Abstract
Lung cancer is one of the most common malignancies worldwide. Despite a significant amount of basic and clinical research, mortality rates remain extremely high, especially for patients affected by advanced stage disease. Recently, new molecules playing several roles in the pathogenesis, diagnosis, and, potentially, clinical management of lung cancer are under investigation, including noncoding fragments of the human genome, also known as noncoding RNAs (ncRNAs). NcRNAs are commonly divided into two categories according to their size. The first category includes small ncRNAs, such as the recently discovered miRNAs, siRNAs, and the classical cellular RNAs (ribosomal, transfer, and other RNAs). Noncoding RNAs greater than 200 nucleotides represent a further category that includes long noncoding RNAs (lncRNAs). LncRNAs have numerous biological and pathophysiological effects. Numerous studies have recently investigated their involvement in the oncogenesis and the progression of pulmonary malignancies. In this chapter, we summarize the current knowledge regarding the role of lncRNAs in the pathogenesis, diagnosis, and clinical management of non-small cell lung cancer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Long noncoding RNAs (lncRNAs) are defined as autonomously transcribed noncoding RNAs that are longer than 200 nucleotides and have minimal coding potential. By contrast, noncoding transcripts with less than 200 nucleotides are defined as small noncoding RNAs (sncRNAs), which include micro-RNAs (miRNAs), small interfering RNAs (siRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and other classes of small RNAs. For a long time, the DNA sequence of lncRNAs was considered “junk DNA.” Since the early 1990s, thousands of lncRNAs have been discovered and investigated; however, their exact number and role in human physiology and pathology are poorly understood. Recently, Hon et al. (2017) integrated multiple transcript collections using FANTOM5 cap analysis of gene expression (CAGE) data to generate a comprehensive atlas of 27,919 human lncRNA genes with high-confidence 5′ ends and expression profiles across 1829 samples from key human primary cell types and tissues.
On the basis of their extension and relation with coding genes on the DNA strands, lncRNAs can be broadly classified as genic (overlapping a protein-coding transcript at one or more nucleotides), nested (contained entirely within protein-coding transcripts), and intergenic (not overlapping a protein-coding transcript); other subtypes describe particular conditions (containing, overlapping, multiple relationships, etc.) (Ransohoff et al. 2018). In accordance with their level of activity, lncRNAs act at the transcriptional, posttranscriptional, and epigenetic level. At the transcriptional level, they have several functions including acting as decoys to disrupt the binding of transcriptional factors with promoters of target genes, altering the localization of transcriptional factors in the genome, competing with endogenous RNA, and forming scaffolds with DNA and proteins. At the posttranscriptional level, they modulate directly or indirectly the effects of micro-RNAs (miRNAs) on target genes and regulate the alternative splicing of mRNA. At the epigenetic level, they interact with proteins involved in histone modifications, regulate DNA methylation in promoter regions, and interact with chromatin modification complexes (Wei and Zhou 2016).
Finally, on the basis of their specific molecular functions, lncRNAs act as molecular signal transducers, decoys, guides for ribonucleoprotein complex, scaffolds, and as sponge to sequester miRNAs (Peng et al. 2018). As signaling molecules, they serve as spatiotemporal indicators of gene regulation that reflect the biological effects of transcription factors (TFs) or signaling pathways; as decoys, they sequester TFs and other proteins away from chromatin or into nuclear subdomains; as guides, they recruit RNA-binding proteins to target genes; and as scaffolds, they recruit several proteins to form complexes with specific biological roles.
The reported functions have been described in several physiologic conditions and pathologies, including cancer (Palmieri et al. 2017). In particular, during the last decade, lncRNAs have been studied in the context of lung cancer, in order to better understand their roles in pulmonary carcinogenesis, and their potential application either as new therapeutic targets or as biomarkers for early diagnosis, prognosis, and therapy monitoring. This chapter provides an overview of the state of the art of current research on lncRNAs in the pathophysiology and clinical management of non-small cell lung cancer.
2 Lung Cancer: Current Status
Lung cancer is one of the most common malignancies and the leading cause of cancer-related deaths worldwide. In 2018, the International Agency for Research on Cancer (IARC) observatory estimated approximately 2,100,000 new cases and more than 1,700,000 deaths, a significant increase in comparison with previous estimations (Paliogiannis et al. 2013). The narrow gap between incidence and mortality rates highlights the challenges with early diagnosis and improving survival rates, especially in patients with advanced stage disease. Cancer Research UK reported recently that the 1-year overall survival rate is 32% for lung cancer patients, while the 5-year survival rate is around 10%. These figures suggest that there is a long way, in terms of basic and clinical research, to improve lung cancer survival rates.
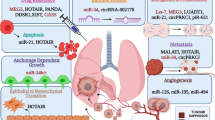
Lung cancer includes a wide range of malignancies, which are broadly divided in small-cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). The former is biologically, pathologically, and clinically different from other subtypes as it is characterized by aggressive behavior, early lymphatic and distant metastasis, and a high responsiveness to chemotherapy (Fig. 1). NSCLC, the focus of this chapter, comprises several further subtypes; the most common are adenocarcinoma (~50%), squamous cell carcinoma (~25%), and large cell carcinomas (~10%). Squamous cell carcinoma was the most common histotype until the 1980s when it was superseded by adenocarcinoma. This might be explained by changes in smoking habits, particularly changes in the characteristics of cigarettes, increased puff volume, increased nitrate levels, and higher smoking incidence in women. Other than their morphological differences (Fig. 1), NSCLC subtypes show consistent differences in biological and clinical behavior, as well as different responses to current therapies, such as the recently introduced targeted agents.
Targeted therapies represent the most important innovation in the treatment of lung cancer over the last few years, also considering that surgical resection is an option in relatively a few cases. In 2004, activating mutations within the kinase domain of the epidermal growth factor receptor (EGFR) gene were discovered in lung adenocarcinomas, suggesting that these tumors were highly sensitive to tyrosine kinase inhibitors (TKIs), a class of agents that selectively inhibits the EGFR molecular pathway (Paliogiannis et al. 2015). TKIs such as gefitinib and erlotinib have been shown to significantly improve the clinical outcomes of approximately 40–60% Asian and 12–16% Caucasian patients with adenocarcinomas harboring EGFR mutations, with survival rates that were nearly twofold when compared to traditional chemotherapy (Shi et al. 2015). However, the increased frequency of resistance to TKIs reduced the initial enthusiasm around these agents, prompting at the same time further research to discover novel molecular targets and molecules with greater therapeutic efficacy and lower resistance rates. For example, additional agents, such as osimertinib, alectinib, and crizotinib, that target genetic alterations of the ALK and ROS1 genes have been introduced in clinical practice; these drugs show a better ability to overcome resistance mechanisms in comparison to older medications. Other molecular targets such as KRAS, BRAF, HER2, MET, and RET, as well as their molecular pathways, are currently being investigated for the development of novel targeted agents (Colombino et al. 2019).
Immunotherapy with immune checkpoint inhibitors (ICIs) has been introduced more recently in NSCLC, further improving survival outcomes in patients with adenocarcinomas a well as in other NSCLC subtypes. Nivolumab, a monoclonal antibody targeting programmed death 1 protein (PD-1), was the first drug approved for advanced NSCLC not responding to platinum-based chemotherapy. Nivolumab showed durable responses in 10 (37%) of 27 confirmed responders with squamous NSCLC and 19 (34%) of 56 with non-squamous NSCLC that had ongoing response after a minimum follow-up of 2 years (Horn et al. 2017). Other agents, pembrolizumab and atezolizumab, are also gradually replacing standard chemotherapy as second- and first-line treatment in pan-negative advanced NSCLC. For ICIs, the expression of PD-L1 is currently considered as a major predictive factor for immunotherapy, despite some limitations in accurately selecting patients who would respond to treatment. Active research is ongoing to overcome such limitations, including the combination of immunotherapy drugs with different mechanisms of action, and their associations with targeted therapies.
3 LncRNAs in the Pathogenesis of NSCLC
A large number of studies reported abnormal patterns of expression of lncRNAs in NSCLC (Table 1). Specifically, these alterations promote molecular pathways that are involved in proliferation and migration, either by influencing second messengers’ activation or triggering the transcription of growth factors. In this setting, lncRNAs are upregulated in neoplastic tissue and, in some studies, also in blood or plasma; their concentration may vary depending on the stage of the disease (presence of metastasis or tumor size). On the other hand, downregulated lncRNAs have also been described in tumor tissue. These lncRNAs promote cellular apoptosis through activation of proapoptotic genes or sequestration of pro-oncogenic molecules. Therefore, similar to protein-coding genes, lncRNAs can be classified as oncogenic or tumor suppressors.
3.1 Main LncRNAs with Oncogenic Functions
3.1.1 MALAT-1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) is one of the first lncRNAs that was associated with lung cancer, especially with lung adenocarcinoma. MALAT-1 is an 8.7 kB intergenic lncRNA (lincRNA) located on chromosome 11q13. Silencing of MALAT-1 in vitro reduces the mobility of lung adenocarcinoma cells. In alveolar basal epithelial cells (A549 cell line) and human NSCLC cells derived from lymph nodes (H1299 cell line), MALAT-1 sponges miR206 promoting cellular invasion and migration (Tang et al. 2018). MALAT-1 upregulation in lung cancer tissue is associated with metastasis progression and poor prognosis, particularly in early stage NSCLC patients with metastasis (Tano et al. 2010). Indeed, in patients with NSCLC and bone or brain metastases, the circulating concentrations of MALAT-1 were found to be higher than in healthy individuals.
MALAT-1 transcription is regulated by p53, which is able to sequester its promoter. Although the activity of MALAT-1 in lung carcinogenesis is not fully understood, two potential mechanisms of action have been described: (a) contribution to an alternative splicing of pre-mRNAs that produces an aberrant expression of genes such as B-MYB transcription factor, and (b) interaction with the demethylated chromobox homolog 4 (CXB4) that controls the relocation of growth-related genes in interchromatin granules. In addition, it has been reported that the CXC motif chemokine ligand 5 (CXCL5), as a downstream gene of MALAT-1, mediated the effects of MALAT-1 on NSCLC migration and invasion (Guo et al. 2015).
3.1.2 HOTAIR
The HOX transcript antisense RNA (HOTAIR), a 2.4 kB antisense lncRNA located in chromosome 12, exhibits altered expression in several human cancers. In NSCLC cancer cells knocked out for HOTAIR in vitro, a decrease in proliferation and metastasis progression has been described. Experiments in fibroblasts showed that HOTAIR acts on homeobox D (a family of transcription factors) genes through epigenetic silencing: these genes are located in chromosome 2, where HOTAIR is driven to by polycomb repressive complex 2 (PCR2), a protein complex able to silence chromatin by methylation of histone 3 on lysine 27 (H3K27). HOTAIR acts as a scaffold binding to PCR2 and to lysine-specific demethylase 1 LSD-1/CoREST/REST complex. The LSD-1/CoREST/REST complex demethylates the lysine 4 of histone 3, resulting globally in chromatin rearrangement (Rinn et al. 2007). Furthermore, it has been demonstrated that HOTAIR promotes the expression of gelatinases and represses the expression of cell-adhesion proteins and metalloproteinases, promoting neoplastic cell motility and metastasis (Zhao et al. 2014); HOTAIR also suppresses p21waf1 (discussed later) and HOXA5, a further protein involved in NSCLC cell migration and invasion. Taken together, these findings clearly support an important role of HOTAIR in NSCLC progression and metastasis. Nakagawa et al. (2013) confirmed that this lncRNA is associated with a shorter disease-free survival in patients affected by NSCLC.
3.1.3 CCAT2
The role of colon cancer-associated transcript 2 (CCAT2), an lncRNA first described in colorectal cancer, in carcinogenesis is not fully understood (Palmieri et al. 2017). CCAT2 is a 1.7 kb intergenic lncRNA located on chromosome 8q24 that is involved in the WNT signaling pathway through an interaction with the transcription factor 7-like 2 (TCF7L2). This results in the upregulation of the expression of MYC and some miRNAs, such as miR-20a, known to regulate cell proliferation. A single nucleotide polymorphism (SNP) of CCAT2, rs6983267, has been associated with CCAT2 overexpression. Zhao et al. (2018) showed that knockdown of CCAT2 in NSCLC cells limited malignant growth and invasion, while artificial overexpression of CCAT2 led to opposite effects. In addition, CCAT2 knockdown significantly decreased the expression of POK erythroid myeloid ontogenic factor (Pokemon) and induced the expression of the p21 tumor suppressor; this suggests that Pokemon overexpression could reverse the decrease of cell viability and cell invasion triggered by CCAT2 silencing. In addition, CCAT2 overexpression has been significantly associated with lung adenocarcinoma but not with squamous cell cancer. Silencing CCAT2 by siRNA has led to inhibition of proliferation and invasion in NSCLC cell lines in vitro (Qiu et al. 2014).
3.1.4 H19
Overexpression of H19, a 2.3 kb lncRNA located on chromosome 11p15 that is expressed only in the maternally inherited chromosome (imprinting phenomenon), is associated with poor prognosis in various cancers. H19 overexpression is associated with hypomethylation of its promoter region; in NSCLC, this characteristic is frequently associated with loss of imprinting. H19 induces cellular proliferation by stimulating the expression of c-Jun and c-Jun N-terminal kinase 1/2 (JNK1/2), and it acts as a sponge by sequestering let-7, a miRNA able to inhibit carcinogenesis by downregulating tumor-promoting proteins (e.g., RAS and MYC) (Kallen et al. 2013). Other pathophysiological mechanisms in lung oncogenesis have been also identified: miR-17/STAT3, miR-484/ROCK2, and miR-196b/LIN28B regulation, SAHH interaction and attenuation, and BPDE-DNA adduct formation. Several studies confirmed that the higher expression of H19 was positively correlated with advanced tumor stage and tumor size, as well as that H19 expression is an independent prognostic factor for overall survival of NSCLC (Chen et al. 2013).
3.1.5 ANRIL
Noncoding RNA in the INK4 locus (ANRIL) is a recently characterized lncRNA that is functionally correlated with the phospholipase D (PLD): the overexpression of ANRIL is associated with inhibition of PLD, with consequent anti-tumorigenic effects, while knockdown of ANRIL suppresses PLD inhibition-induced apoptosis (Kang et al. 2015). ANRIL is a 3.8 bp antisense lncRNA located on chromosome 9p21 which is transcribed from the INK4b-ARF-INK4a gene cluster and has been proven to be upregulated in multiple cancers, such as breast cancer, cervical cancer, nasopharyngeal carcinoma, and thyroid cancer. In tumorigenesis, ANRIL binds to PCR2 and causes a chromatin rearrangement and consecutive silencing of the INK4A-ARF-INK4B gene cluster, which contains tumor-suppressor genes also known as p16, p14, and p15. In addition, ANRIL has been proven to inhibit the expression of P21 and KLF2 and attenuate the transforming growth factor β (TGF-β)/Smad signaling pathway, promoting cancer invasion and metastasis. In recent studies, the expression level of ANRIL was higher in NSCLC tissues and lung cancer cells than in adjacent non-tumor tissues and normal human bronchial epithelial cells (Lu et al. 2016). The higher expression levels of ANRIL in NSCLC were positively correlated with advanced tumor-node-metastasis stage and had negative prognostic implications. Moreover, knockdown of ANRIL expression could inhibit lung cancer cell proliferation, migration, and invasion in vitro. For this reason, ANRIL has been recently investigated as part of promising panels for the diagnosis of NSCLC, which include other lncRNAs and traditional biomarkers such as the carcinoembryonic antigen (CEA) and the cytokeratin 19 fragment (CYFRA 21-1).
3.1.6 LUACT1 (SCAL1)
Lung cancer-associated transcript 1 is known also as smoke- and cancer-related long-chain noncoding RNA 1 (SCAL-1). It is located on chromosome 5 and it is typically tobacco-induced. The transcript includes four exons and three introns. LUACT1 expression is transcriptionally regulated by nuclear factor erythroid 2-related factor (NRF2) and is determined by knockdown through siRNA in NRF2 and kelch-like ECH-associated protein 1 (KEAP1). Induction of LUACT1 has been shown both in vitro and in vivo. It can play the downstream role of NRF2 in regulation of gene expression and intermediate in protection against oxidation stress in epithelial cells of the respiratory system. In a recent study, the lncRNA landscape in lung cancer has been characterized using publicly available transcriptome sequencing data from a cohort of 567 adenocarcinoma and squamous cell carcinoma tumors; functional validation, using both knockdown and overexpression, shows that the most differentially expressed lncRNA was LUACT1 that was sufficient to affect cellular growth independently of other common cancer mutations (White et al. 2014).
3.1.7 SOX2-OT
SOX2 overlapping transcript (SOX2-OT) is an overlapping lncRNA located on chromosome 3 that is highly expressed in embryonic stem cells. Dysregulation of SOX2-OT has been observed in various tumors, including gastric cancer, esophageal cancer, breast cancer, hepatocellular carcinoma, ovarian cancer, pancreatic ductal adenocarcinoma, laryngeal squamous cell carcinoma, cholangiocarcinoma, osteosarcoma, nasopharyngeal carcinoma, glioblastoma, and lung cancer, wherein it typically functions as an oncogene and possibly as a tumor-suppressor gene. Hou et al. (Hou et al. 2014) showed that the expression level of SOX2-OT in 53.01% of human primary lung cancer was twofold higher than that in pair-matched adjacent non-tumor samples. Compared to adenocarcinomas, SOX2-OT expression was significantly higher in squamous cell carcinoma of the lung. Knockdown of SOX2-OT inhibited cell proliferation by decreasing the number of cells in S phase and inducing G2/M arrest. The protein expressions of EZH2 and cyclin B1 and Cdc2 were reduced, and ectopic expression of EZH2 restored the G2/M transition and cyclin B1 and Cdc2 protein expression (Hou et al. 2014). Further studies demonstrated that SOX2-OT expression was obviously higher in NSCLC tissues and serum samples than in normal controls and that SOX2-OT overexpression was associated with poor survival in patients with lung cancer.
3.2 Main LncRNAs with Onco-suppressive Functions
3.2.1 MEG3
Low concentrations of the lncRNA maternally expressed 3 (MEG3) correlate with poor prognosis in NSCLC (Zhou et al. 2012). MEG3 is a 6.9 kb lncRNA located on chromosome 14q32, expressed only in the maternal-inherited chromosome. It is expressed in normal human tissues, especially in brain and the pituitary, and is thought to be a tumor suppressor. Recent studies showed that MEG3 expression is disrupted in various human cancers, such as bladder cancer, glioma, and hepatocellular carcinoma. In lung cancer, MEG3 upregulates p53 expression, inhibiting the exon 3 ubiquitin ligase from preventing p53 transcription. It can also act as a guide for PCR2, bringing it to the regulatory regions of target genes. Interestingly, its overexpression has different effects in in vitro and in vivo experiments: in vitro overexpression induces cell apoptosis, whereas in vivo overexpression inhibits tumorigenesis. A recent report demonstrated that expression of MEG3 in NSCLC cell lines was negatively correlated with miR-205-5p, which enhances cell proliferation and represses apoptosis through targeting low-density lipoprotein (LDL) receptor-related protein-1 (LRP1) (Wang et al. 2017a). Other reports showed that MEG3 expression was decreased in NSCLC tumor tissues compared with normal tissues and associated with advanced pathological stage and tumor size. Moreover, patients with lower levels of MEG3 expression had a relatively poor prognosis.
3.2.2 TUG1
The concentration of the lncRNA taurine-upregulated gene 1 (TUG1) in squamous carcinoma and adenocarcinoma is negatively associated with advanced disease stage and shorter overall survival. TUG1 is a 5.6 kb intergenic lncRNA located on 22q12 chromosome that was originally identified in a genomic screen of taurine-treated mouse retinal cells. TUG1 has been demonstrated to serve crucial regulatory roles in various cancer-associated biological processes. It binds to PRC2 in the promoter region of CELF1 and negatively regulates CELF1 expression. It guides PCR2 into the homeobox B7 region (HOXB7), an oncogene responsible for activating both the PI3K/ERK and MAPK pathways, resulting in an increase in cellular proliferation (Zhang et al. 2014). PCR2 suppresses the expression of HOXB7. TUG1 expression is mediated by wild-type p53; this effect is lost in cases of p53 mutations with R175H missense substitution. TUG1 is found to exhibit aberrant expression in a variety of malignancies. Dysregulation of TUG1 has been shown to contribute to proliferation, migration, cell cycle changes, inhibited apoptosis, and drug resistance of cancer cells which revealed an oncogenic role for this lncRNA, but some reports have shown downregulation of TUG1 in lung cancer samples compared with noncancerous samples. Interestingly, in NSCLC patients, TUG1 downregulation correlated with sex, smoking status, and tumor differentiation grade.
3.2.3 SPRY4-IT1
Sprouty homolog 1 4 intronic transcript 1 (SPRY4-IT1) is derived from an intron of the SPRY4 gene located in chromosome 5q31.3. Its downregulation, due to transcriptional repression mediated by EZH2, a histone methyltransferase able to induce a H3K27 modification, favors cell migration and invasion in vitro. By contrast, its upregulation induces apoptosis and, in mice, reduces metastasis. In a recent study, SPRY4-IT1 expression was observed to be significantly lower, and the expression of EZH2 significantly higher, in lung adenocarcinoma tissues when compared to the adjacent normal tissues (Wen et al. 2018). SPRY4-IT1-suppressed expression in NSCLC is correlated with larger tumor size and lymph node metastasis.
3.2.4 GAS5
Growth arrest specific 5 (GAS5) is an lncRNA located in chromosome 1q25, involved in inducing apoptosis. In recent studies, the expression pattern of GAS5 was investigated in NSCLC specimens and healthy tissues, and its biological functions in the development and progression of NSCLC were assessed. GAS5 expression was downregulated in cancerous tissues compared to adjacent noncancerous tissues and was highly related to tumor size and stage. Furthermore, GAS5 overexpression increased tumor cell growth arrest and induced apoptosis in vitro and in vivo. In addition, siRNA-mediated knockdown of GAS5 promoted tumor cell growth. It has been demonstrated that the ectopic expression of GAS5 significantly upregulates p53 expression and downregulates transcription factor E2F1 expression. In addition, upregulation of GAS5 in NSCLC cells was able to suppress their growth, migration, and invasion via the miR-205/PTEN axis. Therefore, GAS5 is a tumor suppressor in NSCLC which acts through p53-dependent and p53-independent pathways. A recent study showed that GAS5 circulating concentrations are reduced in NSCLC patients but tend to normalize after surgical resection of the tumor (Liang et al. 2016). The authors found that GAS5 expression levels could distinguish NSCLC patients from control patients with 82.2% sensitivity and 72.7% specificity and that the combination of the GAS5 and carcinoembryonic antigen could produce an area of 0.909 (95% confidence interval 0.857–0.962) under the receiver-operating characteristic curve in distinguishing NSCLC patients from control subjects.
3.2.5 BANCR
BRAF-activated noncoding RNA (BANCR) is a 693-bp lncRNA on chromosome 9 that is overexpressed in melanoma cells and crucial for melanoma cell migration. In a recent study, overexpression of BANCR was found to play a key role in epithelial-mesenchymal transition (EMT) through the regulation of E-cadherin, N-cadherin, and vimentin expression (Sun et al. 2014). In this study, BANCR expression was significantly decreased in 113 NSCLC tumor tissues compared with normal tissues. Additionally, reduced BANCR expression was associated with larger tumor size, advanced pathological stage, metastasis distance, and shorter overall survival of NSCLC patients. Finally, reduced BANCR expression was found to be an independent prognostic factor for NSCLC.
3.2.6 TARID
TCF21 antisense RNA inducing demethylation (TARID) has been demonstrated to activate TCF21 expression by inducing promoter demethylation. This occurs because TARID interacts with both the TCF21 promoter and GADD45A (growth arrest and DNA-damage-inducible, alpha), a regulator of DNA demethylation. In a pilot study, TARID was downregulated in NSCLC cells and tissues, but its potential pathophysiological and clinical roles need to be further elucidated (Arab et al. 2014).
3.3 LncRNAs and Endothelial to Mesothelial Transition (EMT)
EMT is described as a reversible phenomenon in which an epithelial cell loses its distinctive characteristics and becomes a mesenchymal cell through the activation of different pathways that culminates in loss of E-cadherin. EMT plays a crucial role in the pathogenesis of NSCLC. It is believed that EMT represents one of the mechanisms of resistance to target therapies in NSCLC patients; moreover, EMT and subsequently mesothelial to endothelial transition (MET) could represent a key process that allows lung cancer cells to metastasize. Numerous lncRNAs have been demonstrated to be involved in EMT, particularly MALAT-1, HOTAIR, and SPRY4-IT1. As discussed earlier, MALAT-1 concentrations in peripheral blood are higher in patients with brain metastases, suggesting a possible role in inducing EMT. In particular, MALAT-1 upregulation increases ZEB1/2 and decreases E-cadherin levels concentrations in these patients.
HOTAIR is able to bind to PCR2, a protein complex needed for H3K27 trimethylation that contains the histone methyltransferase EZH2, which in turn represses E-cadherin gene by H3K27 methylation (Cao et al. 2008). SPRY4-IT1 and BANCR act in similar ways as previously discussed.
4 LncRNAs and Lung Cancer Risk Factors
Altered concentrations of lncRNAs have been described in relation to well-known risk factors for lung cancer, especially cigarette smoking. The most studied is smoking cancer-associated lncRNA 1 (SCAL-1) which is induced by cigarette smoking and is upregulated in NSCLC cell lines (Thai et al. 2013). This lncRNA, located in chromosome 5, is regulated transcriptionally by nuclear factor erythroid 2-related factor (NRF2). As previously discussed, cigarette smoking initially induces upregulation of the active H19 allele. This is likely to progress to loss of imprinting as the burden of smoking increases and as the epithelium undergoes transition from normal to neoplastic. The lncLUACT1, previously described, also correlates with cigarette smoking, while lncRNA DQ786227 and lncRNA LOC728228 are involved in malignant transformation in respiratory cells exposed to benzo(a)pyrene (Gao et al. 2013; Hu et al. 2015). Finally, lncRNA CAR-10 is upregulated in NSCLCs due to a different risk factor for NSCLC—air pollution. This lncRNA is upregulated by a polycyclic aromatic hydrocarbon, dibenz[a,h]anthracene (a pollutant of smoke and oils), that increases the expression of FoxF2. CAR-10 bounds and stabilizes transcription factor Y-box-binding protein 1 (YB-1), leading to upregulation of the EGFR and proliferation of lung cancer cells (Wei et al. 2016).
5 LncRNAs as Diagnostic and Prognostic Biomarkers
5.1 LncRNAs as Diagnostic Biomarkers
The high interest gained by lncRNAs in cancer is also related to their putative role as diagnostic biomarkers for early diagnosis of tumors and/or for differential diagnosis of specific malignancies. This would be particularly relevant in NSCLC in order to improve the currently low survival rates. From this perspective, lncRNAs have some features that make them suitable as potential biomarkers: as previously discussed, the tissue or blood concentrations of some of them change in relation to either the presence of the tumor, its stage, or prognosis; others are tissue-specific, and their concentrations change in particular NSCLC histotypes. Furthermore, they can also be determined in other biological fluids, particularly pleural effusions, frequent in NSCLC patients. The most relevant issue against the implementation of specific lncRNAs detection in clinical practice is represented by their generally low concentrations in biological fluids, which prevents easy determination with standard analytical methods. Concentrations and types of lncRNA detected also vary depending on the biological sample (whole blood, serum, and plasma); to date, there are no NSCLC-associated lncRNA recognized in sputum.
Whole blood concentrations of several lncRNAs are altered in NSCLC patients (Table 1). For the discrimination of NSCLC patients from cancer-free controls, MALAT-1 showed a sensitivity of 56% and a specificity of 96% in cellular fractions of whole blood (Weber et al. 2013). Hu et al. (2015) reported that circulating SPRY4-IT1, ANRIL, and NEAT1 were significantly increased in plasma samples of NSCLC patients. Receiver operating characteristic curve (ROC) analysis revealed that plasma ANRIL provided the highest diagnostic performance with an area under ROC curve value (AUC) of 0.798. Combination of the three factors further increased the diagnostic performance (AUC, 0.876; sensitivity, 82.8%; specificity, 92.3%). Other lncRNAs, particularly NR-026689, XIST, HIF1A-AS1, lncRNA16, UCA1, RP11-397D12.4, AC007403.1, and ERICH1-AS1, have been shown to be increased in NSCLC patients. On the other hand, blood concentrations of onco-suppressive lncRNAs, particularly GAS5, BANCR, and TARID, are generally lower in NSCLC patients.
Currently, none of the molecules described is used in clinical practice due to the technical reasons mentioned above and the need to better establish their predictive capacity (sensibility, specificity, positive and negative predictive values) through adequately designed clinical trials. In some cases, combination of lncRNAs with traditional biomarkers may be effective. Qiu et al. (2014) showed that CCAT2 combined with CEA could predict lymph node metastasis in NSCLC patients. Currently, one clinical trial conducted in China is recruiting patients to evaluate the role of lncRNAs as potential biomarkers for lung cancer diagnosis [NCT03830619].
The association between blood concentrations of some lncRNAs and stage of the disease is also of interest. For example, LINC00313, an intergenic lncRNA, can be detected in serum of patients affected with T2N1-stage lung adenocarcinoma (Li et al. 2015). Another interesting feature of some lncRNAs is their histotype specificity. Even if NSCLC subtypes present specific morphologic, immunohistochemical, and molecular characteristics, the exact diagnosis may be difficult in some cases. Biomarkers detectable in serum specific for NSCLC histotypes would be useful in this context. In a study that compared lncRNAs expressed in lung adenocarcinoma and lung squamous cell carcinoma tissues, LINC01133 was upregulated in lung squamous cell carcinomas, but not in adenocarcinomas (Zhang et al. 2015). Conversely, Qiu et al. (2014) reported that CCAT2 overexpression is significantly associated with lung adenocarcinoma, but not with squamous cell cancer.
5.2 LncRNAs as Prognostic Markers
Most of the lncRNAs described have been found to be related to the prognosis of the disease, despite their obscure role in NSCLC pathogenesis. For example, RP11-21 L23.2, GPR158-AS1, RP11-701P16.5, and RP-11379F4.4 were correlated with poor overall survival, while CTD-2358C21.4, RP11-94 L15.2, KCNK15.AS1, and AC104134.2 were related to better overall survival (Zhou et al. 2015). Also, low expression of some lncRNAs is associated with prognosis: for example, low concentrations of MEG3, GAS-6AS1, and other onco-suppressing molecules correlate with poor overall survival (Han et al. 2013).
6 LncRNAs in NSCLC Therapy
Some lncRNAs have shown to be implied in lung cancer therapy, both for their implications in acquired or non-acquired therapy resistance and for their possible use as therapeutic targets.
6.1 Role of LncRNAs in Resistance to Therapy
6.1.1 Resistance to Chemotherapy
The development of resistance to chemotherapy, for example, cisplatin, is commonly observed in lung cancer. A different expression profile of 1380 lncRNAs was found in vitro in A549 cells and cisplatin-resistant A549/CDDP cells, suggesting that lncRNAs are involved in chemotherapy resistance mechanisms (Yang et al. 2013). Cisplatin acts by inhibiting DNA replication and damaging cell membrane, leading to apoptosis. Resistance may arise because of altered expression of lncRNAs, which can reactivate proliferation pathways and/or repair cisplatin-induced damage. AK126698 targets Wnt, while lncROR targets PI3K/AKT/mTOR, increasing sensitivity to cisplatin-based therapies (Shi et al. 2017). HOTAIR hyper-expression induces cisplatin resistance by downregulating the cyclin-dependent kinase inhibitor 1, a protein associated with cell cycle arrest, and upregulating the expression of stem cell-related biomarkers such as Klf4 (Liu et al. 2016). Furthermore, H19 plays a role in platinum therapy resistance by regulating apoptosis proteins such as BAX, BAK, and FAS (Wang et al. 2017b). Furthermore, patients with downregulation of MEG3 exhibit reduced response to cisplatin therapy, probably because MEG3 is able to modulate the expression of p53, activate the Wnt/β-catenin pathway, and sponge regulatory miRNAs.
Two lncRNAs were found to be associated with altered response to paclitaxel: CNQ1OT1, which exhibits increased expression in lung adenocarcinoma cells that are paclitaxel-sensitive (Ren et al. 2017), and TUG1, by influencing EZH2 in lung squamous cell carcinoma (Niu et al. 2017).
6.1.2 Resistance to Targeted Therapy
Some lncRNAs can affect the efficacy of anti-EGFR TKIs by inducing alterations which allow cancer cells to “escape” their effects, resulting in an acquired resistance to therapy and disease progression. UCA1 expression, upregulated in patients with EGFR-TKIs resistance, activates the AKT-mTOR pathway and stimulates EMT (Cheng et al. 2015). The lncRNA BC087858 also induces EGFR-TKIs resistance by activating the PI3K/AKT and MERK/ERK pathways, as well as inducing EMT (Pan et al. 2016). lncBC0587858 induces EMT by upregulating FOXC1, leading to E-cadherin inhibition and induction of EMT (Xia et al. 2013). MALAT-1 promotes EMT through EMT-associated transcription genes and activation of the Wnt pathway (Samatov et al. 2013).
6.1.3 Resistance to Radiotherapy
Mechanisms of radioresistance are still largely unknown. Some studies suggested a modulation by noncoding RNAs principally in response to DNA damage, radiation-associated cell death, hypoxia, and activation of cancer stem cells. The ncRNAs responsible for these events are predominantly miRNAs; however, some lncRNAs are also involved, particularly lncROR, pR-lncRNA-1, LINC-PINT, and TUSC7. In mice lung cancer models, upregulation of HOTAIR is associated with a decreased radiosensitivity by the inactivation of the β-catenin pathway (Chen et al. 2015). LncRNAs may be also suitable as markers of response to radiotherapy: the lncRNA plasmacytoma variant transcript 1 (PVT1) and the lncGAS5 have been demonstrated to serve as putative biomarkers in predicting radiosensitivity (Wu et al. 2017; Xue et al. 2017).
6.2 LncRNAs as New Therapeutic Targets
The comprehension of the pathogenic roles of lncRNAs in lung carcinogenesis is essential in order to investigate and establish new therapeutic targets. Numerous studies have been published to date (the most important are summarized in this chapter), and several interactions between lncRNAs and the main molecular pathways involved in lung carcinogenesis have been explored, evidencing opportunities for novel therapies. It is in principle possible to modulate the action of lncRNAs by blocking these interactions with siRNAs, antisense oligonucleotides, ribozyme, and aptamers. As previously discussed, these methods have been employed to inhibit oncogenic lncRNAs in several studies, with encouraging results. For example, experimental silencing of MALAT-1 with antisense oligonucleotides in mouse models reduced lung cancer metastasis (Gutschner et al. 2013). Nevertheless, there are currently no clinical trials testing lncRNA-targeting agents suggesting that additional research is warranted.
7 Conclusions and Future Perspectives
NSCLC is currently the leading cause of cancer-related death worldwide. Despite recent advances in the surgical and clinical management, mortality rates remain extremely high and close to incidence rates. Therefore, further research is warranted to improve survival, especially in the advanced stages of the disease. LncRNAs represent an emerging class of noncoding RNAs, which show encouraging results and potential applications in the diagnosis of NSCLC and in predicting the prognosis in subgroups of patients. In particular, numerous lncRNAs have been evaluated as biomarkers for early diagnosis, differential diagnosis, and stage stratification of NSCLC patients with encouraging results. Nevertheless, their clinical applicability and their predictive potential have to be tested with methodologically tailored studies. Furthermore, recent evidence strongly suggests that some lncRNAs or their combinations can predict either sensitivity or resistance to cisplatinum-based chemotherapy and TKI-based treatments, which further impacts prognosis. In this context, lncRNAs might be proposed as biomolecular markers for patient selection and implementation of personalized oncological treatments and for establishing alternative therapeutic strategies in cases of prediction of resistance to these treatments.
Currently, there are no available biomarkers to implement such a task, with the exception of some somatic mutations, such as the T790M EGFR mutation which determines resistance to TKIs in patients with lung adenocarcinoma. These mutations can be absent in the initial diagnosis and develop subsequently during treatment. This dictates the need to monitor the mutational status of driver and resistance-conferring genes during the course of the disease. Liquid biopsy methods are being developed to this regard, but they harbor several limitations, mainly due to technical reasons. In this setting, the use of lncRNAs might be useful for the prediction of sensitivity/resistance to therapies; however, their detection in biological fluids is challenging due to instability, which imposes the use of novel techniques.
Finally, lncRNAs can be molecular therapeutic targets themselves, considering their involvement in several pathophysiological mechanisms of lung cancer. Unfortunately, these small molecules are involved in numerous complex physiological and pathological processes, which currently limits their potential as targets. Further research is warranted to understand the interactions between lncRNAs and other classes of molecules to better elucidate their potential implication in the treatment of NSCLC.
References
Arab K, Park YJ, Lindroth AM et al (2014) Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 55:604–614
Cao Q, Yu J, Dhanasekaran SM et al (2008) Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 27:7274–7284
Chen B, Yu M, Chang Q et al (2013) Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget 4:1427–1437
Chen J, Shen Z, Zheng Y et al (2015) Radiotherapy induced Lewis lung cancer cell apoptosis via inactivating β-catenin mediated by upregulated HOTAIR. Int J Clin Exp Pathol 8:7878–7886
Cheng N, Cai W, Ren S et al (2015) Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 6:23582–23593
Colombino M, Paliogiannis P, Cossu A et al (2019) EGFR, KRAS, BRAF, ALK, and cMET genetic alterations in 1440 Sardinian patients with lung adenocarcinoma. BMC Pulm Med 19:209
Gao L, Mai A, Li X et al (2013) LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol Lett 223:205–210
Guo F, Guo L, Li Y et al (2015) MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol 8:15903–15910
Gutschner T, Hämmerle M, Eissmann M et al (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73:1180–1189
Han L, Kong R, Yin DD et al (2013) Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol 30:694
Hon CC, Ramilowski JA, Harshbarger J et al (2017) An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543:199–204
Horn L, Spigel DR, Vokes EE et al (2017) Nivolumab versus docetaxel in previously treated patients with advanced non-small cell lung cancer: two-year outcomes from two randomized, open label, phase III trials (CheckMate 017 and CheckMate 057). J Clin Oncol 35:3924–3933
Hou Z, Zhao W, Zhou J et al (2014) A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol 53:380–388
Hu G, Yang T, Zheng J et al (2015) Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Mol Carcinog 54:192–204
Kallen AN, Zhou XB, Xu J et al (2013) The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52:101–112
Kang YH, Kim D, Jin EJ (2015) Down-regulation of phospholipase D stimulates death of lung cancer cells involving up-regulation of the long ncRNA ANRIL. Anticancer Res 35:2795–2803
Li M, Qiu M, Xu Y et al (2015) Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol 36:9969–9978
Liang W, Lv T, Shi X et al (2016) Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine (Baltimore) 95:e4608
Liu MY, Li XQ, Gao TH et al (2016) Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J Thorac Dis 8:3314–3322
Lu Y, Zhou X, Xu L et al (2016) Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther 9:3077–3084
Nakagawa T, Endo H, Yokoyama M et al (2013) Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun 436:319–324
Niu Y, Ma F, Huang W et al (2017) Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 16:5
Paliogiannis P, Attene F, Cossu A et al (2013) Lung cancer epidemiology in North Sardinia, Italy. Multidiscip Respir Med 8:45
Paliogiannis P, Attene F, Cossu A et al (2015) Impact of tissue type and content of neoplastic cells of samples on the quality of epidermal growth factor receptor mutation analysis among patients with lung adenocarcinoma. Mol Med Rep 12:187–191
Palmieri G, Paliogiannis P, Sini MC et al (2017) Long non-coding RNA CASC2 in human cancer. Crit Rev Oncol Hematol 111:31–38
Pan H, Jiang T, Cheng N et al (2016) Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget 7:49948–49960
Peng W, Wang J, Shan B et al (2018) Diagnostic and prognostic potential of circulating long non-coding RNAs in non small cell lung cancer. Cell Physiol Biochem 49:816–827
Qiu M, Xu Y, Yang X et al (2014) CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol 35:5375–5380
Ransohoff JD, Wei Y, Khavari PA (2018 Mar) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19(3):143–157
Ren K, Xu R, Huang J et al (2017) Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol 80:243–250
Rinn JL, Kertesz M, Wang JK et al (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323
Samatov TR, Tonevitsky AG, Schumacher U (2013) Epithelial-mesenchymal transition: focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Mol Cancer 12:107
Shi Y, Li J, Zhang S et al (2015) Molecular epidemiology of EGFR mutations in asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology - mainland China subset analysis of the PIONEER study. PLoS One 10:e0143515
Shi H, Pu J, Zhou XL et al (2017) Silencing long non-coding RNA ROR improves sensitivity of non-small-cell lung cancer to cisplatin resistance by inhibiting PI3K/Akt/mTOR signaling pathway. Tumour Biol 39:1010428317697568
Sun M, Liu XH, Wang KM et al (2014) Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 13:68
Tang Y, Xiao G, Chen Y et al (2018) LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anti-Cancer Drugs 29:725–735
Tano K, Mizuno R, Okada T et al (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584:4575–4580
Thai P, Statt S, Chen CH et al (2013) Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol 49:204–211
Wang P, Chen D, Ma H et al (2017a) Long non-coding RNA MEG3 regulates proliferation and apoptosis in non-small cell lung cancer via the miR-205-5p/LRP1 pathway. RSC Adv 7:49710–49719
Wang Q, Cheng N, Li X et al (2017b) Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget 8:2558–2567
Weber DG, Johnen G, Casjens S et al (2013) Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small-cell lung cancer. BMC Res Notes 6:518
Wei MM, Zhou GB (2016) Long non-coding RNAs and their roles in non-small-cell lung Cancer. Genomics Proteomics Bioinformatics 14:280–288
Wei MM, Zhou YC, Wen ZS et al (2016) Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget 7:59556–59571
Wen X, Han XR, Wang YJ et al (2018) Effects of long noncoding RNA SPRY4-IT1-mediated EZH2 on the invasion and migration of lung adenocarcinoma. J Cell Biochem 119:1827–1840
White NM, Cabanski CR, Silva-Fisher JM et al (2014) Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 15:429
Wu D, Li Y, Zhang H et al (2017) Knockdown of Lncrna PVT1 enhances Radiosensitivity in non-small cell lung Cancer by sponging Mir-195. Cell Physiol Biochem 42:2453–2466
Xia L, Huang W, Tian D et al (2013) Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology 57:610–624
Xue Y, Ni T, Jiang Y et al (2017) Long noncoding RNA GAS5 inhibits tumorigenesis and enhances radiosensitivity by suppressing miR-135b expression in non-small cell lung cancer. Oncol Res 25:1305–1316
Yang Y, Li H, Hou S et al (2013) The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One 8:e65309
Zhang EB, Yin DD, Sun M et al (2014) P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 5:e1243
Zhang J, Zhu N, Chen X (2015) A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol 36:7465–7471
Zhao W, An Y, Liang Y et al (2014) Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci 18:1930–1936
Zhao C, Qiao C, Zong L et al (2018) Long non-coding RNA-CCAT2 promotes the occurrence of non-small cell lung cancer by regulating the Wnt/β-catenin signaling pathway. Oncol Lett 16:4600–4606
Zhou Y, Zhang X, Klibanski A (2012) MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol 48:R45–R53
Zhou M, Guo M, He D et al (2015) A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med 13:231
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Paliogiannis, P., Scano, V., Mangoni, A.A., Cossu, A., Palmieri, G. (2020). Long Noncoding RNAs in Non-Small Cell Lung Cancer: State of the Art. In: Jurga, S., Barciszewski, J. (eds) The Chemical Biology of Long Noncoding RNAs. RNA Technologies, vol 11. Springer, Cham. https://doi.org/10.1007/978-3-030-44743-4_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-44743-4_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-44742-7
Online ISBN: 978-3-030-44743-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)