Abstract
Automatic pancreas segmentation from Computed Tomography (CT) images is a prerequisite of clinical practices such as cancer detection, yet challenging due to the variability in shape. To address this challenge, we propose a Hierarchical Convolutional Neural Network (H-CNN) to fuse multi-scale features, which could remedy the lost image details in progressive convolutional and pooling layers. In our proposed H-CNN, a hierarchical fusion block is designed to fuse low-level and high-level features across different layers. The H-CNN is evaluated on NIH pancreas dataset and outperforms the current state-of-art methods by achieving 86.59% ± 4.33% in terms of DSC. The experimental results confirm the effectiveness of the proposed H-CNN.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Pancreas segmentation from CT images is an important step in computer-aided diagnosis and treatment such as cancer detection [1]. In practice, to reduce the damage to other adjacent tissues and save human surgery, it is worthwhile [2] to explore an automated and precise method for pancreas segmentation from medical images. As segmentation from CT images is still a challenging task in pancreas diagnoses [3], we focus on pancreas segmentation from CT images in this paper.
1.1 Challenges and Motivations
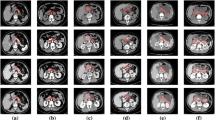
There are two main challenges for automated CT pancreas segmentation. Firstly, the greatly irregular boundary of pancreas across different diseases (as shown in Fig. 1). Secondly, the inherent image noise and distortions in CT images. Convolutional Neural Networks (CNNs) [4] are formed by consecutive convolutional layers [4] and pooling layers [5], which have shown excellence in image segmentation. However, since pooling layers [6] in CNNs inevitably loss information details when applied to CT images, it is difficult to precisely delineate the variant boundary of the pancreas. To overcome the limitation [7, 9] of CNNs, there are effective methods proposed. For example, Deeplab [8] designed dilated convolution to replace pooling layer, which can enlarge the receptive field without down-scaling feature maps. Other methods such as SegNet [10] progressively up-sample the convolved feature maps from previous layers to improve the image details. These methods made use of the enlarged convolved features from penultimate layers to retain more local image details.
The aforementioned methods showed improvements for three-channel color images, yet failing to achieve great performance on single-channel CT images to segment the pancreas. To more effectively make use of the convolved features from different layers in CNNs, Yu et al. [11] have proved the effectiveness of combining multi-scale features to retain local image details which are important to delineate boundary. Therefore, in this paper, we take the advantages of CNNs in a deep fusion of multi-scale features to remedy boundary information and improve pancreas segmentation from CT images.
1.2 Related Work
Earlier pancreas segmentation methods for CT images, can be mainly grouped into probabilistic atlas and statistical shape modeling [13, 14]. For example, Suzuki et al. [12] incorporated the spatial interrelations into a statistical atlas for pancreas segmentation. However, as it is difficult to find a model which covers all possible variabilities, these shape-based methods commonly fail to solve the challenge that the variant boundary shape of different pancreas.
More recently, some investigators have proposed CNNs based methods for pancreas segmentation. As the highly convolved features are produced from a set of layers in CNNs, they can be treated as high-level features. Roth et al. [17] learned high-level features from a holistically-nested network to further refine them with a random forest. Ronneberger et al. [18] proposed a popular model (U-Net) for medical image segmentation. Milletari et al. [19] extended U-Net into a 3D model (V-Net) and achieved improvement. Zhou et al. [20] suggested to find a rough pancreas region and refined the region through learning an FCN based fixed-point model iteratively. Although high-level features can contain more semantic information, relying on these features may limit the segmentation performance of CNNs – because of the lost boundary information obtained from low-level layers in CNNs.
1.3 Contribution
The main contribution of this paper is fusing multi-scale features for pancreas segmentation from CT images. Firstly, alternative to generating segmentation maps from high-level features, our H-CNN hierarchically extracts and fuse high-level and low-level features to address the challenge of irregular pancreas boundary. The proposed Hierarchical Fusion Block (H-block) can hierarchically refine different level features, especially the proposed H-block can capture context from a larger image region to better make use of features. Secondly, we used residual connections [21] in H-block to propagate gradients throughout different layers.
2 Method
As shown in Fig. 2, H-CNN is built upon encoder-decoder architecture. The encoder sub-network is a VGG16-type network to extract image features in different resolution. Then, the Hierarchical Fusion Block (H-block) in the decoder sub-network fuses high-level features convolved from the encoder with low-level features to retain local details.
Overview of the proposed H-CNN for pancreas segmentation. (a): the architecture of H-CNN. The H-CNN adopts VGG16 for feature extraction. Finally, H-blocks perform multi-scale feature fusion on the top of VGG. (b): detailed structure of a Hierarchical fusion block which fuses multi-scale features to remedy the local details from the feature extraction stage.
2.1 Convolution-Pooling Block
Convolution-pooling blocks (CP-block) lies in the encoder sub-network, where each of them has two convolutional layers and one pooling layers. The convolutional layers convolve input image to extract features, while the pooling layers enlarge the receptive field and reduce the sensitivity of features to shift and distortion.
2.2 Hierarchical Fusion Block
Due to the lost local details in encoder sub-network are important for boundary delineation, we propose a hierarchical fusion block (H-block) to fuse multi-scale features so as to remedy the local details. As shown in Fig. 2(b), each H-block has three main components: multi-scale fusion block, hierarchical convolution block and residual convolution block.
Multi-scale Fusion.
This block first up-samples the high-level features for input adaptation, which generates feature maps at the same feature dimensions as the low-level features. Then, all feature maps are fused by concatenation.
Hierarchical Convolution Block.
The output features then are fed into the hierarchical convolution block. The proposed hierarchical convolution block aims to capture features from a larger image region. In particular, this part has a set of convolution blocks, each having a dilated convolution layer and a convolution layer. The dilated convolution layer could generate features from an enlarged receptive filed without losing feature details. Note that each dilated convolution is followed by a convolution layer which servers as cross-channel interaction and information aggregation. The output features of all hierarchical convolution blocks are fused together with the input features through summation.
3 Data and Experiment
3.1 Data and Evaluation Metrics
The NIH pancreas segmentation dataset [3] containing 82 CT samples, is used to evaluate the proposed model. The resolution of each sample is 512× 512× D, where \( {\text{D}} \in \left[ {181,\,466} \right] \). Manual ground-truths for the samples are also supplied.
Dice Srensen Coefficient (DSC) and Volumetric overlap error (VOE) are two common evaluation metrics in pancreas segmentation [17, 22]. In this paper, we used these two metrics to evaluate our model. Denote P and G as the segmentation result and ground-truth mask, DSC is formulated as \( DSC\left( {{\text{P}},{\text{G}}} \right) = \frac{{2\,*\,\left| {P\, \cap \,G} \right|}}{\left| P \right|\, + \,\left| G \right|} \). The value of DSC ranges in [0, 1], where a good segmentation method should have a high DSC. VOE is defined as \( VOE\left( {{\text{P}},{\text{G}}} \right) = 1 - \frac{{\left| {P\, \cap \,G} \right|}}{{\left| {P\, \cup \,G} \right|}} \), which represents the error rate of segmentation result.
3.2 Implementation
All experiments were run on an NVIDIA TITAN GPU to boost the training. For the data augmentation, we utilized rotation (90°, 180° and 270°) and flip in all three planes, to increase the number of training samples. We then trained H-CNN by SGD optimizer with a 10 mini-batch and a base learning rate to be 0.001 via polynomial decay in a total 80000 iterations. Following the training protocol [9], we performed 4-fold cross-validation to validate our model. The H-CNN is compared with four state-of-art pancreas segmentation methods, including Fixed-Point [20], Hierarchical FCN [23, 24], Holistically-Nested [17] and DeepOrgan [3].
4 Results
Overall Performance.
The experimental results (DSC) of H-CNN and comparison methods on the NIH pancreas segmentation dataset are listed in Table 1. The comparison with previous methods showed that our method achieved a better segmentation result. To quantify the improvements in terms of statistical significance, we tested the p-value whose value ≤ 0.05 indicates a significant difference.
Evaluation of Hierarchical Fusion Block (H-Block).
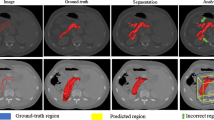
After high-level features obtained from the encoder sub-network, we fused multi-scale features via H-block. To provide a clear pattern about the effect of this block, we removed the H-block and directly used original decoder structure in U-Net to produce the final segmentation maps. As shown Fig. 4, the proposed H-block can optimize pancreas segmentation compared to the baseline model U-Net. To quantify these improvements in terms of statistical significance we performed student t-tests, where the p-value ≤ 0.05 (DSC: p-value = 0.004 and VOE: p-value = 0.017).
5 Discussion
H-CNN Segmentation.
The advantage of using H-CNN for pancreas lies in designing a Hierarchical Fusion Block (H-block) which fuses multi-scale features to remedy the lost local details brought by pooling. Although the location of pancreas can be predicted using the high-level features obtained from the down-sampling procedure, the pancreas boundary cannot be precisely delineated. Some CNNs for segmentation such as FCN and U-Net, also fuse low-level and high-level features. However, these networks just simply fuse corresponding features directly from encoder with up-sampled decoder output through skip connection, which may not efficiently use different level features. By contrast, the proposed Hierarchical Fusion block (H-block) can hierarchically refine different level features, especially the proposed H-block can capture context from a larger image region to better make use of features.
H-CNN and the State-of-Art Methods.
DeepOrgan segmented pancreas by classifying the candidate regions with random forest. Hierarchical FCN purely used high-level features for segmentation. However, as these two methods purely relied on high-level features which failed to delineate complex boundary (Fig. 3). Holistically-Nested FCN and Fixed-point methods are not end-to-end models, thus, the trained models may not be suboptimal. By contrast, our H-CNN fused multi-scale features to more precisely delineate the pancreas segmentation.
6 Conclusion
In this paper, we proposed a CNN based model, H-CNN, for CT pancreas segmentation. Motivated by the high relevance of low-level features and boundary delineation, we fused low-level images cues and high-level convolved features to delineate the pancreas boundary. Our H-CNN outperformed the existing popular CNN models.
References
Hidalgo, M.: Pancreatic cancer. N. Engl. J. Med. 362(17), 1605–1617 (2010)
Beger, H.G., Matsuno, S., Cameron, J.L.: Diseases of the Pancreas: Current Surgical Therapy. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-28656-1
Roth, H.R., et al.: DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 556–564. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24553-9_68
Shin, H.-C., et al.: Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 35(5), 1285–1298 (2016)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Lee, C.-Y., Gallagher, P., Tu, Z.: Generalizing pooling functions in CNNs: mixed, gated, and tree. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 863–875 (2017)
Springenberg, J.T., Dosovitskiy, A., Brox, T., Riedmiller, M.: Striving for simplicity: the all convolutional net. arXiv preprint arXiv:1412.6806 (2014)
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2017)
Noh, H., Hong, S., Han, B.: Learning deconvolution network for semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1520–1528 (2015)
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Yu, F., Wang, D., Shelhamer, E., Darrell, T.: Deep layer aggregation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2403–2412 (2018)
Suzuki, Y., et al.: Automated segmentation and anatomical labeling of abdominal arteries based on multi-organ segmentation from contrast-enhanced CT data. In: Drechsler, K., et al. (eds.) CLIP 2012. LNCS, vol. 7761, pp. 67–74. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-38079-2_9
Shimizu, A., Kimoto, T., Kobatake, H., Nawano, S., Shinozaki, K.: Automated pancreas segmentation from three-dimensional contrast-enhanced computed tomography. Int. J. Comput. Assist. Radiol. Surg. 5(1), 85 (2010)
Wolz, R., Chu, C., Misawa, K., Fujiwara, M., Mori, K., Rueckert, D.: Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans. Med. Imaging 32(9), 1723–1730 (2013)
Dmitriev, K., Gutenko, I., Nadeem, S., Kaufman, A.: Pancreas and cyst segmentation. In: Medical Imaging 2016: Image Processing. International Society for Optics and Photonics, p. 97842C (2016)
Grady, L.: Random walks for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 11, 1768–1783 (2006)
Roth, H.R., Lu, L., Farag, A., Sohn, A., Summers, R.M.: Spatial aggregation of holistically-nested networks for automated pancreas segmentation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 451–459. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_52
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Milletari, F., Navab, N., Ahmadi, S.-A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV). IEEE, pp. 565–571 (2016)
Zhou, Y., Xie, L., Shen, W., Wang, Y., Fishman, E.K., Yuille, A.L.: A fixed-point model for pancreas segmentation in abdominal CT scans. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10433, pp. 693–701. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66182-7_79
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Mahapatra, D., et al.: Automatic detection and segmentation of Crohn’s disease tissues from abdominal MRI. IEEE Trans. Med. Imaging 32(12), 2332–2347 (2013)
Roth, H.R., et al.: Hierarchical 3D fully convolutional networks for multi-organ segmentation. arXiv preprint arXiv:1704.06382 (2017)
Ren, J., Wang, D., Jiang, J.: Effective recognition of MCCs in mammograms using an improved neural classifier. Eng. Appl. Artif. Intell. 24(4), 638–645 (2011)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, Z., Zheng, J. (2020). Deep Neural Network for Pancreas Segmentation from CT Images. In: Ren, J., et al. Advances in Brain Inspired Cognitive Systems. BICS 2019. Lecture Notes in Computer Science(), vol 11691. Springer, Cham. https://doi.org/10.1007/978-3-030-39431-8_39
Download citation
DOI: https://doi.org/10.1007/978-3-030-39431-8_39
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-39430-1
Online ISBN: 978-3-030-39431-8
eBook Packages: Computer ScienceComputer Science (R0)