Abstract
Cancer is the second leading cause of death globally and is responsible for an estimated 9.6 million deaths in 2018. Approximately 70% of deaths from cancer occur in low and middle-income countries. One defining feature of cancer is the rapid creation of abnormal cells that grow uncontrollably causing tumor. Gliomas are brain tumors that arises from the glial cells in brain and comprise of 80% of all malignant brain tumors. Accurate delineation of tumor cells from healthy tissues is important for precise treatment planning. Because of different forms, shapes, sizes and similarity of the tumor tissues with rest of the brain segmentation of the Glial tumors is challenging. In this study we have proposed fully automatic two step approach for Glioblastoma (GBM) brain tumor segmentation with Cascaded U-Net. Training patches are extracted from 335 cases from Brain Tumor Segmentation (BraTS) Challenge for training and results are validated on 125 patients. The proposed approach is evaluated quantitatively in terms of Dice Similarity Coefficient (DSC) and Hausdorff95 distance.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cancer is a term used for diseases in which abnormal cells divide without control and can invade other tissues. It spreads beyond their usual boundaries, and which can then invade adjoining parts of the body. It can affect almost any part of the body. Lack of awareness, suboptimal medical infrastructure, fewer chances of screening, and low doctor-patient ratio are the prime reasons for the rise in various cancers [3].
According to Central Brain Tumor Registry of the United States (CBTRUS), 86,970 new cases of primary malignant and non-malignant brain tumors are expected to be diagnosed in the United States in 2019 [1]. An estimated 16,830 deaths will be attributed to primary malignant brain tumors in the US in 2018. In developing country India alone, total deaths due to cancer in 2018 are 7,84,821 [2]. More than 50% of the cases in India are diagnosed in stage 3 or 4, which decreases the patient’s chances of survival. Reports say India has highest mortality-to-incidence ratio in the whole world.
Late-stage presentation and inaccessible diagnosis and treatment are common in various types of cancer. In 2017, only 26% of low-income countries reported having pathology services generally available in the public sector. More than 90% of high-income countries reported treatment services are available compared to less than 30% of low-income countries. Only 1 in 5 low- and middle-income countries have the necessary data to drive cancer policy. Annotation of brain tumors in MRI images is time consuming task for the radiologists. It has been observed that there is high inter-rater variation when the same tumor is marked by different radiologists [16]. Thus, we aimed at developing a fully automatic algorithm which accurately segment the Glial brain tumor without any manual intervention. Also, on average, segmentation of the intra-tumor parts for a single patient is completed in 60 seconds. An automatic segmentation algorithm will be useful as a reference and will save the radiologist time to attend more patients. This is of most importance in developing countries with large population.
Top row: sub-tumor parts like Edema, Necrotic and Enhancing tumor. Bottom row: appearance of Sub tumor parts in different MR modalities: FLAIR, T1, T2, T1ce [8]
Gliomas are the most frequent primary brain tumors in adults and account for 70% of adult malignant primary brain tumors. Glioma arises from glial cells and infiltrate the surrounding tissues such as white matter fibre tracts with very rapid growth [16]. Patients with High Grade Gliomas, also known as Glioblastoma tumors have average survival time of one year. Patient undergoes MRI scan for imaging of the brain tumor and treatment planning. Various intra-tumor parts like Enhancing Tumor (ET), Tumor Core (TC)/Necrosis and Edema appears differently in different MR modalities as shown in Fig. 1.
Brain tumor segmentation is a challenging task because of its non-rigid and complex shape, variation in size and position from patient to patient. These challenges make classical segmentation techniques such as thresholding, edge detection, region growing, classification and clustering ineffective at accurate delineation of complex boundaries between tumor and healthy tissues. Brain tumor segmentation methods are broadly classified into four categories as: Threshold based, Region based, Pixel classification based and Model based techniques with pros and cons over each other [4, 6]. Many approaches to brain tumor segmentation have been implemented over decades but there is no winning theory.
Recently, methods based on Deep Convolutional Neural Networks are outperforming over all traditional machine learning methods in various domains like medical image segmentation, image classification, object detection and tracking etc. [7, 18]. The computational power of GPUs has enabled researchers to design deep neural network models with convolutional layers which are computationally expensive in all the domains [5, 14, 15]. Ronneberger et al. [17] proposed U-Net architecture for biomedical image segmentation with limited images. This paper is major breakthrough in the field of medical image segmentation like liver, brain, lung etc. Inspired from the above literature we developed two step approach based on Deep Convolutional Neural Network 2D U-Net model. Various heterogeneous histologic sub-regions like peritumoral edema, enhancing tumor and necrosis were accurately segmented in spite of thin boundaries between intra-tumor parts.
2 Patients and Method
2.1 Database
In this study, we trained model on popular Brain Tumor Segmentation Challenge (BraTS) 2019 dataset [9,10,11,12,13]. BraTS is the popular challenge organised at Medical Image Computing and Computer Assisted Intervention (MICCAI) conference since 2012. Organisers collected dataset from various hospitals all over the Globe with various scanners and acquisition protocols which made the task challengingFootnote 1. The BraTS dataset comprised of 335 training patients out of which 259 are with High Grade Glioma (HGG) and 76 with Low Grade Glioma (LGG). MRI data of each patient was provided with four channels as FLAIR, T1, T2, T1ce with volume size 240 \(\times \) 240 \(\times \) 155. Also, for each case annotations marked by expert radiologists were provided for Whole Tumor (WT), Enhancing Tumor (ET), Tumor Core (TC) and Edema. Validation dataset of 125 patients was provided without ground truths. In addition to this 166 patients data was provided for testing purpose. Evaluation of the proposed method is done by submitting the segmentation results on online evaluation portalFootnote 2.
2.2 Preprocessing
BraTS dataset is already skull-stripped and registered to 1 mm \(\times \) 1 mm \(\times \) 1 mm isotropic resolution. Bias fields were corrected with N4ITK tool [19]. All four MR channel is normalised to zero mean and unit variance.
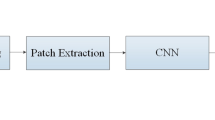
2.3 Patch Extraction
Researchers have proposed several ways of training U-Net for biomedical image segmentation like training with complete multi-modal images and with images patches. In our database there was high class imbalance among the different intra-tumor labels to be segmented along with the non-tumor region. In order to address this, we extracted patches from all the four channels of MRI data. We explicitly augmented patches of underrepresented class by applying various affine transformation like scaling, rotation, translation. Training patches of size 64 \(\times \) 64 were extracted from the FLAIR, T1, T2, T1ce for training.
2.4 Cascaded U-Net
The proposed two step approach is shown in Fig. 2. We have cascaded two U-Net architecture for WT segmentation and intra-tumor segmentation respectively. In step one first U-Net is trained to segment the Whole tumor. Four 64 \(\times \) 64 patches from T1, T2, T1ce, FLAIR are concatenated together and given input along with the corresponding WT mask to the first layer of U-Net as shown in Fig. 3. Second U-Net is trained with WT patch along with corresponding intra-tumor patch as input to give segmentation output for enhancing tumor and tumor core. It should be noted that the area without ET and TC in whole tumor is Edema.
We modified 2D U-Net with large number of feature channels in down-sampling and up-sampling layers as shown in Fig. 3. The architecture consists of a contracting path to capture context and a symmetric expanding path that enables precise localization. At the first layer four 64 \(\times \) 64 multichannel MR volume data was given input for training along with the corresponding ground truth. The number of features maps increases in the subsequent layers to learn the deep tumor features. These are followed by Leaky ReLU activation function and the features were down-sampled in encoding layer. Similarly, in decoding layer after convolution layers and Leaky ReLU activation function, features maps were up-sampled by factor of 2. Features maps from encoding layers were concatenated to corresponding decoding layer in the architecture. At the output layer, the segmentation map predicted by the model was compared with the corresponding ground truth and the error was back propagated in the intermediate U-Net layers. The output layer is a 1 \(\times \) 1 convolution with one filter for the first stage i.e. WT segmentation, and three filters for the second stage i.e. ET, TC and Edema segmentation. The learning rate \((\alpha )\) was initialised to 0.001. After every epoch learning rate was decreased linearly by the factor of \(10^{-1}\) which avoid convergence of the model to local minima. The model was trained for 100 epochs since validation loss did not improved beyond that. Further, for better optimization a momentum strategy was included in the implementation. This used a temporally averaged gradient to damp the optimization velocity.
3 Result and Discussion
The quantitative evaluation of the proposed model was done on BraTS 2019 challenge dataset. No ground truths are provided for validation dataset and predicted results are to be uploaded on online evaluation portal for fair evaluation. The sample result on BraTS challenge dataset is shown in Fig. 4 with Glial tumors. Edema, Enhancing Tumor and Tumor Core segmented by our approach are shown with Green, Blue and Red colours respectively. The performance in terms of dice similarity index is shown in Table 1.
The proposed architecture was implemented using Keras and Tensorflow library which supported the use of GPUs. GPU implementation greatly accelerated the implementation of deep learning algorithms. The approximate time to train the model was 48 hours on 16 GB NVIDIA P100 GPU using cuDNN v5.0 and CUDA 8.0 with 128 GB RAM. The prediction on validation data took less than 60 seconds for a single patient with four MR channels data, each of dimension 240 \(\times \) 240 \(\times \) 155.
There was high class imbalance in the dataset. Around more than 98% pixels belonged to background/healthy class, and rest were labelled as Edema, enhancing tumor and necrotic tumor. Training of the model was challenging because of this class imbalance as there were chances of overfitting to the healthy class. This overfitting of the model to healthy tissue would lead to misclassification of necrotic pixels to healthy pixels. This problem was overcome by augmenting the data with under represented regions. We increased the training data with augmentation techniques like rotation, scaling, shifting etc. This had improved the performance of the model with better class balance. Patches from the boundary region of the tumor were added explicitly for better training of the model. Thus, the segmentation accuracy at the tumor boundaries improved because of additional patches.
4 Conclusion
In conclusion, we presented two step cascaded brain tumor segmentation approach with 2D U-Net architecture based on Deep Convolutional Neural Networks. The encoder-decoder type ConvNet model for pixel-wise segmentation is proposed. We incorporated information from all four MR channels for segmentation which could delineate the tumor boundaries more accurately. We considered different training schems with variable patch sizes, data augmentation methods, activation functions, loss functions and optmizers. This automated approach will definitely provide second opinion to the radiologists for automatic segmentation of brain tumor within minutes. This will enable radiologists for quick reporting and deal with more patients where there is poor patient to doctor ratio.
References
Central brain tumor registry of the united states (2018). http://www.cbtrus.org/factsheet/factsheet.html
Central brain tumor registry of the united states (2018). http://cancerindia.org.in/cancer-statistics/
World health organization fact-sheets (2018). https://www.who.int/news-room/fact-sheets/detail/cancer
Angulakshmi, M., Lakshmi Priya, G.: Automated brain tumour segmentation techniques a review. Int. J. Imaging Syst. Technol. 27(1), 66–77. https://doi.org/10.1002/ima.22211
Baheti, B., Gajre, S., Talbar, S.: Detection of distracted driver using convolutional neural network. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), pp. 1145–11456, June 2018. https://doi.org/10.1109/CVPRW.2018.00150
Baid, U., Talbar, S.: Comparative study of k-means, gaussian mixture model, fuzzy c-means algorithms for brain tumor segmentation. In: International Conference on Communication and Signal Processing 2016 (ICCASP 2016). Atlantis Press (2016)
Baid, U., et al.: Deep learning radiomics algorithm for gliomas (drag) model: a novel approach using 3D UNET based deep convolutional neural network for predicting survival in gliomas. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 369–379. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_33
Baid, U., Talbar, S., Talbar, S.: Brain tumor segmentation based on non negative matrix factorization and fuzzy clustering. In: Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies, BIOIMAGING, BIOSTEC 2017, Porto, Portugal, 21–23 February 2017, vol. 2, pp. 134–139 (2017). https://doi.org/10.5220/0006250701340139
Bakas, S., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 4, 170117 (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Segmentation labels and radiomic features for pre operative scans of the TCGA-GBM collection. Cancer Imaging Arch. 170117 (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Segmentation labels and radiomic features for pre operative scans of the TCGA-LGG collection. Cancer Imaging Arch. 170117, (2017). https://doi.org/10.1038/sdata.2017.117
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. CoRR abs/1811.02629 (2018). http://arxiv.org/abs/1811.02629
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. ArXiv abs/1811.02629 (2018)
Hariharan, B., Arbelaez, P., Girshick, R., Malik, J.: Object instance segmentation and fine-grained localization using hypercolumns. IEEE Trans. Pattern Anal. Mach. Intell. 39(4), 627–639 (2017)
Kamnitsas, K., et al.: Efficient multi-scale 3D CNN with fully connected crf for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/TMI.2014.2377694
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Smistad, E., Falch, T.L., Bozorgi, M., Elster, A.C., Lindseth, F.: Medical image segmentation on GPUs - a comprehensive review. Med. Image Anal. 20(1), 1–18 (2015). https://doi.org/10.1016/j.media.2014.10.012
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010). https://doi.org/10.1109/TMI.2010.2046908
Acknowledgment
This publication is an outcome of the R & D work undertaken project under the Visvesvaraya PhD Scheme funded by Ministry of Electronics & Information Technology, Government of India, being implemented by Digital India Corporation with reference number: PhD-MLA/4(67/2015-16).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Baid, U., Shah, N.A., Talbar, S. (2020). Brain Tumor Segmentation with Cascaded Deep Convolutional Neural Network. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2019. Lecture Notes in Computer Science(), vol 11993. Springer, Cham. https://doi.org/10.1007/978-3-030-46643-5_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-46643-5_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-46642-8
Online ISBN: 978-3-030-46643-5
eBook Packages: Computer ScienceComputer Science (R0)