (and other SOS pathway mutants). The results inferred that ABI2 is a negative regulator of CIPK24 in SOS pathway (Ohta et al. 2003).
11.6.3 Phosphatases in Regulating Guard Cell: The Best-Characterized ABA Signaling Pathway
In Arabidopsis, ABI1 and ABI2 encode PP2Cs that negatively regulate ABA signaling including stomatal closure. In guard cells, open stomata 1 (OST1/SRK2E) has been identified as an important component of ABA signal transduction, and mutations in the gene SRK2E/OST1/SnRK2.6 impair stomatal closure (Yoshida et al. 2002). The ost1 mutants have ABA-insensitive stomata (Mustilli et al. 2002). It is known that ABI1 and ABI2 encode PP2C-type protein phosphatases and negatively regulate ABA signaling events including stomatal closure (Mustilli et al. 2002). It was demonstrated that ABI1 interacted with OST1 and plays a critical role in the stomatal opening (Yoshida et al. 2006).
The classical ABA signaling is followed in this case. The absence of ABA keeps the PP2C bound to SnRK2 and inactivated (Umezawa et al. 2010). Upon exposure of plants to drought stress, ABA accumulates leading to the closure of stomata. In response to dehydration stress, ABA binds to PYR/PYL receptor and forms a complex with PP2Cs (ABI1 and others) (Umezawa et al. 2010). As soon as there is ABA, PP2C releases OST1 (an SnRK), and the activated OST1 can phosphorylate channels [slow anion channel1 (SLAC1), quickly activating anion channel 1 (QUAC1), GORK] and respiratory burst oxidase homolog F (RBOH F) (Balmant et al. 2016). The channels mediate anion release from stomata causing a depolarization of the membrane of stomata. The depolarization (of the membrane) and KAT1 phosphorylation block K+ influx into the stomata (Balmant et al. 2016). The phosphorylation of RBOHF produces ROS in the cell, and it functions as second messengers for ABA signaling and activates Ca2+ channels and also OST1 (aka SnRK2E) acts upstream of ROS production (Pei et al. 2000; Mustilli et al. 2002). The ROS in the cell leads to Ca2+ spike in the cell. The Ca2+ spike is picked up by Ca2+ sensors, which further helps in stomatal closure. Thus the PP2C serves as a switch to serve as a control of stomatal physiology (Balmant et al. 2016) (summarized in Fig. 11.3). This model gives us a complete picture of the events involved in the modulation of stomata under stress. ABA signaling, K+ transport, ROS, and Ca2+ signaling all play an important role to modulate stomata, and phosphatase (PP2C) stands at the top to control this pathway. By keeping the SnRK2 (OST1) dephosphorylated, phosphatase acts as a key controller of the pathway.
The PP2C phosphatases play a major role in modulating stomatal physiology. Under normal condition, PP2C binds to SnRK (OST1) and keeps the stomata open. Under stress (drought), ABA binds to the receptors and removes PP2C from SnRK. SnRK then through a series of phosphorylation events activates the RBOHF, SLAC1, and QUAC1. Activation of RBOHF produces ROS, which in turn activates Ca2+ channels, thus resulting in activation of CDPKs. CDPKs can further activate SLAC1 through phosphorylation. The anions being pushed out of stomata by SLAC1 and QUAC1 result in depolarization of membrane and activation of GORK which pushes out K+ from stomata causing the stomatal closure
11.7 Conclusions
The different classes of plant phosphatases are yet to be classified fully. The future research should focus on characterizing the neglected classes more so that our understanding of the plant phosphatases is increased holistically. What we do know for sure is that phosphatases play a key role in fine-tuning the physiological pathways at the molecular level. There are innumerable examples that are testimony to this fact. The role of phosphatases in the core ABA signaling pathway is well established. Several of the protein phosphatases like PP2Cs are Ser/Thr phosphatases working as vital components of ABA signal transduction. This is the best-characterized pathway for plant phosphatases to date. We have discussed in the chapter some major K+ signaling pathways where the role of phosphatases has been proven, and the AKT1 pathway is an example of this. In all probability, the GORK channel may be modulated by the CPK21 and PP2CA pair. Some of the other important K+ transport elements still need their phosphatase pair to be identified. We know that CBL1/9-CIPK23 module can regulate HAK5, but the phosphatase to counter the module’s action in vivo is still unknown. The new kinase and phosphatase pair of CIPK9 and AP2C1 awaits the identification of their target transport element. Taking into account that both are involved in plant K+ response, it is possible that the target will be involved in K+ transport. We need to think where the phosphatase binds to when regulating a target. We know from the AIP and AKT1 model that the phosphatase binds to ankyrin domain of AKT1. But there is an alternate complex model proposed by Lan and colleagues (Lan et al. 2011) as discussed earlier in the chapter which should be kept into consideration for future explanations.
Besides, we have clues that phosphatases are involved in modulating development, ion accumulation, biotic stress pathway, and others. For the SOS pathway, we have the kinase and phosphatase pair identified. But overall very few actual pairs (kinase and phosphatase) are known. We are certain that abiotic stress stimuli perturb the expression of phosphatase genes. So in the next step, high-throughput interactome studies should be designed that enable us to find out the physical targets of phosphatases.
References
Aleman F, Nieves-Cordones M, Martinez V, Rubio F (2011) Root K(+) acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol 52:1603–1612
Andreeva AV, Kutuzov MA (2004) Widespread presence of “bacterial-like” PPP phosphatases in eukaryotes. BMC Evol Biol 4:47
Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8:509
Balmant KM, Zhang T, Chen S (2016) Protein phosphorylation and redox modification in stomatal guard cells. Front Physiol 7:26
Batistic O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61:211–222
Behera S, Long Y, Schmitz-Thom I, Wang XP, Zhang C, Li H, Steinhorst L, Manishankar P, Ren XL, Offenborn JN, Wu WH, Kudla J, Wang Y (2017) Two spatially and temporally distinct Ca(2+) signals convey Arabidopsis thaliana responses to K(+) deficiency. New Phytol 213:739–750
Behl R, Raschke K (1987) Close coupling between extrusion of H(+) and uptake of K (+) by barley roots. Planta 172:531–538
Bhaskara GB, Nguyen TT, Verslues PE (2012) Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol 160:379–395
Bheri M, Pandey GK (2019a) PP2A phosphatases take a giant leap in the post-genomics era. Curr Genomics 20:154–171
Bheri M, Pandey GK (2019b) Protein phosphatases meet reactive oxygen species in plant signaling networks. Env Exp Bot 161:26–40
Bohm J, Messerer M, Muller HM, Scholz-Starke J, Gradogna A, Scherzer S, Maierhofer T, Bazihizina N, Zhang H, Stigloher C, Ache P, Al-Rasheid KAS, Mayer KFX, Shabala S, Carpaneto A, Haberer G, Zhu JK, Hedrich R (2018) Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Curr Biol 28:3075–3085.e3077
Bolton MD (2009) Primary metabolism and plant defense—fuel for the fire. Mol Plant-Microbe Interact 22:487–497
Boscari A, Clement M, Volkov V, Golldack D, Hybiak J, Miller AJ, Amtmann A, Fricke W (2009) Potassium channels in barley: cloning, functional characterization and expression analyses in relation to leaf growth and development. Plant Cell Environ 32:1761–1777
Brauer EK, Ahsan N, Dale R, Kato N, Coluccio AE, Pineros MA, Kochian LV, Thelen JJ, Popescu SC (2016) The Raf-like kinase ILK1 and the high affinity K+ transporter HAK5 are required for innate immunity and abiotic stress response. Plant Physiol 171:1470–1484
Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262:539–544
Cheeseman JM, Hanson JB (1979) Energy-linked potassium influx as related to cell potential in corn roots. Plant Physiol 64:842–845
Cheng X, Liu X, Mao W, Zhang X, Chen S, Zhan K, Bi H, Xu H (2018) Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L.). Int J Mol Sci 19:3969
Chérel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB (2002) Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14:1133–1146
Costa A, Luoni L, Marrano CA, Hashimoto K, Koster P, Giacometti S, De Michelis MI, Kudla J, Bonza MC (2017) Ca2+-dependent phosphoregulation of the plasma membrane Ca2+-ATPase ACA8 modulates stimulus-induced calcium signatures. J Exp Bot 68:3215–3230
Cuellar T, Azeem F, Andrianteranagna M, Pascaud F, Verdeil JL, Sentenac H, Zimmermann S, Gaillard I (2013) Potassium transport in developing fleshy fruits: the grapevine inward K+ channel VvK1.2 is activated by CIPK-CBL complexes and induced in ripening berry flesh cells. Plant J 73:1006–1018
Cuellar T, Pascaud F, Verdeil JL, Torregrosa L, Adam-Blondon AF, Thibaud JB, Sentenac H, Gaillard I (2010) A grapevine shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J 61:58–69
Das AK, Helps NR, Cohen PT, Barford D (1996) Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 15:6798–6809
Degrave A, Siamer S, Boureau T, Barny MA (2015) The AvrE superfamily: ancestral type III effectors involved in suppression of pathogen-associated molecular pattern-triggered immunity. Mol Plant Pathol 16:899–905
Demidchik V, Maathuis FJ (2007) Physiological roles of nonselective cation channels in plants: from salt stress to signaling and development. New Phytol 175:387–404
Demidchik V, Straltsova D, Medvedev SS, Pozhvanov GA, Sokolik A, Yurin V (2014) Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J Exp Bot 65:1259–1270
Dreyer I, Blatt MR (2009) What makes a gate? The ins and outs of Kv-like K+ channels in plants. Trends Plant Sci 14:383–390
Dreyer I, Uozumi N (2011) Potassium channels in plant cells. FEBS J 278:4293–4303
Dubeaux G, Neveu J, Zelazny E, Vert G (2018) Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell 69:953–964.e955
Duby G, Hosy E, Fizames C, Alcon C, Costa A, Sentenac H, Thibaud JB (2008) AtKC1, a conditionally targeted shaker-type subunit, regulates the activity of plant K+ channels. Plant J 53:115–123
Durian G, Rahikainen M, Alegre S, Brosche M, Kangasjarvi S (2016) Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front Plant Sci 7:812
Fuchs I, Stolzle S, Ivashikina N, Hedrich R (2005) Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 221:212–221
Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signaling pathway. Nature 462:660–664
Gao HB, Chu YJ, Xue HW (2013) Phosphatidic acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol Plant 6:1692–1702
Gapper C, Dolan L (2006) Control of plant development by reactive oxygen species. Plant Physiol 141:341–345
Garcia-Mata C, Wang J, Gajdanowicz P, Gonzalez W, Hills A, Donald N, Riedelsberger J, Amtmann A, Dreyer I, Blatt MR (2010) A minimal cysteine motif required to activate the SKOR K+ channel of Arabidopsis by the reactive oxygen species H2O2. J Biol Chem 285:29286–29294
Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferriere N, Thibaud JB, Sentenac H (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94:647–655
Geiger D, Becker D, Vosloh D, Gambale F, Palme K, Rehers M, Anschuetz U, Dreyer I, Kudla J, Hedrich R (2009) Heteromeric AtKC1-AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem 284:21288–21295
Gentry MS, Brewer MK, Vander Kooi CW (2016) Structural biology of glucan phosphatases from humans to plants. Curr Opin Struct Biol 40:62–69
Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011
Grabov A (2007) Plant KT/KUP/HAK potassium transporters: single family—multiple functions. Ann Bot 99:1035–1041
Gupta R, Huang Y, Kieber J, Luan S (1998) Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J 16:581–589
Gust AA, Felix G (2014) Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr Opin Plant Biol 21:104–111
Halford NG (2009) New insights on the effects of heat stress on crops. J Exp Bot 60:4215–4216
Han EH, Petrella DP, Blakeslee JJ (2017) ‘Bending’ models of halotropism: incorporating protein phosphatase 2A, ABCB transporters, and auxin metabolism. J Exp Bot 68:3071–3089
Han M, Wu W, Wu WH, Wang Y (2016) Potassium transporter KUP7 is involved in K(+) acquisition and translocation in Arabidopsis root under K(+)-limited conditions. Mol Plant 9:437–446
He H, Su J, Shu S, Zhang Y, Ao Y, Liu B, Feng D, Wang J, Wang H (2012) Two homologous putative protein tyrosine phosphatases, OsPFA-DSP2 and AtPFA-DSP4, negatively regulate the pathogen response in transgenic plants. PLoS One 7:e34995
Hedrich R (2012) Ion channels in plants. Physiol Rev 92:1777–1811
Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratge-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, Kudla J (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 21:1116–1130
Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280:918–921
Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138:1184–1194
Hobiger K, Friedrich T (2015) Voltage sensitive phosphatases: emerging kinship to protein tyrosine phosphatases from structure-function research. Front Pharmacol 6:20
Hong JP, Takeshi Y, Kondou Y, Schachtman DP, Matsui M, Shin R (2013) Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol 54:1478–1490
Huber SC, Huber JL, McMichael RW (1994) Control of plant enzyme activity by reversible protein phosphorylation. Int Rev Cytol 149:47–98
Ivashikina N, Becker D, Ache P, Meyerhoff O, Felle HH, Hedrich R (2001) K(+) channel profile and electrical properties of Arabidopsis root hairs. FEBS Lett 508:463–469
Johansson I, Wulfetange K, Poree F, Michard E, Gajdanowicz P, Lacombe B, Sentenac H, Thibaud JB, Mueller-Roeber B, Blatt MR, Dreyer I (2006) External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J 46:269–281
Johnson LN, O’Reilly M (1996) Control by phosphorylation. Curr Opin Struct Biol 6:762–769
Johnson LN, Snape P, Martin JL, Acharya KR, Barford D, Oikonomakos NG (1993) Crystallographic binding studies on the allosteric inhibitor glucose-6-phosphate to T state glycogen phosphorylase b. J Mol Biol 232:253–267
Kanwar P, Sanyal SK, Tokas I, Yadav AK, Pandey A, Kapoor S, Pandey GK (2014) Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 56:81–95
Kataya AR, Heidari B, Hagen L, Kommedal R, Slupphaug G, Lillo C (2015b) Protein phosphatase 2A holoenzyme is targeted to peroxisomes by piggybacking and positively affects peroxisomal beta-oxidation. Plant Physiol 167:493–506
Kataya AR, Heidari B, Lillo C (2015a) Protein phosphatase 2A regulatory subunits affecting plant innate immunity, energy metabolism, and flowering time—joint functions among B’eta subfamily members. Plant Signal Behav 10:e1026024
Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB (2006) A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J 46:400–413
Kerk D, Templeton G, Moorhead GB (2008) Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol 146:351–367
Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schultke S, Lee SC, Kudla J, Luan S (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52:473–484
Kim MJ, Ruzicka D, Shin R, Schachtman DP (2012) The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol Plant 5:1042–1057
Kleist TJ, Spencley AL, Luan S (2014) Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front Plant Sci 5:187
Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Konert G, Trotta A, Kouvonen P, Rahikainen M, Durian G, Blokhina O, Fagerstedt K, Muth D, Corthals GL, Kangasjarvi S (2015) Protein phosphatase 2A (PP2A) regulatory subunit B’gamma interacts with cytoplasmic ACONITASE 3 and modulates the abundance of AOX1A and AOX1D in Arabidopsis thaliana. New Phytol 205:1250–1263
Krzywinska E, Bucholc M, Kulik A, Ciesielski A, Lichocka M, Debski J, Ludwikow A, Dadlez M, Rodriguez PL, Dobrowolska G (2016) Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase. BMC Plant Biol 16:136
Krzywińska E, Kulik A, Bucholc M, Fernandez MA, Rodriguez PL, Dobrowolska G (2016) Protein phosphatase type 2C PP2CA together with ABI1 inhibits SnRK2.4 activity and regulates plant responses to salinity. Plant Signal Behav 11:e1253647
Lacombe B, Pilot G, Gaymard F, Sentenac H, Thibaud JB (2000) pH control of the plant outwardly-rectifying potassium channel SKOR. FEBS Lett 466:351–354
Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C (1996) Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J 9:195–203
Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S (2011) Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol Plant 4:527–536
Lassowskat I, Bottcher C, Eschen-Lippold L, Scheel D, Lee J (2014) Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Front Plant Sci 5:554
Lebaudy A, Very AA, Sentenac H (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581:2357–2366
Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci U S A 106:21419–21424
Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci U S A 104:15959–15964
Lefoulon C, Boeglin M, Moreau B, Very AA, Szponarski W, Dauzat M, Michard E, Gaillard I, Cherel I (2016) The Arabidopsis AtPP2CA protein phosphatase inhibits the GORK K+ efflux channel and exerts a dominant suppressive effect on Pphosphomimetic-activating mutations. J Biol Chem 291:6521–6533
Leran S, Edel KH, Pervent M, Hashimoto K, Corratge-Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B (2015) Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8:ra43
Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci U S A 103:12625–12630
Li W, Xu G, Alli A, Yu L (2018) Plant HAK/KUP/KT K(+) transporters: function and regulation. Semin Cell Dev Biol 74:133–141
Liu LL, Ren HM, Chen LQ, Wang Y, Wu WH (2013) A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol 161:266–277
Luan S (1998) Protein phosphatases and signaling cascades in higher plants. Trends Plant Sci 3:271–275
Luan S (2002) Tyrosine phosphorylation in plant cell signaling. Proc Natl Acad Sci U S A 99:11567–11569
Luan S (2003) Protein phosphatases in plants. Annu Rev Plant Biol 54:63–92
Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14:37–42
Maathuis FJ (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12:250–258
Maathuis FJ, Sanders D (1994) Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc Natl Acad Sci U S A 91:9272–9276
Maathuis FJM, Sanders D (1993) Energization of potassium uptake in Arabidopsis thaliana. Planta 191:302–307
MacKintosh C, Coggins J, Cohen P (1991) Plant protein phosphatases. Subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochem J 273(Pt 3):733–738
Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61:443–462
Moorhead GB, De Wever V, Templeton G, Kerk D (2009) Evolution of protein phosphatases in plants and animals. Biochem J 417:401–409
Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18:448–460
Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by Abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14:3089–3099
Nieves-Cordones M, Miller AJ, Aleman F, Martinez V, Rubio F (2008) A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5. Plant Mol Biol 68:521–532
Nieves-Cordones M, Rodenas R, Chavanieu A, Rivero RM, Martinez V, Gaillard I, Rubio F (2016) Uneven HAK/KUP/KT protein diversity among angiosperms: species distribution and perspectives. Front Plant Sci 7:127
Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50:935–949
Nongpiur R, Soni P, Karan R, Singla-Pareek SL, Pareek A (2012) Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal Behav 7:1230–1237
Ohta M, Guo Y, Halfter U, Zhu JK (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci U S A 100:11771–11776
Ota IM, Varshavsky A (1993) A yeast protein similar to bacterial two-component regulators. Science 262:566–569
Pais SM, Gonzalez MA, Tellez-Inon MT, Capiati DA (2009) Characterization of potato (Solanum tuberosum) and tomato (Solanum lycopersicum) protein phosphatases type 2A catalytic subunits and their involvement in stress responses. Planta 230:13–25
Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, Luan S (2007) CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res 17:411–421
Pandey GK, Kanwar P, Pandey A (2014) Global comparative analysis of CBL-CIPK gene families in plants. Springer, New York
Pawson T (1995) Protein modules and signaling networks. Nature 373:573–580
Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature 406:731–734
Prince DC, Drurey C, Zipfel C, Hogenhout SA (2014) The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol 164:2207–2219
Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, Schachtman DP (2008) The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot 59:595–607
Ragel P, Raddatz N, Leidi EO, Quintero FJ, Pardo JM (2019) Regulation of K+ nutrition in plants. Front Plant Sci 10:281
Ragel P, Rodenas R, Garcia-Martin E, Andres Z, Villalta I, Nieves-Cordones M, Rivero RM, Martinez V, Pardo JM, Quintero FJ, Rubio F (2015) The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol 169:2863–2873
Rahikainen M, Pascual J, Alegre S, Durian G, Kangasjärvi S (2016) PP2A phosphatase as a regulator of ROS signaling in plants. Antioxidants (Basel) 5:E8
Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R (2002) AtKC1, a silent Arabidopsis potassium channel alpha -subunit modulates root hair K+ influx. Proc Natl Acad Sci U S A 99:4079–4084
Ren XL, Qi GN, Feng HQ, Zhao S, Zhao SS, Wang Y, Wu WH (2013) Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J 74:258–266
Rubio F, Fon M, Rodenas R, Nieves-Cordones M, Aleman F, Rivero RM, Martinez V (2014) A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiol Plant 152:558–570
Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150:1345–1355
Santa-Maria GE, Oliferuk S, Moriconi JI (2018) KT-HAK-KUP transporters in major terrestrial photosynthetic organisms: a twenty years tale. J Plant Physiol 226:77–90
Sanyal SK, Pandey A, Pandey GK (2015) The CBL-CIPK signaling module in plants: a mechanistic perspective. Physiol Plant 155:89–108
Sanyal SK, Rao S, Mishra LK, Sharma M, Pandey GK (2016) Plant stress responses mediated by CBL-CIPK phosphorylation network. Enzymes 40:31–64
Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58:47–69
Scherzer S, Bohm J, Krol E, Shabala L, Kreuzer I, Larisch C, Bemm F, Al-Rasheid KA, Shabala S, Rennenberg H, Neher E, Hedrich R (2015) Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc Natl Acad Sci U S A 112:7309–7314
Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, Buchala A, Cardinale F, Meskiene I (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19:2213–2224
Shabala L, Zhang J, Pottosin I, Bose J, Zhu M, Fuglsang AT, Velarde-Buendia A, Massart A, Hill CB, Roessner U, Bacic A, Wu H, Azzarello E, Pandolfi C, Zhou M, Poschenrieder C, Mancuso S, Shabala S (2016) Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of barley (Hordeum vulgare) to salinity stress. Plant Physiol 172:2445–2458
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plant 133:651–669
Shankar A, Agrawal N, Sharma M, Pandey A, Girdhar KPM (2015) Role of protein tyrosine phosphatases in plants. Curr Genomics 16:224–236
Sharma T, Dreyer I, Riedelsberger J (2013) The role of K+ channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front Plant Sci 4:224
Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci U S A 95:975–980
Shi Y (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139:468–484
Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46:1350–1357
Silver DM, Kotting O, Moorhead GB (2014) Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci 19:471–478
Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK (2010) Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11:435
Singh A, Pandey A, Srivastava AK, Tran LP, Pandey GK (2015) Plant protein phosphatases 2C: from genomic diversity to functional multiplicity and importance in stress management. Crit Rev Biotechnol 36:1023–1035
Singh A, Yadav AK, Kaur K, Sanyal SK, Jha SK, Fernandes JL, Sharma P, Tokas I, Pandey A, Luan S, Pandey GK (2018) Protein phosphatase 2C, AP2C1 interacts with and negatively regulates the function of CIPK9 under potassium deficient conditions in Arabidopsis. J Exp Bot 69:4003–4015
Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol 113:909–918
Tang RJ, Liu H, Yang Y, Yang L, Gao XS, Garcia VJ, Luan S, Zhang HX (2012) Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res 22:1650–1665
Tang RJ, Zhao FG, Garcia VJ, Kleist TJ, Yang L, Zhang HX, Luan S (2015) Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci U S A 112:3134–3139
Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, Gendron JM, Jonassen EM, Lillo C, DeLong A, Burlingame AL, Sun Y, Wang ZY (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13:124–131
Tian H, Lv B, Ding T, Bai M, Ding Z (2017) Auxin-BR interaction regulates plant growth and development. Front Plant Sci 8:2256
Trotta A, Wrzaczek M, Scharte J, Tikkanen M, Konert G, Rahikainen M, Holmstrom M, Hiltunen HM, Rips S, Sipari N, Mulo P, Weis E, von Schaewen A, Aro EM, Kangasjarvi S (2011) Regulatory subunit B’gamma of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant Physiol 156:1464–1480
Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci U S A 104:1075–1080
Tsujii M, Kera K, Hamamoto S, Kuromori T, Shikanai T, Uozumi N (2019) Evidence for potassium transport activity of Arabidopsis KEA1-KEA6. Sci Rep 9:10040
Uhrig RG, Kerk D, Moorhead GB (2013b) Evolution of bacterial-like phosphoprotein phosphatases in photosynthetic eukaryotes features ancestral mitochondrial or archaeal origin and possible lateral gene transfer. Plant Physiol 163:1829–1843
Uhrig RG, Labandera AM, Moorhead GB (2013a) Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci 18:505–513
Uhrig RG, Moorhead GB (2011) Two ancient bacterial-like PPP family phosphatases from Arabidopsis are highly conserved plant proteins that possess unique properties. Plant Physiol 157:1778–1792
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the Core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51:1821–1839
van Kleeff PJM, Gao J, Mol S, Zwart N, Zhang H, Li KW, de Boer AH (2018) The Arabidopsis GORK K(+)-channel is phosphorylated by calcium-dependent protein kinase 21 (CPK21), which in turn is activated by 14-3-3 proteins. Plant Physiol Biochem 125:219–231
Verslues PE, Zhu JK (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33:375–379
Very AA, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54:575–603
Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ, Zhang H, Tao WA, Zhu JK (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc Natl Acad Sci U S A 110:11205–11210
Wang XP, Chen LM, Liu WX, Shen L, Wang FL, Zhou Y, Zhang Z, Wu WH, Wang Y (2016) AtKC1 and CIPK23 synergistically modulate AKT1-mediated low potassium stress responses in Arabidopsis. Plant Physiol 170:2264–2277
Wang Y, Wu WH (2010) Plant sensing and signaling in response to K+-deficiency. Mol Plant 3:280–287
Wang Y, Wu WH (2017) Regulation of potassium transport and signaling in plants. Curr Opin Plant Biol 39:123–128
Waraich EA, Ahmad R, Halim A, Aziz T (2012) Alleviation of temperature stress by nutrient management in crop plants: a review. J Soil Sci Plant Nutr 12:221–244
Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184:517–528
Xu C, Jing R, Mao X, Jia X, Chang X (2007) A wheat (Triticum aestivum) protein phosphatase 2A catalytic subunit gene provides enhanced drought tolerance in tobacco. Ann Bot 99:439–450
Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125:1347–1360
Xu J, Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20:56–64
Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9:550
Yang Q, Liu K, Niu X, Wang Q, Wan Y, Yang F, Li G, Wang Y, Wang R (2018) Genome-wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula. Sci Rep 8:12841
Yang W, Zhang W, Wang X (2017) Post-translational control of ABA signaling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol J 15:4–14
Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43:1473–1483
Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281:5310–5318
Yu RM, Wong MM, Jack RW, Kong RY (2005) Structure, evolution and expression of a second subfamily of protein phosphatase 2A catalytic subunit genes in the rice plant (Oryza sativa L.). Planta 222:757–768
Yu RM, Zhou Y, Xu ZF, Chye ML, Kong RY (2003) Two genes encoding protein phosphatase 2A catalytic subunits are differentially expressed in rice. Plant Mol Biol 51:295–311
Yu X, Han J, Wang E, Xiao J, Hu R, Yang G, He G (2019) Genome-wide identification and homoeologous expression analysis of PP2C genes in wheat (Triticum aestivum L.). Front Genet 10:561
Zhao S, Zhang ML, Ma TL, Wang Y (2016) Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28:3005–3019
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324
Acknowledgments
P.B.K. is thankful for the CSIR Emeritus Scientist Fellowship through Grant No. 38(1325)/12/EMR-II) from the Council of Scientific and Industrial Research, New Delhi. Research in GKP’s lab is funded by Department of Biotechnology, Science and Engineering Research Board, University Grant Commission (UGC), and Delhi University (R&D grant) India. Financial support from the DBT-RA Program in Biotechnology and Life Sciences is gratefully acknowledged by S.K.S.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Sanyal, S.K. et al. (2020). Role of Protein Phosphatases in Signaling, Potassium Transport, and Abiotic Stress Responses. In: Pandey, G.K. (eds) Protein Phosphatases and Stress Management in Plants. Springer, Cham. https://doi.org/10.1007/978-3-030-48733-1_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-48733-1_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-48732-4
Online ISBN: 978-3-030-48733-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)