Abstract
Brain tumor segmentation plays an important role in clinical diagnosis for neurologists. Different imaging modalities have been used to diagnose and segment brain tumor. Among all modalities, MRI is preferred because of its non-invasive nature and better visualization of internal details of the brain. However, MRI also comes with certain challenges like random noise, various intensity levels, and in-homogeneity that makes detection and segmentation a difficult task. Manual segmentation is extremely laborious and time consuming for the physicians. Manual segmentation is also highly dependent on the physician’s domain knowledge and practical experience. Also, the physician may not be able to see details at the pixel level and may only notice the tumor if it is more prominent and obvious. Therefore, there is a need for brain tumor segmentation techniques that play major role in perfect visualization to assist the physician in identifying different tumor regions. In this paper, we present recent advancements and comprehensive analysis of MRI-based brain tumor segmentation techniques that used conventional machine learning and deep learning methods. We analyze different proposed conventional and state-of-the-art methods in chronological order using Dice similarity, specificity, and sensitivity as performance measures.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The goal of automated or semi-automated brain tumor segmentation methods is to detect and accurately segment the abnormal regions within the brain. Brain tumors are mainly divided into two grades: Low Graded Glioma (LGG) and High Graded Glioma (HGG) [2]. LGG is less aggressive in nature while HGG is more aggressive in nature. Patients diagnosed with LGG have 5 years while patients diagnosed with HGG have 1 year of life expectancy on the average respectively [17]. The possible ways for treating the brain tumor are chemotherapy, radiations, and surgery. In clinician practice, neurologist manually segments the abnormal regions at each slice of MR imaging modalities [12, 23, 33]. This manual segmentation is subjective, prone to error, time consuming, costly and highly dependent on the neurologist’s experience. Therefore, automated or semi-automated framework for accurate brain tumor segmentation is highly demanded. Such system could help in timely detection of tumor and also guides treatment planning. In this regards, different researchers attempted different Machine Learning (ML) techniques in order to develop system, which can address these demands, act as intelligent assistant and also enhance the neurologist efficiency [23].
In the light of literature, different ML techniques have been used for brain tumor segmentation task that can be divided into two categories: Conventional ML based techniques and Deep Learning (DL) based techniques. All conventional ML techniques follow below mentioned typical steps: pre-processing, feature extraction, feature selection, training classifier and post processing. At pre-processing step, different image processing techniques are applied in order to reduce the noise and improve the quality of the features, which directly affect the segmentation result. At features extraction step, domain pertinent handcrafted features are computed that directly map the domain knowledge. At feature selection step, different dimensionality reduction techniques are applied in order to reduce feature vector dimension that results in reduced training time, no overfitting, and increased classification accuracy. At training step, the selected classifier is trained using the selected feature vector. Finally, at post-processing step, the results of the classifier are further processed to achieve better results.
Recently, deep learning based techniques achieved record shattering state-of-the-art results on verity of computer vision and biomedical image segmentation tasks [1]. Taking inspiration from these results, different biomedical researchers suggest novel CNN architectures for brain tumor segmentation using MR images. These CNN architectures are based on 2D or 3D convolutional kernels and obtained state-of-the-art results [3, 4, 8, 20, 21].
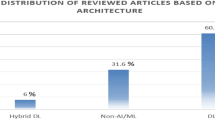
In this paper, we have comprehensively summarized recent ML techniques that are employed specifically for brain tumor segmentation task from year 2010 to 2017. We have categorized ML techniques into two sub-categories: conventional and DL techniques, which can be seen in Fig. 1. Each sub-category is further divided based on the technique being used. We analyzed different proposed conventional and DL state-of-the-art methods that are using Dice similarity, accuracy, error rate, specificity, and sensitivity as performance measures.
The rest of the paper is organized as follows: Sect. 2 presents in detail different machine learning techniques used for brain tumor segmentation, which are further categorized into conventional and DL techniques. In Sect. 3, we evaluate and discuss the methods that are mentioned in Sect. 2. Lastly in Sect. 4, we conclude the discussion.
2 Conventional and Deep Learning Techniques
The number of research publications dedicated to brain tumor segmentation has been grown significantly in the last few decades. The observation captured the increasing demands for fully or semi-automated segmentation methods that could provide complementary information and play an insolent assistant role that aid the neurologist in enhancing quality of treatment. In order to address these requirements, different neuro-computing researchers proposed different ML techniques and achieved promising results.
2.1 Conventional Techniques
The conventional machine learning (ML) techniques employed for brain tumor segmentation consist of Support Vector Machine (SVM), Random Forest (RF), k-Mean and fuzzy C-Mean. These ML techniques are further classified into two categories: Classification and Clustering algorithms.
2.2 Classification
Most of the classification based ML techniques perform central pixel classification to segment brain tumor using MR images. Following are the classification based techniques that are used for brain tumor segmentation:
SVM Based Techniques
Arikan et al. [7] proposed semi-automated hybrid method of SVM and interactive seed selection based method for brain tumor segmentation. At pre-processing step, they used anisotropic diffusion filter for noise removal in the MR images. Then, random seeds selected from pre-processed MR images to train SVM classifier. For performance evaluation, they used publicly available Medical Image Computing and Computer-Assisted Intervention (MICCAI) Brain Tumor Segmentation benchmark (BRATS) dataset. They selected four patients from MICCIA BRATS-2015 dataset to evaluate the performance of their proposed method. Their proposed method achieved average Dice Similarity (DS) of about 81% compared with ground truth.
Vaishnavee et al. [31] coped with brain tumor segmentation challenge using SVM and Self Organizing Map (SOM) techniques. At pre-processing step, histogram equalization was performed. The four features (i.e. mean, intensity, number of occurrences and variance) were computed to train classifier. At second step, they employed SOM clustering in order to localize and segment abnormal brain clusters. Moreover, they also performed the classification of brain MR images into different intra-tumor classes. In order to perform this classification in sub-tumor types, Gray Level Co-occurrence Matrix (GLCM) texture features were computed that were then followed by Principle Component Analysis (PCA) as dimensionality reduction step. Proximal Support Vector Machine (PSVM) classified the MR image in one of three classes: either normal, benign, or malignant.
Rathi et al. [25] addressed brain tumor segmentation as pixel classification task. Their approach computed three features: intensity (i.e. mean, variance, standard deviation, median intensity, skewness, and kurtosis), texture (i.e. contrast, correlation, entropy, energy, homogeneity, cluster shade, sum of square variance) and shape (i.e. circularity, irregularity, area, perimeter and shape index). PCA and Linear Discriminant Analysis (LDA) were used to reduce the dimensionality of the computed feature matrix. The SVM was trained to classify each pixel as white matter, gray matter, cerebrospinal fluid (CSF), tumor or non-tumor. The experiment was conducted on 140 brain MR images taken from Internet Brain Segmentation Repository (IBSR).
Zhang et al. [35] fused multi-spectral MR images and addressed the challenges in much efficient and effective way. Their work resulted in reduce computational cost, faster inference and less segmentation error. In their framework, they redefined feature extraction, feature selection and introduced additional adaptive training step. Three features (Intensities, texture information and wavelet transform) were computed in different kernel windows that propagate across multi-spectral images. During the training stage, SVM was retrained until the feature set got consistent. The SVM pixel classification was then forwarded to region growing algorithm, which further performed the refinement of the tumor boundaries. Their obtained result justified the fusion of multi-spectral MRI images and the two step classification.
Reddy et al. [26] computed three features (Local Binary Pattern (LBP), Histogram of Oriented Gradients (HOG) and Mean Intensity (MI)) in neighborhood pixels. These three features were concatenated to train SVM as pixel level discriminative classifier. Then, the result of SVM was smoothed with Gaussian filter in order to generate the confidence surface. The confidence surface contained the likelihood of each pixel as tumor or non-tumor. The incorporation of the confidence surface with level set and region growing segmentation algorithm achieved promising dice similarity score and outperformed the original version of both algorithms without confidence surface as prior.
RF Based Techniques
Following are the RF based techniques proposed for brain tumor segmentation.
Ellwaa et al. [11] proposed a fully automated MRI-based brain tumor segmentation method, which used iterative random forest. The accuracy was improved by iteratively selecting the patient that possessed the detailed information from dataset, which was used to train random forest classifier. The method was validated on BRATS-2016 dataset. The criteria of selecting the patient with highest information helped them to achieve promising results.
Lefkovits et al. [16] proposed a RF based discriminative model for brain tumor segmentation using multi-modal MR images. The discriminative model was used to establish the optimal parameters. These determined optimum parameters were used to fine-tune RF. The fine-tuned RF obtained the optimized tumor segmentation. They used BRATS-2012 and BRATS-2013 datasets for validation of their framework. Their method showed comparable results to other proposed methods on BRATS 2012 and BRATS-2013 datasets.
Meier et al. [19] proposed a RF based discriminative model using highly discriminative features. At pre-processing step, MedPy was used for intensity standardization, which was used to harmonize the sequence intensity profile. The voxel wise features were extracted for training the random forest, which was based on intensities values. The classifier was trained easily and consistently performed well on a large range of parameters.
Reza et al. [27] proposed an improved brain tumor segmentation method that computed textural features on multi-modal MR images. They used BRATS-2013 and BRATS-2014 datasets to validate their method. Mean and median (statistical validation) were calculated to check the efficiency of the proposed method. They performed pixel level classification using random forest classifier. In order to obtain further improved results, they used binary morphological filters on obtained segmented MR images to get precise contours. Moreover, the smallest objects were removed using connected components algorithm during post-processing. The holes in the tumor regions were also detected and filled, using connected neighbor intensities.
Abbasi et al. [5] proposed an automated 3D model for brain tumor detection and segmentation. For preprocessing, bias field correction and histogram matching was used and then ROIs (Region of Interests) were identified and separated from the background of the FLAIR image. They used HOG and LBP for learning features. The RF was used to segment brain tumor on BRATS 2013 dataset.
2.3 Clustering
Clustering based techniques are also used for the detection and localization of abnormalities within brain MR images. Following are most recent researches based on clustering technique for brain tumor segmentation using MR images.
Singh et al. [29] proposed an unified approach of fuzzy C-Mean and level-set segmentation algorithm to delineate the brain tumor and region of interest (ROI). Their proposed method comprised of two steps. At first step, fuzzy c-mean applied to divide the brain MR images into different clusters. At second step, level-set algorithm applied to initial contour in order to delineate the abnormal regions in MR images. Their cascaded strategy that comprised of distinct approaches obtained promising results.
Eman et al. [6] proposed a novel combination of k-Mean and fuzzy C-Mean (KIFCM) for brain tumor segmentation. Their proposed method comprised of five steps: pre-processing, clustering, extraction, contouring, and segmentation. At pre-processing step, median filter employed to reduce random noise and brain surfer was used to extract the brain area. At clustering step, KIFCM applied to obtain the clustering results. At extraction and contouring step, they first applied threshold and then smoothed the thresholded output with de-noising operation. Finally, level-set was employed to contour the tumor area in MR image. Their proposed method was computationally efficient and outperformed k-Mean, Expectation Maximization and fuzzy C-Mean individually.
Kaya et al. [14] proposed a PCA based technique for brain tumor segmentation using only T1-weighted MR images. Five common PCA based techniques were used to reduce the dimensionality of feature vector in k-Means and fuzzy C-Mean clustering algorithm. The reconstruction error and Euclidean distance errors was used to evaluate their method. The Probabilistic PCA outperformed others four PCA method.
Verma et al. [32] proposed a method for brain tumor segmentation, which was based on mountain clustering technique. They improved the mountain clustering algorithm in their proposed research; their method was comparable with well-known k-Means and fuzzy C-Mean clustering techniques based on the cluster entropy. Their method performed well and presented minimum average entropy (0.000831) against k-Means (0.000840) and fuzzy C-Mean (0.000839) algorithm.
Selvakumar et al. [28] proposed a combined fuzzy C-Mean and k-Mean clustering method to segment the brain tumor pictured in MR images. In preprocessing phase, skull striping and de-nosing operations were performed. At the segmentation step, first brain tumor was segmented uing k-Mean, then the segmented result was proceeded to fuzzy C-Mean to perform final segmentation of tumor regions. Their proposed method extracted the features using these two clustering algorithm that was further used for approximate reasoning. Their method showed the shape and exact spatial location of the tumor, which enhanced to diagnosing of infected tumorous brain region. They used amount of the area as evaluation metric that was calculated from clusters to find the stage.
2.4 Deep Learning (DL) Techniques
We classified the CNN based techniques for brain tumor segmentation in two categories: 2D-CNN and 3D-CNN.Which are discussed below.
2D-CNN Techniques
Chang et al. [9] proposed a fully CNN based architecture for brain tumor segmentation with Hyperlocal Local Features Concatenation (HLFC). The fully convolutional network had five convolutional layers with nonlinear activation functions. Bilinear interpolation unsampled the output of last convolution layer to the original input sized image. Hyperlocal feature concatenation reintroduced the original data in the network by concatenation operation across channels, which was used to produce the segmentation map with two narrow size (3 \( \times \) 3) convolutional filters after concatenating hyperlocal features and bilinear interpolated image. They validated the proposed method on BRATS-2016 dataset. Their method was computationally efficient and able to complete segmentation task on an optimized GPU in less than one second.
The patch wise learning of CNN model ignore the contextual information of the whole image. To overcome this issue, Lun et al. [18] presented a CNN based fully automated method for brain tumor segmentation using MR images. Their method incorporated with multi modalities MR images and used global features based CNN model. Local regions were in the form of central pixel labeled patch, which was extracted by cropping image with 30 \( \times \) 30 kernel. These patches were used for training the model. They adopted a re-weighting scheme for loss layer in CNN to elevate the imbalance label problem. Their method was validated on BRATS-2015 dataset. Their global features based re-weighting method outperformed the prior patch wise learning method.
Pereira et al. [23] proposed an automated CNN based method for brain tumor segmentation using four MR weighted modalities (T1, T1c, T2 and FLAIR). They used fixed size 3 \( \times \) 3 convolutional kernels. The fixed size convolutional kernel enabled them to develop a deeper CNN with more non-linearities applied and fewer weightes. For intensity correction, they used N4ITK method. They proposed separated architectures for High Graded Glioma (HGG) and Low Graded Glioma(LGG). They validated their method on BRATS-2013 and BRATS-2015 dataset. The deeper CNN model with fixed smaller size convolutional kernels obtained highest dice similarity on all sub-tumoral compartments.
Xiao et al. [33] segmented brain tumor in MR images using DL. They integrated the stacked denoising auto-encoder into segmentation procedure. In preprocessing phase, the patches of each MRI were extracted and to obtained the gray level sequence of image patches. The deep learning based classification model use extracted gray level image patches as input and performed classification. Then classification results were mapped on to the binary image. In post-processing phase, they used morphological filters to smoothened the edges and filling the gaps in tumor region. After post processing, they got fined tumor segmentation results.
Havaei et al. [12] proposed a cascaded two-pathway CNN architecture by extracting smaller and larger sized patches at the same time respectively. The cascaded CNN processed smaller and larger contextual details of the central pixel simultaneously. Patches of size 33 \( \times \) 33 and 65 \( \times \) 65 were extracted for local and global pathway of CNN to classify the label of the central pixel respectively. Their novel two pathways CNN architecture processed the local and global detail of the central pixel and obtained near state-of-the-art results.
Davy et al. [10] proposed a fully automated brain tumor segmentation approach based on local structure prediction with CNN and k-Means. Similarly, Rao et al. [24] extracted multi plane patches around each pixel and trained four different CNNs each taking input patches from a separate MRI modality image. Outputs of the last hidden layers of those CNNs are then concatenated and used as feature maps to train a random forest (RF) classifier.
3D-CNN Techniques
Kamnitsas et al. [13] proposed 3D-CNN architecture for brain tumor segmentation, named DeepMedic. DeepMedic was previously presented for lesion segmentation. Moreover, their 3D-CNN architecture was extended with residual connection to investigate the lesion effects. BRATS-2015 dataset was used for validating their method. Their results showed that the DeepMedic with residual connection performed well then the simple DeepMedic CNN architecture.
Kayalibay et al. [15] worked on segmentation of medical imaging data using 3D-CNN. Their proposed method was the combination of 3D convolutional filters and CNN. They applied their method to segment brain tumor in MR images. Moreover, they also employed their method to segment bones in hand MR images. Their method was trained and tested on each modality separately. The fused the modality wise CNN output which helped to obtained promising results. Their method obtained comparable dice similarity score on the BRATS-2013 and BRATS-2015.
Yi et al. [34] segmented glioblastoma using three-dimensional CNN. The convolutional layers in their architecture were combined with the Difference of Gaussian (DoG) filters to perform the 3-dimensional convolution operation. They used BRATS-2015 dataset that contained 274 tumor samples. They used dice similarity to measure the quality of the segmentation of their proposed method.
Urban et al. [30] proposed a 3D-CNN architecture for the multi-modal MR images for glioma segmentation. Multi-modality 3D patches, basically cubes of voxels, extracted from the different brain MRI modalities are used as inputs to a CNN to predict the tissue label of the center voxel of the cube. The proposed network achieved promising results across three tumors but due to the 3D-CNN that was based on 3D patches the overall network computational burden was more when compared to 2D patches based 2D-CNN methods.
Nie et al. [22] proposed fully automated 3D-CNN based technique for brain tumor segmentation using T1, Diffusion Tensor Imaging (DTI) and functional MRI (fMRI) MR modality. Different pre-processing technique applied on each MR modality i.e. intensity normalization were applied on T1, tensor modelling applied on DTI and for fMRI frequency specific BOLD (blood oxygen level- dependent) fluctuation power were calculated. They used 3D-CNN as features extractor and SVM performed the final prediction.
3 Performance Evaluation and Discussion
We created a separate table for each category that is proposed for brain tumor segmentation. Each table further presents the detailed results of techniques used under same category. For instance, Tables 1, 2, 3, and 4 presents the detailed results of techniques based on SVM, RF, 2D-CNN, and 3D-CNN respectively. Furthermore, the tables also summarize their research technique, dataset and performance measurement matrices that is used to quantify technique’s effectiveness. Different technique are using different performance measure criteria. Some of these are dice similarity, precision, accuracy, total error, and entropy.
Conventional ML techniques relied heavily on the handcrafted features that attempt to model the domain knowledge. On the other hand, DL techniques have the unique capability to learn increasingly complex hierarchy of features in extremely unsupervised fashion, which were impossible to represent with handcrafted features. These additional features basis enabled the DL based technique to outperform conventional ML techniques, Particularly, Convolutional Neural Network (CNN) performed well on a verity of vision and biomedical image segmentation tasks.
Another ML technique that is extensively used for brain tumor segmentation are clustering based techniques.These technique are semi-automated and requires neurologist intervention in order to generate the fully clustered tumor regions. However, these technique are heavily dependent spatial information distribution of MR images (i.e. intensity, texture etc.). Such dependencies often cause to deviate the result of the clustering based techniques.
3D-CNN based techniques attempt to model three dimensional details of the three dimensional MR images using 3D convolutional kernels and obtained promising results. However, there is a disadvantage of using 3D-CNN for medical image segmentation task. Because in medical field, there is always a challenge of scarcity of labeled data. In order to address this challenge, 2D-CNN architectures that process 2D patches are introduced. The 2D-CNN based techniques obtained comparable results to 3D-CNN. However, 2D-CNN only process the local contextual information that result in decreased accuracy compared to 3D-CNN.
4 Conclusion
In this paper, we presented the comprehensive review of ML based techniques used for brain tumor segmentation. We classified the ML techniques into two broad categories: conventional and deep learning based techniques. The conventional techniques are further classified based on ML algorithms that are being used. While deep learning based technique are classified into 2D-CNN and 3D-CNN techniques. However, CNN based technique outperformed conventional ML based technique and obtained state-of-the-art results on brain tumor segmentation. 3D-CNN based techniques are far demanding computationally and inappropriate for biomedical image segmentation task. On the other hand, 2D-CNN architectures are specifically designed for medical segmentation task that address the limited available data challenge and are less demanding computationally. Furthermore, 2D-CNN method also achieved comparable results to 3D-CNN.
References
The 9 Deep Learning Papers You Need To Know About (Understanding CNNs Part 3) - Adit Deshpande - CS Undergrad at UCLA (’19)
Tumor Types - National Brain Tumor Society
MICCAI 2014 challenge on multimodal brain tumor segmentation. In: Proceedings of the MICCAI-BRATS, Boston, Massachusetts (2014)
MICCAI challenge on multimodal brain tumor segmentation. Proceedings MICCAI-BRATS Workshop, 2016 (2016)
Abbasi, S., Tajeripour, F.: Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient. Neurocomputing 219, 526–535 (2017)
Abdel-Maksoud, E., Elmogy, M., Al-Awadi, R.: Brain tumor segmentation based on a hybrid clustering technique. Egypt. Inf. J. 16(1), 71–81 (2015)
Arikan, M., Fröhler, B., Möller, T.: Semi-automatic brain tumor segmentation using support vector machines and interactive seed selection. In: Proceedings of the MICCAI-BRATS, Boston, Massachusetts, pp. 1–3 (2014)
Farahani, K., Kalpathy-Cramer, J., Kwon, D., Menze, B.H., Reyes, M.: MICCAI 2015 challenge on multimodal brain tumor segmentation. In: Proceedings of the MICCAI-BRATS, Munich, Germany (2015)
Chang, P.D.: Fully convolutional neural networks with hyperlocal features for brain tumor segmentation. In: Proceedings MICCAI-BRATS Workshop, pp. 4–9 (2016)
Havaei, M., et al.: Brain tumor segmentation with deep neural networks. In: proceedings of BraTS-MICCAI (2014)
Ellwaa, A., et al.: Brain tumor segmantation using random forest trained on iteratively selected patients. International Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Lecture Notes in Computer Science, pp. 129–137. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-55524-9_13
Havaei, M., et al.: Brain tumor segmentation with deep neural networks. Med. Image Anal. 35, 18–31 (2017)
Kamnitsas, K., et al.: Deepmedic on brain tumor segmentation. BrainLes 2016. LNCS, vol. 10154. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-55524-9_14
Kaya, I.E., Pehlivanlı, A.Ç., Sekizkardeş, E.G., Ibrikci, T.: PCA based clustering for brain tumor segmentation of T1w MRI images. Comput. Methods Programs Biomed. 140, 19–28 (2017)
Kayalibay, B., Jensen, G., van der Smagt, P.: CNN-based Segmentation of Medical Imaging Data. arXiv preprint arXiv:1701.03056 [cs.CV], January 2017
Szilagyi, L., Lefkovits, L., Lefkovits, S.: Brain tumor segmentation with optimized random forest. In: Proceedings MICCAI-BraTS Workshop, pp. 30–34 (2016)
Louis, D.N., et al.: The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131(6), 803–820 (2016). https://doi.org/10.1007/s00401-016-1545-1
Lun, T.K., Hsu, W.: Brain tumor segmentation using deep convolutional neural network. In: Proceedings of BRATS-MICCAI (2016)
Wiest, R., Reyes, M., Meier, R., Knecht, U.: CRF-based brain tumor segmentation: alleviating the shrinking bias. In: Crimi, A., Menze, B., Maier, O., Reyes, M., Winzeck, S., Handels, H. (eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. LNCS. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-55524-9_10
Menze, B., Jakab, A., Bauer, S., Reyes, M., Prastawa, M., Van Leemput, K.: MICCAI 2012 challenge on multimodal brain tumor segmentation. In: Proceedings of the MICCAI-BRATS, Nice, France (2012)
Menze, B., Jakab, A., Reyes, M., Prastawa, M.: MICCAI 2013 challenge on multimodal brain tumor segmentation. In: Proceedings of the MICCAI-BRATS, Nagoya, Japan (2013)
Nie, D., Zhang, H., Adeli, E., Liu, L., Shen, D.: 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 212–220. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_25
Pereira, S., Pinto, A., Alves, V., Silva, C.A.: Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 35(5), 1240–1251 (2016)
Rao, V., Sarabi, M.S., Jaiswal, A.: Brain tumor segmentation with deep learning. In: Proceedings of MICCAI Multimodal Brain Tumor Segmentation Challenge (BraTS), pp. 56–59 (2015)
Rathi, V.P., Palani, S.: Brain tumor MRI image classification with feature selection and extraction using linear discriminant analysis. arXiv preprint arXiv:1208.2128 (2012)
Reddy, K.K., Solmaz, B., Yan, P., Avgeropoulos, N.G., Rippe, D.J., Shah, M.: Confidence guided enhancing brain tumor segmentation in multi-parametric MRI. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 366–369. IEEE (2012)
Reza, S., Iftekharuddin, K.M.: Improved brain tumor tissue segmentation using texture features. In: Proceedings of MICCAI-BRATS Challenge on Multimodal Brain Tumor Segmentation (2014)
Selvakumar, J., Lakshmi, A., Arivoli, T.: Brain tumor segmentation and its area calculation in brain MR images using K-mean clustering and FUZZY C-mean algorithm. In: International Conference on Advances in Engineering, Science and Management (ICAESM), pp. 186–190. IEEE (2012)
Singh, P., Bhadauria, H.S., Singh, A.: Automatic brain MRI image segmentation using FCM and LSM. In: 3rd International Conference on Reliability, Infocom Technologies and Optimization (ICRITO) (Trends and Future Directions), pp. 1–6. IEEE (2014)
Urban, G., Bendszus, M., Hamprecht, F., Kleesiek, J.: Multi-modal brain tumor segmentation using deep convolutional neural networks. In: Proceedings of MICCAI BraTS (Brain Tumor Segmentation) Challenge, Winning Contribution, pp. 31–35 (2014 )
Vaishnavee, K.B., Amshakala, K.: An automated MRI brain image segmentation and tumor detection using SOM-clustering and proximal support vector machine classifier. In: 2015 IEEE International Conference on Engineering and Technology (ICETECH), pp. 1–6. IEEE (2015)
Verma, N.K., Gupta, P., Agrawal, P., Cui, Y.: MRI brain image segmentation for spotting tumors using improved mountain clustering approach. In: Applied Imagery Pattern Recognition Workshop (AIPRW), pp. 1–8. IEEE (2009)
Xiao, Z., et al.: A deep learning-based segmentation method for brain tumor in MR images. In: 2016 IEEE 6th International Conference on Computational Advances in Bio and Medical Sciences (ICCABS), pp. 1–6. IEEE, October 2016
Yi, D., Zhou, M., Chen, Z., Gevaert, O.: 3-D convolutional neural networks for glioblastoma segmentation. arXiv preprint arXiv:1611.04534 [cs.CV], November 2016
Zhang, N., Ruan, S., Lebonvallet, S., Liao, Q., Zhu, Y.: Kernel feature selection to fuse multi-spectral MRI images for brain tumor segmentation. Comput. Vis. Image Underst. 115(2), 256–269 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Khan, H., Alam Zaidi, S.F., Safi, A., Ud Din, S. (2020). A Comprehensive Analysis of MRI Based Brain Tumor Segmentation Using Conventional and Deep Learning Methods. In: Brito-Loeza, C., Espinosa-Romero, A., Martin-Gonzalez, A., Safi, A. (eds) Intelligent Computing Systems. ISICS 2020. Communications in Computer and Information Science, vol 1187. Springer, Cham. https://doi.org/10.1007/978-3-030-43364-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-43364-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-43363-5
Online ISBN: 978-3-030-43364-2
eBook Packages: Computer ScienceComputer Science (R0)