Abstract
Learned vocal communication emerges from the coordination of sensory and motor learning, reflects the function of a distributed but integrated neural circuit, and unfolds across several timescales, often occurring in maturing animals. Because nearly all brain organization and function originates from patterns of genomic activation, it is crucial to understand principles of how the genome works in order to understand how learned vocal communication arises. In this chapter, the fact that genome functions have high evolutionary conservation will be leveraged to provide a conceptual guide for how research using a species of songbird, the zebra finch, can deepen and expand clinical findings from humans. Additionally, this chapter provides examples for how studies in the zebra finch can uncover fundamental processes of learned vocal communication that are of value for understanding human speech and language. Examples include the organization of specialized neural circuits, responses to social communication experiences, activation of motor plans, and consideration of how the age and sex of the individual intersect with vocal communication skills, all of which have potential to inform on vocal learning mechanisms in humans. Together, our current state of knowledge advances the idea that humans and songbirds do not simply share superficial parallels; rather, they share deep biological properties to accomplish the complex, multi-level processes required for learning and producing meaningful communication patterns.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Behavior
- Critical period
- Epigenetics
- Genome
- Histone
- Learning and memory
- mTOR
- Neural development
- Sensitive period
- Sex differences
- Song
- Songbird
- Zebra finch
8.1 Introduction
This chapter relates the dynamics occurring at the genomic and whole-animal levels to deepen the appreciation for how complex and important natural behaviors, such as learned vocal communication, emerge from biological substrates (also see Sakata and Woolley, Chap. 1). Here, several dimensions of genomic function, which include the sequence, regulation, and function of RNA, protein products, and epigenetic modifications, are considered in the context of learned vocal communication.
The genome is not static, thus the relationships between the genome, the brain, and behavior are interdependent. This chapter does not comprehensively describe any specific example of the genome-brain-behavior interrelationship but does consider vocal learning mechanisms in light of biological dimensions that influence vocal communication. Those major dimensions include sex, age, prior experience, social context, individual brain areas, and neural circuits. The stories presented herein demonstrate how to initiate discoveries to deepen mechanistic understanding of what biological and experiential factors influence vocal learning across species. Examples come almost exclusively from one species of songbird, the zebra finch (Taeniopygia guttata), as it shares multiple key features of vocal learning with humans and has the most comprehensive data across biological dimensions.
8.2 Parallels in Human and Zebra Finch Vocal Learning
8.2.1 Behavioral Similarities
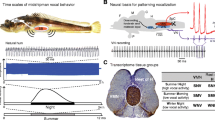
The behavioral similarities between human speech and language acquisition and song learning in songbirds have been described before (Doupe and Kuhl 1999) and in this volume (Sakata and Woolley, Chap. 1; Sakata and Yazaki-Sugiyama, Chap. 2). A few broad strokes describing the process of zebra finch developmental song learning here will serve to ground later discussions of the genome within this context (Fig. 8.1).
Juvenile zebra finches learn to produce song much like humans acquire speech. Shown is a timeline of post-hatch (P) development in the zebra finch. Birds hatch on P1 and are considered adults at P90. Male zebra finches memorize the song of an adult tutor during social interactions; the ability for tutor song memorization is normally restricted to a critical period that spans P30–65. There is a period of ~30 days when the young males can memorize tutor song. Through a process of sensorimotor error correction, the young males use feedback to shape their own song to eventually largely resemble the tutor’s song structure. As adults, each male sings one crystalized song that is based on his experiences with the tutor but is unique to him. In this way, song is culturally transmitted. (also see Sakata and Woolley, Chap. 1; Sakata and Yazaki-Sugiyama, Chap. 2)
Zebra finches live in a rich social environment throughout their lives; multi-family colonies can have over one-hundred members (Zann 1996). Song is a tool to communicate in this complex environment. Only male zebra finches can sing. Each male sings one stereotyped song his entire adult life, which can be 80% similar to another bird’s song but is unique. The combination of song uniqueness and stability facilitates individual recognition within the colony across time. Males sing as part of their courtship display (called directed song) and after a female chooses a male, the pair forms a tight, exclusive, and long-lasting mate bond. Interestingly, male zebra finches also sing in nonreproductive contexts. This undirected song is thought to function as rehearsal to maintain song stereotypy (see Podos and Sung, Chap. 9).
Males acquire their song during posthatch development. They have one critical period, defined as a restricted phase when a specific experience has profound and lasting effects on a particular brain system and behavior (more on critical periods in Sect. 8.6). During the critical period for song, they can form an auditory representation of an adult tutor song in a process termed tutor song memorization. Using their memory of the tutor’s song as a kind of template, young males undergo a process of sensory-motor error correction during which they alter their initial, immature vocalizations such that they come to resemble the syllable structure and order of the tutor’s song. From the multi-modal integration of sensory, sensorimotor, and motor learning, each male enters adulthood with a single, unique, and highly stereotyped song (Gobes et al. 2017; London 2017).
8.2.2 Functional Similarities in Neural Circuits
The neural circuitry for learned vocal communication does not superficially appear equivalent in songbirds and humans. The human cortex has a typical mammalian laminar structure whereas songbird brains are organized into nuclei. However, the differences in macroscopic organization belie the remarkable conservation in both form and function of neural circuits across the species.
Like humans, songbirds have brain areas specialized for the learning and production of vocal communication (Fig. 8.2) (Petkov and Jarvis 2012; Pfenning et al. 2014). This includes brain areas for processing complex auditory stimuli that are integrated with the social context (the auditory forebrain or auditory lobule, AL; see Table 8.1 for all abbreviations) (details in Woolley and Woolley, Chap. 5), a cortico-basal ganglia-thalamic-cortical loop for fine-grained sensorimotor practice and performance (HVC, Area X, DLM, LMAN)(see Murphy, Lawley, Smith, and Prather, Chap. 3; Leblois and Perkel, Chap. 4), and areas that drive precise syringeal motor outputs coordinated with tongue and respiratory patterns (HVC, RA, nXIIts, nRA)(see Elie and Theunissen, Chap. 7). Indeed, direct functional analogies between song and language areas have been proposed (Bolhuis et al. 2010; Phenning et al. 2014), and patterns of gene expression have revealed that, although laminar structure is not a characteristic of avian brains, the genes that characterize cortical layers in mammals are expressed in songbird brains (Dugas-Ford et al. 2012; Karten 2013). Thus, songbirds and humans may share deeper features for vocal learning than the word “parallel” suggests. Future discussions may find a more suitable word that moves beyond the implication that vocal learning in humans and songbirds is outwardly similar but occurs without any shared mechanistic underpinnings.
Songbirds have a specialized neural circuit for learned vocal communication that has equivalencies in humans. Shown is a schematic of a pseudosagittal section through an adult male zebra finch brain. Gray outlines large regions of the brain and the regional names are labeled in gray. Colored circles and arrows depict the location and connections among major nodes of the song circuit. These avian brain areas function as in human circuits for speech and language. Dark red nodes are telencephalic nuclei similar to human cortical regions, the blue node designates the basal ganglia (Area X), purple outlines a thalamic relay nucleus, and green denotes hindbrain nuclei (nXIIts for syringeal control). The circuit is interconnected, and commonly divided into the posterior motor pathway (solid arrows), the anterior forebrain pathway (dashed arrows), and the auditory forebrain pathway (dotted arrows)(see Sakata and Yazaki-Sugiyama, Chap. 2; Woolley and Woolley, Chap. 5 for further discussions of these circuits) AL, the auditory forebrain, also called the auditory lobule; DLM, medial portion of the dorsolateral thalamic nucleus; HVC (used as a proper name); LMAN, lateral magnocellular nucleus of the anterior nidopallium; nXIIts, tracheosyringeal nucleus of the twelfth cranial nerve; RA, robust nucleus of the arcopallium
8.3 Why Study Genomes?
What are the elements of the genome that provide both the program to reliably organize neural systems receptive to experience and the dynamic, experience-triggered responses required for processes like learned vocal communication? One of the fascinating features of the genome is that there are multiple timescales that influence how it functions. On the longest scale, there are evolutionary pressures. Evolutionary influences are reflected in the genomic DNA as sequence changes in specific regions of stability that can give clues to function when compared across generations or species. On the shortest scale, transcription can be regulated within minutes of an experience, and experiences accumulate in epigenetic modifications to the DNA and histones (proteins around which the genomic DNA wraps). Collectively, these features of the genomic DNA and histone proteins serve as a kind of biological archive of an individual, representing selection pressures placed on prior generations and the accumulation of lifetime experiences to date. The following section provides a brief overview of chromatin, the combination of genomic DNA and histone proteins, spanning the time frames relevant to the emergence of complex learned behavior (Fig. 8.3).
Chromatin (the combination of genomic DNA and histone proteins) is a significant bridge between patterns of behavior and their neurobiological substrates. The role of natural selection in shaping the genetics of an individual (the nucleotides that comprise the sequence of genomic DNA) is well-appreciated. Genomic DNA codes for proteins and RNAs that create neural systems that support behavior (receptivity, green arrows). Genomic function can also be altered via neural responses to the environment (responsiveness, blue arrows). These same interdependencies exist on the timescale of a lifetime (dashed arrows)
8.3.1 Genomic Sequence as the Central Dogma of Molecular Biology
The sequence of genomic DNA describes the genetics of an individual. Evolutionary-scale selection pressures influence DNA sequence, which manifests in signatures specific to particular species and even individuals of a particular familial lineage. DNA sequence is important because it encodes the RNAs and proteins that construct the cells that comprise brain areas and networks; the set of RNAs and proteins required for cellular structure and function is one way to define the output of the genome (Fig. 8.3).
The Central Dogma of Molecular Biology states that genomic DNA is transcribed into messenger RNA (mRNA), which is transported from the nucleus to the cytoplasm for translation into protein (Fig. 8.4). The Central Dogma explains production of proteins that include the building blocks of cell morphology, the enzymes for cellular metabolism, and creation of signaling molecules, including neurotransmitters, the receptors for cell-cell signaling, the transcription factor proteins that regulate gene expression, and the hormones that can signal whole-body states to the brain. The focus on protein-coding portions of the genome has led to major breakthroughs in how brains are organized during maturation, how experience can be rapidly signaled through neural circuits, and how cells and synapses are remodeled to encode experience as memory. Researchers are collecting massive datasets of mRNAs with the aim of profiling sets of processes that occur in the brain, and experiments guided by the Central Dogma continue to elucidate neural processes. Much of the sequence of the genome, however, does not code for proteins, suggesting that research must look beyond coding regions to understand genome function.
The multilevel and interdependent regulation of genomic function provides the complement of proteins for the organization of neural circuits and for their ability to respond after experience. A major output of the genome is various proteins. The top row shows the linear relationship between genomic DNA, mRNA, and protein as described in the central dogma of molecular biology. In part because of whole genome sequencing, the number of interacting features of the process of regulating the genome has grown in complexity. The bottom row shows examples of our more advanced understanding of genomic activation at the level of DNA, RNA, and protein. Amino acids: D, aspartic acid; H, histidine; L, leucine; M, methionine; S, serine; V, valine (portions modified from Genome Research Limited; https://www.yourgenome.org/facts/what-is-the-central-dogma)
8.3.2 Moving Beyond the Central Dogma to a More Complex View of the Genome
The sequencing and assembly of whole animal genomes, including those of the human and zebra finch (Venter et al. 2001; Warren et al. 2010), forced the revelation that the sequence of an individual’s protein-coding DNA alone would not elucidate the causal relationships between an individual’s genetics and how his/her brain functions to support behavior. DNA sequence that does not code for a protein, once considered “junk” DNA, is also important because much of this DNA is essential to regulate transcription (Fig. 8.4) (The ENCODE Project Consortium, 2012). For example, regulatory regions of the genomic DNA that do not get synthesized into proteins are essential for understanding how transcription is directed and can have species-specific functional consequences without significant alteration in the protein-coding sequence (Hammock and Young 2005; Gilad et al. 2006).
There are also many types of RNAs that are categorized as noncoding because, unlike mRNAs, they are not translated into proteins. The most abundant noncoding RNAs (ncRNAs) are ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) that directly contribute to protein translation. There are also small and large ncRNAs that specifically and combinatorially regulate the availability of mRNAs and, therefore, the population of proteins (Wang et al. 2012; Hollins and Cairns 2016). Understanding the diversity and functional roles of ncRNAs in the brain continues to grow rapidly. New discoveries will likely be essential for understanding how the genome organizes the neural circuits for vocal learning and regulates the dynamic genomic response to sensory and motor experiences that shape vocal communication patterns (Nguyen et al. 2018; Marty and Cavaillé 2019)
Further, we now know that an individual’s environment works through the brain to alter the structure–not the sequence–of the genome. Structural changes in the genome are mediated by epigenetic mechanisms (see Sect. 8.5.3). Epigenetic modifications include methylation of the genomic DNA itself, methylation of RNA, and post-translational modifications (PTMs) of histone proteins (Strahl and Allis 2000; Fu et al. 2014a; Allis and Jenuwein 2016). Both DNA and histone modifications locally alter the probability that the associated protein-coding gene will be transcribed and, therefore, meaningfully shift the output of the genome. RNA methylation alters the stability and structure of RNAs that influence how available they are for function (Zhao et al. 2016). Epigenetic modifications are almost exclusively accumulated within an individual’s lifetime and thus represent a more immediate process than natural selection to affect the relationship between genomic function, brain, and behavior. Additionally, the effects of chromatin modifications are known to alter long-distance three-dimensional conformations of genomic DNA. This three-dimensional folding provides another way that chromatin structure influences transcription by bringing distal portions of genomic DNA, for example a regulatory region, in close proximity to a protein-coding gene where it can alter transcription (Lin et al. 2018).
8.3.3 Chromatin Is the Biological Hinge Point for Neural Receptivity and Responsivity
Learned behaviors such as vocal communication require an organized neural circuit that can be remodeled; the brain must be receptive to, and properly responsive to, experience. Chromatin can be regarded as a master regulator of cellular function and, therefore, is a hub for mechanisms of receptivity and responsiveness (Fig. 8.3). Chromatin regulates the synthesis of sets of RNAs and proteins, which build cells that assemble into circuits that form nodes in neural systems that can drive learned behavior. Receptivity results from the establishment of those neural systems. Responses to environmental stimuli or signals generated by the individual’s own behavior trigger receptive neural systems, and the neural plasticity required for learning and memory results from chromatin function. Sections 8.3.1 and 8.3.2 provided a brief overview of some of the major chromatin components that are currently understand as regulators of the output of the genome. An integrated view of the response from cell membrane signaling to the nucleus and the mechanisms by which the genome’s response remodels cells for plasticity has been well-conceptualized as a “genomic action potential” (Clayton 2000).
8.3.4 Summary of Chromatin in Brain and Behavior
Understanding of the complexity of genomic regulation continues to grow. As described in Sect. 8.1, the genome is the basis for nearly all of the structural and functional components of a brain, and genomic features are highly conserved evolutionarily. Each new discovery provides additional power to meaningfully investigate what makes songbirds and humans capable of the complex behavior of vocal learning as bound by shared chromatin-related processes. No one chromatin feature is likely to explain how an individual acquires learned vocalizations. Rather, vocal learning is almost certainly a combination of genes, molecular signaling activation patterns, ncRNAs, and epigenetic mechanisms that determine neural organization and function. There is an obvious need to keep investigating each of these features independently and how they may interact.
8.4 The Case of FoxP2
Given the features common to both human speech acquisition and zebra finch song learning, discoveries linking genomic features with speech abilities in humans can be mechanistically dissected in zebra finches. The ability to perform invasive measures and manipulations in the zebra finch is invaluable for uncovering fundamental features of vocal learning in humans, too. Investigations into the function of the gene for the Forkhead box protein P2 (FoxP2) demonstrate how fruitful such interspecies investigations can be.
8.4.1 Identification and Analysis of Human FoxP2
Mutations in the FoxP2 gene were first discovered through classic pedigree analysis of a family known as KE. Members of the KE family displayed a speech disorder characterized by language disruptions and deficits in controlling the fine movements of the tongue and lips needed to produce speech clearly. Pedigree inspection suggested a single-gene etiology with autosomal dominance, and subsequent genetic analysis identified a narrow genomic region that contained the FoxP2 gene (Fisher et al. 1998; Lai et al. 2001). An independent case of speech disruption convincingly connected mutations in FoxP2 to disorders in speech (MacDermot et al. 2005).
The type of gene mutation that affected KE family members was a single point mutation, which alters one amino acid in the entire protein structure. The human FoxP2 protein is over 700 amino acids long, so one change seems to have a disproportionate impact. The reason such a small genetic change can have such a dramatic effect on complex behavior is because the FoxP2 protein is a transcription factor. Transcription factors bind to gene regulatory regions of the DNA to influence the expression level of the associated genes. Each transcription factor can regulate many protein-coding genes. The KE family’s FoxP2 mutation was in the segment of the gene that codes for its DNA-binding domain; therefore, the transcriptional regulatory function of FoxP2 amplifies the impact of its mutation in the KE family (Nudel and Newbury 2013).
Curiously, the FoxP2 protein sequence is almost 100% conserved across species. That fact made the discovery of two amino acids that were seemingly unique to humans compared to nonhuman primates even more compelling (Enard et al. 2002). The species difference raised the possibility that these two changes conferred language ability to humans. Notably, these amino acids are not the same as those associated with speech disorders. Instead, the human-specific amino acids are found within coding exon 7. This region of exon 7 does not encode for a known protein functional domain, suggesting that the effect of the amino acid changes may broadly influence the three-dimensional structure of FoxP2 in ways that affect its function other than its direct ability to bind DNA. Despite the insights gained from the myriad of clinical and comparative FoxP2 studies, a number of questions about the importance and mechanisms of FoxP2 function remained unanswered from these endeavors.
8.4.2 Mechanistic Questions About FoxP2 Function Addressed in Songbirds
Findings in humans set up three big research questions about FoxP2 functions in vocal communication that are best answered with a nonhuman animal model:
-
1.
Do species-typical FoxP2 sequences dictate the ability for learned vocal communication?
-
2.
What does the pattern of FoxP2expression tell us about its function in learned vocal communication?
-
3.
What are the downstream genes regulated by FoxP2 that may explain its influence on learned vocal communication?
The first question was tackled by doing a comparative analysis of FoxP2 transcript sequences across multiple species, including vocal learners (humans and songbirds) and vocal nonlearners (mice and birds that are not songbirds) (Haesler et al. 2004; Scharff and Haesler 2005). These analyses yielded two important insights. First, sequence comparison showed a remarkably high degree of predicted protein sequence conservation across species: >98% (Teramitsu et al. 2004; Haesler et al. 2004). In comparative analyses, greater sequence similarity provides evidence that a specific stretch of sequence has functional significance. Data indicate that mammals and birds last shared a common ancestor more than 300 million years ago; thus, the nearly identical protein sequence across the four groups of vertebrates tested suggests that the function of FoxP2 is also unchanged (O’Leary et al. 2013; Prum et al. 2016). Second, the amino acids that distinguished human FoxP2 from other primate gene sequences—the individual changes that were postulated to confer vocal learning capability—were not observed in songbirds. In other words, the sequence of the FoxP2 gene does not systematically sort with vocal learning ability; genetics did not indicate that specific variants of the FoxP2 gene predict vocal learning in a species.
The second question focuses on ways to infer the function of FoxP2 in vocal learning by examining its gene expression patterns in brain areas required for song. For example, one of the structural brain differences in affected KE members compared to other members was found in the basal ganglia (Vargha-Khadem et al. 1998). The basal ganglia have several functions, including the learning and production of finely tuned motor skills like those required for speech (see Leblois and Perkel, Chap. 4). In humans, it is not possible to definitively determine if neural differences in affected KE family members were the cause of the speech impairments or if they arose after years of impaired speech production. However, it is possible to construct an argument for a causal relationship in zebra finches. Indeed, FoxP2 is expressed in the songbird basal ganglia (Area X; Fig. 8.2), starting at the earliest stages of neural development (Teramitsu et al. 2004). It is expressed in the major cell type of the striatal portion of the basal ganglia in humans and songbirds: the medium spiny neurons (Haesler et al. 2004; Kreitzer 2009). Further, its transcription is rapidly reduced in Area X when males sing (Scharff and Adam 2013), indicating that FoxP2 may be involved in the process of developmental song learning.
These studies laid essential groundwork to implicate FoxP2 in the same component of vocal communication in zebra finches as was affected in the KE family. One causal test would be to reduce FoxP2 production in Area X and ask if the resulting song phenotype was disrupted; reducing the abundance FoxP2 in the zebra finch would be functionally akin to a human gene mutation that minimizes the function of FoxP2. FoxP2 knockdown was first reported in 2007 and, indeed, reducing FoxP2 levels in Area X of juvenile males during the phase of song learning when they depend on it for sensorimotor learning led to deficits in song structure (Haesler et al. 2007).
In addition, because of the patterns of regulated transcription when birds sing, overexpression could disrupt vocal learning (Murugan et al. 2013; Heston et al. 2018) as there are multiple systems that follow a “Goldilocks scenario” whereby too much or too little of a cellular process prevents the signal-to-noise ratio required to convey information. With expression of designer receptors exclusively activated by designer drugs (DREADDs), it is possible to inhibit or potentiate cell firing using a nonbiological ligand (Urban and Roth 2015; Roth 2016). For existing DREADDs, the intended ligand was an antibiotic called clozapine N-oxide (CNO), although it can be reverse metabolized into clozapine, resulting in unintended effects that were not always accounted for in early studies. However, DREADDs expressed in Area X revealed a complicated relationship with LMAN in the execution of moment-to-moment variability in adult song production (Heston et al. 2018). Generally, these types of manipulations, which cannot be done in humans, were essential to support the hypothesis that FoxP2 mutations can have causal effects on basal ganglia function that lead to deficits in vocal production patterns.
The baseline and singing-regulated pattern of FoxP2 transcription, as well as results from its manipulated expression patterns and data on Area X function, were consistent with the notion that FoxP2 contributes to vocal production. Thus, it was necessary to address the third question that requires identification of the genes that FoxP2 transcriptionally regulates. This is important because perturbations in the availability of these factors would be most directly related to deficits in basal ganglia function and vocal production.
Several experimental strategies can be employed to discover individual genes or sets of genes regulated by a transcription factor. One strategy is to survey the genome for the short DNA regulatory sequences that FoxP2 proteins recognize as locations for binding and then determine which protein-coding genes are associated with those regulatory regions. One of the individual genes identified in this way was Contactin Associated Protein Like 2 (CNTNAP2)(Fisher and Scharff 2009). CNTNAP2 is an intriguing protein because it is an adhesion molecule that affects cellular properties that direct cell-to-cell communication (Fisher and Scharff 2009). In the zebra finch, the FoxP2-CNTNAP2 interaction was confirmed in brain areas relevant to vocal communication and was regulated in ways consistent with a role in song production (Panaitof et al. 2010; Condro and White 2014; Adam et al. 2017). A second strategy is to manipulate FoxP2 in zebra finches and identify genes differentially expressed as a result. Using a combination of experimental conditions, testable hypotheses regarding the shifting gene networks can be formulated. This strategy has revealed transcriptional networks in Area X that are perturbed by alterations in FoxP2 DNA binding and that may be specifically involved in developmental song-motor learning (Burkett et al. 2018). Other strategies that revealed miRNAs that regulate FoxP2 mRNAs (see Sect. 8.7 for more information) added an epigenetic layer of modulation onto the genetic and genomic processes by which FoxP2 influences vocal learning (Shi et al. 2013; Fu et al. 2014b).
8.4.3 FoxP2 Summary
As the case study of FoxP2 demonstrates, studies in songbirds provide integrative mechanistic data on a variety of questions that would be difficult to acquire in humans. For example, work in songbirds can reveal when a gene is expressed during development, if its expression is restricted to specific cell types, and how its expression is localized across the neural network for vocal learning. Songbird research also allows causal manipulations that link gene expression to behavior. Each of these general advantages are part of the FoxP2 story. In short, studies in zebra finches can confirm but also expand our knowledge base of how genes can affect the acquisition and regulation of complex behaviors such as vocal communication.
8.5 Genome-Brain-Behavior Interdependencies in Songbirds Inform On Human Communication
Songbirds have demonstrated value for identifying neural and experiential mechanisms that influence learned vocal communication. For example, the zebra finch model permits meaningful investigation into both developmental and adult processes and the separation of auditory and motor components of learned vocal communication. Importantly, these studies can be combined with different readouts of genomic function in the context of cells, circuits, and the whole animal. Further, the nuclear structure of the songbird brain confers advantages for investigation because functional areas can be identified with the naked eye. Visible structures allow for specific and reliable anatomical localization of genomic features to test how they segregate among behavioral components of vocal learning. The following subsections describe the difference between genetics, the static DNA sequence, and genomics, the dynamic regulation of transcriptional output of the genome. In the following subsections, chromatin is positioned as the center of both upstream and downstream molecular processes that regulate genome function. In addition, sex differences, epigenetics, and immediate early gene expression are used as a backdrop to understand the interplay between genes, brain, and behavior in vocal learning.
8.5.1 Sex Differences as a Path to Mechanism (Receptivity)
Behaviors typically acquired during development, such as vocal communication, depend on the construction of a neural network that is sufficiently organized to encode the experiences that guide vocal output patterns. Biological sex is one organizing process that affects the brain and begins very early in maturation; therefore, biological sex may provide some key insights into mechanisms that create the neural network for vocal learning.
In humans, sex differences in language disorders are documented, although they are more difficult to parse mechanistically than in songbirds. Often speech and language deficits are associated with broader syndromes, such as autism spectrum disorders (Halladay et al. 2015), attention deficit hyperactivity disorder (Cohen et al. 2000), and schizophrenia (Walder et al. 2006), which can complicate investigation. Further, sex differences can be blurred by the broad range of individual variation and can be influenced by environmental factors such as differential application of special education intervention measures (Barbu et al. 2015; Kvande et al. 2018). These factors mean that differences in speech and language abilities between boys and girls are often difficult to parse by gender alone. Additionally, there are some distinctions in how language function is represented in the brains of adult men and women, but determining if the structural distinctions are a cause or effect of potential sex differences in speech and language is nearly impossible (Wallentin 2009; Etchell et al. 2018). Ultimately, the effect of sex on vocal communication phenotypes is not clear.
In zebra finches, however, there is one of the starkest sex differences described in brains. The sex difference is apparent with the naked eye, and it encompasses the motor and sensorimotor nuclei of the song circuit (remember, females cannot produce song, but males can) (Nottebohm and Arnold 1976; Wade and Arnold 2004). These differences make zebra finches valuable for discovering mechanistic underpinnings of sex differences with the potential to uncover organizational principles of vocal learning circuits.
The dogma of sexual differentiation, as defined from mammalian studies, is essentially that gonadal steroids do it all: a gene on the sex chromosomes determines whether or not testes develop, and testicular secretions form a masculine brain and body (Arnold and Schlinger 1993; Arnold et al. 2004). Gonadally derived steroids are undoubtedly powerful mediators of maturation, but they do not explain all of sexual differentiation. In fact, understanding how the genome is regulated in the brain has revealed how the dogma fails to fully explain how brains are organized. Notably, steroids can be locally synthesized in the brain to influence specific functions (see Remage-Healey, Chap. 6), and the complement of sex chromosomes themselves alter the phenotype of brain cells, in part because sex chromosome-linked genes are transcribed in the brain (Arnold et al. 2004; London et al. 2009b). Because extreme differences can be useful for initial discovery steps, the zebra finch affords a unique opportunity to consider mechanisms of vocal circuit organization that can then be applied to more subtle systems like those in humans.
8.5.1.1 Direct Genetic Effects: Sex Chromosome Gene Expression
Each sex chromosome can code for unique protein variants of the same gene. Because males and females differ in their complement of sex chromosomes—mammalian males are XY and females are XX; avian males are ZZ and females are ZW—genetic differences can directly influence the brain via neural expression of genes localized to sex chromosomes. Importantly, the sex chromosome complement that is expressed in rodent brains affects essential cellular features, including the abundance of neurons with specific neurochemical phenotypes (Carruth et al. 2002; Arnold and Chen 2009). Quite possibly, similar processes occur in humans.
Perhaps the most striking demonstration of sex chromosome gene expression in the brain comes from a naturally occurring gynandromorphy: an individual that is nearly perfectly split hemispherically as male and female (Agate et al. 2003). In a zebra finch gynandromorph, transcription of a gene variant found on the W chromosome, ASW, was restricted to the side of the bird that matched the female plumage. Further, the version of the protein kinase gene PKCi localized to the Z chromosome was more highly expressed in the right hemisphere, matching the side where male-typical plumage was present (there is minimal Z-inactivation and therefore male:female dosage of Z-linked genes is typically close to 2:1) (Agate et al. 2003; Itoh et al. 2007).
Because the entirety of the gynandromorph’s brain would have been exposed to the same environment of circulating gonadal hormones, this individual provided a rare opportunity to test for direct genetic effects on song circuit sexual dimorphism. Indeed, when the volumes of major singing nuclei were measured, they were larger in the right hemisphere (greater Z gene-expressing and male typical plumage) than in the left, as compared to the more symmetrical volumes found in normal males. Interestingly, however, nuclei in the left (W gene-expressing female hemisphere) were also partially masculinized, indicating that while direct genetic effects likely determined the majority of the song nuclei volumes, there may be additional, local signaling molecules such as neurally synthesized steroids (see Sect. 8.5.1.2) that could act on both hemispheres.
8.5.1.2 Effects of Regulated Gene Expression: Autosomal Chromosomes
The singing circuitry is masculinized by the steroid estradiol. All steroids are synthesized from cholesterol molecules through a series of enzymatic conversions. The spatiotemporal distribution of steroidogenic enzymes, therefore, determines the steroid-producing capacity of particular regions. In developing and adult zebra finches, circulating levels of estradiol are indistinguishable in males and females, and estradiol is produced within the brain, indicating that the enzyme required for the last step of estrogen synthesis is present in the brain (Adkins-Regan et al. 1990; Schlinger and Arnold 1992). However, the song circuitry is still masculinized in genetic males gonadectomized early in development, leading to the hypothesis that all five major enzymes needed to convert cholesterol to estradiol are in the brain (Arnold 1975; Arnold and Schlinger 1993). Steroids synthesized from enzymes expressed within the brain itself are termed neurosteroids to distinguish them from steroids originating in the periphery (London et al. 2009b).
Sex differences in the zebra finch song circuit are detectable as early as nine days after hatching (posthatch day 9, P9)(Gahr and Metzdorf 1999; Kim et al. 2004), and estradiol is most masculinizing during the first week of posthatch life (Adkins-Regan et al. 1994). Consistent with neurosteroid contributions to these early processes of sexual differentiation, the genes for steroidogenic enzymes are expressed at P1 and P5 (London and Schlinger 2007). Interestingly, transcription occurs within the cells along the lateral ventricle, especially where neurogenesis is particularly prolific (Dewulf and Bottjer 2005), indicating that neurosteroids may be affecting brain organization at its earliest stages. Genes for steroidogenic enzymes continue to be expressed in song nuclei at later developmental ages and are transcribed in different combinations across brain areas (London et al. 2006). Each steroid can have multiple effects and drive fundamental elements of brain organization in other systems (London 2016); thus, control of neurosteroid production via genomic regulation may influence sex differences in vocal communication abilities and may hold some potential for understanding sex differences in communication disorders in humans.
8.5.2 Dynamic Experience-Dependent Processes for Vocal Learning
Learning is by definition a dynamic process and, therefore, cannot be completely explained by the static genetic sequence of an individual. Long term memory formation requires new transcription and translation as a result of an experience. There are several ways to examine mechanisms that modulate patterns of transcription and translation and to detect patterns of genomic activation that correspond to experience-dependent neural processing. The following subsections provide an overview for how two of these strategies, immediate early gene (IEG) expression and patterns of epigenetic modifications, can promote our understanding of gene, brain, and behavioral interdependencies.
8.5.2.1 Immediate Early Genes as a Tool for Anatomical and Functional Discovery
Transcription of IEGs is regulated by pre-existing transcription factor proteins that can be activated within milliseconds after cell firing. Immediate early genes, therefore, are among the first new mRNAs and proteins generated after a cell has fired, and they can be used to identify cells and brain areas that were active during an experience. Their transcription depends on cellular activation, but the absence of IEG expression does not mean that a cell has not fired. Instead, IEGs represent the activation of selective molecular processes triggered by cell firing. Their expression provides two levels of information: the cell has fired and a specific molecular process was initiated (Tischmeyer and Grimm 1999; Minatohara et al. 2016). ZENK, an IEG with multiple names (zif268, egr-1, ngfi-a, krox-24) has been most comprehensively studied in zebra finches (Mello et al. 2004). This gene was described as a necessary component of behavioral learning in other animal systems and has been leveraged to probe features of vocal learning and production (Bozon et al. 2002; Alberini 2009). Sections 8.5.2.1.1 and 8.5.2.1.2 provide an overview of adult song-recognition learning and developmental song learning in the zebra finch and how IEG expression studies lend insight into the molecular cascades initiated in learning and plasticity.
8.5.2.1.1 Sensory Learning in Adults: Song Recognition Learning
Male and female adult zebra finches learn to recognize the songs of others, which helps them to distinguish individuals within their colony. In adults, IEGs such as ZENK are rapidly transcribed after a bird hears songs of other zebra finches that are unfamiliar to them (Mello et al. 2004). Interestingly, the numbers of cells that induce ZENK transcription are reduced as exposure to the same song is repeated (Dong and Clayton 2008, 2009). The characteristics of this process are consistent with canonical definitions of habituation, which is a form of nonassociative learning (Thompson and Spencer 1966; Rankin et al. 2009).
Two regions in the brain show ZENK transcription upon hearing novel conspecific songs: the caudomedial nidopallium (NCM) and caudal mesopallium (CM). Both NCM and CM are major components of the auditory forebrain, which is a composite brain area outside of the traditional song circuit for motor control (Vates et al. 1996). Interestingly, NCM and CM are tightly interconnected with an adjacent primary auditory cortical area, Field L. However, unlike NCM and CM, neurons in Field L do not express ZENK in response to hearing novel conspecific songs (Mello et al. 2004). This underscores the molecular specificity of the IEG response. Because NCM and CM receive their information from Field L, neurons in Field L must be firing for NCM and CM to be activated, yet the molecular cascade to transcribe ZENK is not triggered in Field L cells. With one exception, dusp-1, data to date indicate that hearing complex and biologically meaningful sounds initiates genomic regulation in NCM and CM, but not Field L (Horita et al. 2010). Of course, future studies may provide additional examples of IEGs transcribed in primary auditory areas and greater complexity of IEG-mediated auditory processing will be revealed.
The initial discovery of song-induced IEG expression led to further insights about the mechanistic complexity of sensory song learning in adults. For example, the numbers of cells in NCM and CM expressing IEGs positively correlate with the biological relevance of the particular song being heard, including directed versus undirected song (Mello et al. 2004), mate versus unfamiliar male (Woolley and Doupe 2008), and higher-order structural song complexity (Lin et al. 2014). All of these findings were consistent with the idea that IEG activation was a feature of NCM and CM neurons responding to higher-order features of the song, not simply responding to the basic acoustic features. This was confirmed by studies that demonstrated that changing the physical and social contexts in which song was experienced could alter the magnitude of the genomic response in NCM and CM (Kruse et al. 2004; Vignal et al. 2005) and by experiments that demonstrated how prior social contacts influenced the response to hearing song (Woolley and Doupe 2008; Lin et al. 2014). These findings also indicated that NCM and CM processing is more complicated than pure auditory processing. Instead, contextual features modulate the processing of song stimuli (see Woolley and Woolley, Chap. 5). How and why this can occur is unknown.
8.5.2.1.2 Sensory Learning in Juveniles: Tutor Song Memorization
As juveniles, males memorize their tutor’s song, and this sensory learning serves as the foundation of their own song structure (see Sakata and Yazaki-Sugiyama, Chap. 2). There is also evidence that young females learn the song of their dad, though this is more difficult to assess because females cannot sing (Braaten et al. 2006; Braaten et al. 2008).
The baseline density of ZENK expression in NCM and CM was as high at the age of onset of the critical period for tutor song memorization as it was in adults who had heard biologically relevant song (Jin and Clayton 1997; Roper and Zann 2006). Therefore, it was possible that ZENK expression was necessary for tutor song memorization to occur. In the early 2000s, it was not technically possible to directly “knock-down” a single gene in the songbird brain, but it was possible to disrupt an upstream protein signal that was necessary for ZENK transcription, ERK (extracelluar signal regulated kinase) (Cheng and Clayton 2004). Combining a transient disruption of ZENK induction in NCM and CM during a juvenile male’s tutor experiences prevented him from producing high fidelity copies of the tutor’s song (London and Clayton 2008). This experiment thus made two contributions: (1) molecular regulation of experience-dependent IEG expression was causally linked with sensory learning, and (2) the NCM/CM regions were identified as essential anatomical loci for tutor song memorization.
8.5.2.1.3 Motor Learning and Production
A large portion of the song circuit integrates sensory information with motor output during developmental song learning and drives song production across the lifespan of the individual (see Sakata and Yazaki-Sugiyama, Chap. 2; Murphy, Lawley, Smith, and Prather, Chap. 3). A lot of attention is given to the highly stereotyped nature of adult song, but IEG studies have provided some interesting mechanistic clues about how subtle changes in song structure, especially across maturation and in different social contexts, may occur.
When adult males sing, IEGs are expressed within the components of the circuit that control song production (Jarvis and Nottebohm 1997; Kimpo and Doupe 1997). Singing adults do not need to hear for ZENK to be expressed in the motor circuitry: deafened birds show the same pattern of induction across motor circuitry as hearing birds when they sing undirected song (Jarvis and Nottebohm 1997). Thus, at least by maturity, the motor circuit appears to generate song independent of the signaling required for NCM and CM sensory processing in juveniles.
Singing does not induce the same distribution of ZENK across the motor circuitry in juvenile males compared to adult males (Jin and Clayton 1997). In particular, ZENK mRNA is restricted to the posterior portion of RA (robust nucleus of the arcopallium) in adults but is expressed throughout RA in P35 males who have just begun the process of vocal learning (Fig. 8.1). There is an intriguing shift in synaptic inputs from LMAN and HVC onto neurons in RA as the birds develop their song; perhaps the synaptic anatomy and genomic function are functionally connected, and the developmental change in connectivity alters the distribution of molecular signaling in RA (Mooney and Konishi 1991; Aronov et al. 2008). If so, this would be an interesting example of how IEG induction reveals specific molecular processes underlying neural function.
Additionally, within adults, the social context in which the male sings changes the neural pattern of ZENK expression. After an adult male sings directed song to a female, ZENK expression in LMAN, Area X, and RA is lower than after the bird sings undirected song, even if he is surrounded by other birds but not singing directly to any of them (Jarvis et al. 1998). Not everyone is comfortable integrating information across different experiments within a study, but it is possible that, unlike what occurs in adults who sing undirected song, directed song induces ZENK in NCM and CM, and this induction during directed song is prevented by deafening (Jarvis et al. 1998). There may be additional modulatory signals that combine with auditory input during directed song to initiate cascades that promote ZENK transcription in these higher-order sensory processing areas. Lastly, it is important to note that there may be additional molecular regulation on IEG function in song control nuclei after transcription has occurred as ZENK protein distributions do not always recapitulate ZENK mRNA patterns (Whitney et al. 2000). More work tracking mRNA and protein dynamics will be needed to understand what, if any, the functional ramifications of this disconnect are.
8.5.2.2 Epigenetics in Adult Song
As introduced in Sect. 8.3.2, epigenetic mechanisms are defined as those that change the function of the genome without altering the sequence of the genomic DNA. There are several types of epigenetic mechanisms: ncRNAs, modifications to DNA (DNA methylation), modifications to RNA (RNA methylation), and a diversity of modifications to histone proteins. All of these processes have the effect of modulating the abundance and mixture of mRNAs available for translation in a cell. Like other chromatin features, epigenetic modifications appear to be highly conserved evolutionarily in terms of function. The understanding of epigenetic mechanisms active in the song circuit is still nascent but will likely grow in the coming years.
The following sections will review recent research that investigated ncRNAs and histone PTMs in adults to demonstrate that these approaches have value in testing mechanisms of recognition learning and vocal production (epigenetic modifications in juvenile zebra finches are discussed in Sect. 8.6). There is much remaining to be discovered about how epigenetic mechanisms influence vocal learning and production.
8.5.2.2.1 miRNAs
The number of transcripts available in the cell for translation can be lowered by miRNAs via binding to short recognition sequences in mRNAs (O’Brien et al. 2018; Gebert and MacRae 2018). Predicting the effect of one miRNA is difficult because each mRNA often includes multiple miRNA recognition sequences, there can be more than one type of miRNA that recognizes the same mRNA, and there may be more than one site per mRNA for a particular miRNA (Kim et al. 2016). Upon the miRNA binding to the mRNA, the mRNA is cleaved by a protein complex recruited by the bound miRNA, preventing the mRNA from being translated into protein (Shukla et al. 2011; O’Brien et al. 2018).
Many more miRNAs have been predicted in the zebra finch than have been functionally studied (Luo et al. 2012). Initial reports, however, indicate that they are involved in auditory processing and song production in adults and may regulate sex-specific responses as well as the availability of FoxP2 (Gunaratne et al. 2011; Shi et al. 2013). For now, these reports demonstrate that there is great potential for additional miRNA investigations. As data are acquired from future studies, miRNAs will likely become recognized as important regulators of the dynamic processes underlying vocal communication.
8.5.2.2.2 Histone Post-Translational Modifications
Like other proteins, histones can be post-translationally modified by the addition of molecular side chains such as phosphate, acetyl, and methyl groups. The specific type of PTM, the number of PTMs, the amino acid that receives the PTM, and the specific histone protein with PTMs all alter chromatin structure, which in turn shifts the probability that the associated DNA regions will be transcribed (Strahl and Allis 2000).
The addition and removal of histone PTMs is performed by a set of enzymes that are often referred to as writers and erasers. The full diversity of histone modifications that act in adult songbirds is not known, but at least one eraser that removes acetyl groups (a histone deacetylase, HDAC) influences adult song recognition. Prior work in rodents demonstrated that accumulation of histone acetylations via HDAC3 deletion enhanced learning and memory such that a subthreshold learning experience was transformed into one that coded a memory that lasted at least 24 hrs (McQuown and Wood 2011; McQuown et al. 2011). In adult zebra finches, pharmacological inhibition of HDAC3 in the auditory forebrain after a subthreshold song playback experience also resulted in neural measures indistinguishable from a more robust experience known to support song recognition learning (Phan et al. 2017). This is consistent with a pattern seen in other systems, including in human auditory processing, in which increased histone acetylation improves learning, although perceptual training alone can have similar effects in humans (Gervain et al. 2013; Van Hedger et al. 2015).
After adult males have sung, epigenetic modifications may also influence the transcriptional probability of genes involved in song stability. One type of histone acetylation (on Lysine 27 of histone H3: H3K27ac) is used to identify regions that can be actively transcribed. Using a procedure called chromatin immunoprecipitation followed by DNA-seq (ChIPseq), analysis of H3K27ac-associated DNA confirmed that there is a set of genes that can be transcribed selectively in each of four major song control nuclei: HVC, LMAN, RA, and Area X (Whitney et al. 2014). Collectively, approximately 2000 genes were found to be actively regulated upon singing across these regions. Discoveries like these open the door to many more investigations about how epigenetic mechanisms can stably and dynamically influence the acquisition and production of vocal communication.
8.5.3 Conclusion of Dynamic Experience-Dependent Processes
Investigations at various levels of genomic function in the zebra finch continue to uncover features of brain circuit organization and function. The advantages of being able to dissociate how genomic regulation influences sensory functions compared to motor functions, evaluate how maturation is controlled, and assess how the genome associates with behavioral measures of learned vocal communication make zebra finches a powerful resource for revealing meaningful genomic function.
8.6 Critical Period Informs on Mechanisms that Modulate Learning Plasticity
One of the most striking features of song learning in the zebra finch is that the process of tutor song memorization is defined by a critical period, as described previously (Fig. 8.1). Further, that same experience has no measurable effect on behavior before or after the critical period. While the critical period “open” may be set by genetically determined maturational processes, the “close” depends on experience, and preventing the relevant experience extends the age at which brain and behavior remain open to its effects (Figs. 8.1, 8.5) (e.g., Knudsen 2004; Takesian and Hensch 2013). Because brain areas with critical periods undergo extreme fluctuations in experience-dependent plasticity, they are excellent models in which to disentangle chromatin mechanisms of maturation from those required for experience-dependent learning.
Multiple timescales of song learning can be measured with appropriate levels of neurobiology. (a) A schematic depiction of a timeline highlighting the critical period for tutor song memorization and major elements of developmental song learning. A paradigm of acute song playbacks (left, in red circle), in which juveniles were either played song or left in silence, revealed sex and age differences in mTOR cascade activation, quantified by the density of S6 phosphorylation (pS6), in the caudomedial nidopallium (NCM) and caudal mesopallium (CM) (histograms on right; asterisk indicates statistical significance p < 0.05). (b) Juvenile males reared under a tutor (reared with an adult male; example song depicted) during P30–65 have their critical period closed, while song-isolated juveniles (reared with adult females that do not sing but do produce calls, which have similar acoustic features to some song syllables, example depicted) have an extended critical period (extension is denoted by dashed red line). Without additional tutoring, tutored males sing faithful copies of the tutor’s song as adults and isolate-reared males sing an abnormally structured song. (c) Measures of epigenetic histone post-translational modifications in the auditory forebrain from tutored and isolate-reared males showed distinct proportions of repressive and active chromatin. (d) These data lead to a hypothesis linking tutor experience with critical period closing, mediated by epigenetic mechanisms. (portions of this figure were modified from Ahmadiantehrani and London 2017a; London 2017; Ahmadiantehrani et al. 2018; Kelly et al. 2018)
In the zebra finch, there is evidence that the critical period regulates the social sensory process of tutor song memorization rather than the motor rehearsal component of developmental song learning (see Sakata and Yazaki-Sugiyama, Chap. 2). Notably, preventing young males from hearing song between P30–65 extends the age at which tutoring successfully shapes the juvenile’s song structure (London 2017); but birds reared without tutoring still sing, with essentially no alteration in the patterns of gene expression in song control nuclei (Mori and Wada 2015). Interestingly, males can be raised with adult females that produce innate calls with acoustic structures similar to some types of song syllables, but this auditory and social experience is not sufficient to close the critical period (Fig. 8.5) (Eales 1985, 1987). Because tutor song memorization relies on genomic and molecular processes in NCM and CM during tutor experiences (London and Clayton 2008; Ahmadiantehrani and London 2017a), it is possible that an important neural locus for the critical period is in the auditory forebrain.
Evidence for genomic regulation as a determining factor in the onset and offset of the critical period exists in the auditory forebrain across biological scales (Fig. 8.5). Single-gene expression patterns were the first indication that baseline and experience-dependent changes occurred in the auditory forebrain at ages relevant to the critical period (Jin and Clayton 1997). In fact, the profile of auditory forebrain RNAs are different at baseline (in silence) and after hearing song when compared between males prior to and after the critical period (London et al. 2009a). Additionally, hearing song activates a particular molecular signaling cascade, the mechanistic target of rapamycin (mTOR), in male NCM and CM at P30, the onset of the critical period (Roper and Zann 2006), but not one week earlier and not in females who cannot sing at either age (Fig. 8.5) (Ahmadiantehrani and London 2017a). Manipulations of mTOR signaling in juvenile males experiencing a tutor prevent high fidelity tutor song copying (Ahmadiantehrani and London 2017a). The same mTOR manipulations do not have the equivalent effect on adult song recognition learning as they do on tutor song memorization (Ahmadiantehrani et al. 2018). Interestingly, mTOR regulates the initiation of protein synthesis (Hoeffer and Klann 2010), suggesting that proteins controlled by mTOR activation may underlie the onset and specificity of the critical period for tutor song memorization (Ahmadiantehrani and London 2017a; Ahmadiantehrani et al. 2018).
Importantly, critical periods are distinguished from age-limited learning by the fact that it is experience, not age, that closes the phase of experience-dependent neural plasticity. Knudsen (2004) reviews the logic of this tie between experience, behavior, and neural plasticity in several systems. Therefore, it is important to dissociate age from prior tutor experience to understand how tutor song memorization prevents subsequent tutor experience from being learned. Epigenetic mechanisms that alter the balance of repressed and active chromatin, especially surrounding genes involved in transcription and translation, are associated with closed and extended critical periods in P67 male auditory forebrain (Fig. 8.5) (Kelly et al. 2018). The data support the hypothesis that tutor song memorization leads to an accumulation of repressive chromatin that limits transcriptional and cellular responses following a tutor encounter that occurs after P67 (Fig. 8.5). Because memory formation requires new transcription and translation, this limited genomic response prevents additional tutor song memorization. On the other hand, tutor song isolation during the critical period leads to relatively more abundant regions of active chromatin. Active chromatin has high potential for transcription, which would then permit the complement of new proteins required for memory formation to be synthesized even when the male experiences a tutor past the normal close of the critical period (Fig. 8.5).
In general, language acquisition is believed to be most effective earlier in development, but whether or not there are one or more critical periods that control vocal learning in humans is still contested (Van Hedger et al. 2015; Werker and Hensch 2015). Zebra finch studies that causally relate neural mechanisms with vocal learning capability may therefore provide some insights into these issues as well as broader criteria for learning.
8.7 The Value of Comparative Studies
A diversity of song learning styles exists across the 5000 extant species of songbirds (Clayton et al. 2009). There is great value in leveraging the experiments nature performed, and discovery can come from comparative studies. Just like comparisons of FoxP2 sequences between humans and nonhuman primate species suggested functional properties of the protein in vocal learning and production, the comparison of genomic features across birds with unique behavioral vocal learning and production traits can identify possible connections between genomes and behavior. The continued accumulation of genomic and transcriptomic sequences from a variety of species will bolster these investigations.
Comparative studies can address several questions. For example, are there genetic or genomic features that differentiate species of birds that can learn vocalizations from those that cannot? Second, are genomes regulated similarly across species as they transition within a lifetime from being able to learn to being closed to learning? Like zebra finches, parasite birds (birds that lay their eggs in the nests of another host species) might have restricted learning abilities to support learning of their own species’ song rather than their host species’ song. Alternatively, some species continue to engage in vocal learning through adulthood (see Sakata and Woolley, Chap. 1). For example, starlings can acquire song every day. Do mechanisms of the zebra finch critical period also characterize the open and closed phases of learning in other species? Third, some animals have vocal plasticity, permitting them to adjust acoustic features, such as frequency, based on their environment, but they do not have stable communication patterns as a result of experience with other individuals (i.e., vocal learning). Might the genome uncover features intermediate between those with inflexible vocal patterns and those with vocal learning? Finally, similarities and differences in genomic regulation between the different nuclei of the song circuit could elucidate key neural processes that distinguish learning from nonlearning brain areas and distinguish mechanisms that are needed for sensory, sensorimotor, and motor learning.
8.8 The Future of Genome-Brain-Behavior Investigations
The following sections outline a few avenues of research that can provide important insights into genome function with regard to complex behaviors like vocal communication.
8.8.1 Genomic Identification of Cellular Specifications
All cells of an individual have the same genomic sequence: how genomic function is regulated determines the transcription of genes characteristic to distinct cell types. The coarsest categorization of cell types in the brain is neurons and glia, but there are many subtypes still being discovered.
Cell types are informative for function because, for example, they can be excitatory or inhibitory, have different electrical properties, distinct sets of membrane receptors, and a range of metabolic activities. Vocal learning occurs during development, and cell types shift across the lifetime, which may be a partial explanation for restricted vocal learning capabilities despite continued experience. Finally, the different brain nuclei in the song circuit are functionally specialized, and the entire circuit is specialized compared to the rest of the brain, suggesting specific complements of cell types within each region.
Much more discovery-based research needs to be done, perhaps by taking advantage of epigenetic features such as H3K27ac-mediated epigenetic approaches for identifying regulatory regions considered enhancers. However, beautiful molecular work has been done dissecting and describing the cellular populations within Area X, a complicated composite brain area for vocal plasticity (Person et al. 2008). Another approach was a widespread initiative termed the Zebra Finch Expression Brain Atlas (ZEBrA) that catalogued patterns of gene expression across the entire adult male brain (http://www.zebrafinchatlas.org/). The result is a resource of over 600 genes (and growing) and the anatomical location of their expression that can be used to identify genes potentially specialized for particular facets of vocal learning. This type of resource pairs well with community-wide initiatives that describe the levels of gene expression across various brain areas, ages, and conditions and provide gene-based insights into neural functions (Replogle et al. 2008). Finally, methods that measure the entire population of extant RNAs from a brain area at a particular time may be useful for identifying cellular traits that relate song nuclei to human speech and language regions (Pfenning et al. 2014).
8.8.2 Gene Editing and Genetic Manipulation Technologies
Transitioning from descriptive to causal experimental design requires the ability to do manipulations. The zebra finch has the advantage that experiential manipulations have known functional consequence on the future pattern of song and the ability to learn song past P65. Genetic manipulation has been immensely powerful in revealing mechanisms in rodents and other animals, and the use of such manipulations is advancing in birds.
There are now three reports of transgenic zebra finches that have cells with altered genomes throughout their bodies. One set of transgenic birds expresses green fluorescent protein (GFP), which is useful for cellular anatomy (Agate et al. 2009). Another line of transgenic birds was created with manipulated activation levels of a transcription factor called CREB: these birds displayed deficits in developmental song learning (Abe et al. 2015). And finally, there are transgenics with a mutated Huntington gene (HTT) that display motor song performance issues interpreted as consistent with motor deficits in human Huntington’s disease (Liu et al. 2015). Although currently they are highly inefficient to create and can be difficult to breed, transgenic zebra finches will be powerful tools moving forward as their creation becomes easier.
An alternative to germ-line creation of transgenics is local gene manipulations using either in vivo electroporation or viral vectors for construct delivery (Heston and White 2015; Ahmadiantehrani and London 2017b). This method has the advantage of not directly manipulating function in brain regions other than the one of interest. There are currently several ways to deliver gene constructs to manipulate brain cell function. It is possible to overexpress a gene or to mutate or eliminate a gene using CRISPR/Cas9 technology (Sander and Joung 2014). Alternatively, cells can be inhibited or activated on very short timescales by expressing optogenetic channels that use light as a ligand or on longer timescales via DREADD receptors, which were designed to have antibiotics as their ligand (Boyden 2011; Smith et al. 2016). These strategies can be combined with other elements of manipulation constructs that confer temporal and cell-type specificity (Hisey et al. 2018; Xiao et al. 2018), and they are beginning to reveal how areas of the traditional sensorimotor singing circuitry operate (Roberts et al. 2012; Tanaka et al. 2018; Xiao et al. 2018). There is much more to be done to optimize and popularize strategies for genetic manipulation, but as knowledge about genome-brain-behavior connections becomes more sophisticated, it will be essential to have these tools available to researchers asking the variety of questions that are collectively required to compile a comprehensive understanding of how genomic features contribute to vocal learning. As a last note, pharmacological agents can be beneficial alternatives to genetic manipulation especially because their effects are transient, permitting carefully timed disruptions in function during experience and in controlled situations.
8.9 Summary
Previously, researchers thought that knowing an individual’s genetic sequence would unlock his/her individual biology. It is now clear that genetics is not equivalent to biological determinism; rather, the complexity of our lives alters how our genomes function, and it is the interplay between chromatin, brain, and experience that ultimately produces measurable behavioral patterns such as speech and language. This chapter summarized the basic features of chromatin structure and function and their relationship to vocal learning in humans and songbirds. Strategies to probe the chromatin-brain-behavior interdependencies were explored and discussed with respect to understanding how a complex behavior such as vocal learning emerges. An underlying goal was to demonstrate how cross-species investigations—human to avian and across avian species—can be fruitful when open-minded researchers take advantage of the unique and shared properties of genomes to create meaningful research comparisons. Investigations that focus on various components of genomic function will advance recognition of the fundamental shared features across species that go deeper than superficial parallels.
References
Abe K, Matsui S, Watanabe D (2015) Transgenic songbirds with suppressed or enhanced activity of CREB transcription factor. Proc Natl Acad Sci USA 112(24):7599–7604. https://doi.org/10.1073/pnas.1413484112
Adam I, Mendoza E, Kobalz U, Wohlgemuth S, Scharff C (2017) CNTNAP2 is a direct FoxP2 target in vitro and in vivo in zebra finches: complex regulation by age and activity. Genes Brain Behav 16(6):635–642. https://doi.org/10.1111/gbb.12390
Adkins-Regan E, Abdelnabi M, Mobarak M, Ottinger MA (1990) Sex steroid levels in developing and adult male and female zebra finches (Poephila guttata). Gen Comp Endo 78(1):93–109. https://doi.org/10.1016/0016-6480(90)90051-M
Adkins-Regan E, Mansukhani V, Seiwert C, Thompson R (1994) Sexual differentiation of brain and behavior in the zebra finch: critical periods for effects of early estrogen treatment. J Neurobiol 25(7):865–877. https://doi.org/10.1002/neu.480250710
Agate RJ, Grisham W, Wade J, Mann S, Wingfield J, Schanen C, Palotie A, Arnold AP (2003) Neural, not gonadal, origin of brain sex differences in a gynandromorphic finch. Proc Natl Acad Sci U S A 100(8):4873–4878. https://doi.org/10.1073/pnas.0636925100
Agate RJ, Scott BB, Haripal B, Lois C, Nottebohm F (2009) Transgenic songbirds offer an opportunity to develop a genetic model for vocal learning. Proc Natl Acad Sci U S A 106(42):17963–17967. https://doi.org/10.1073/pnas.0909139106
Ahmadiantehrani S, London SE (2017a) Bidirectional manipulation of mTOR signaling disrupts socially mediated vocal learning in juvenile songbirds. Proc Natl Acad Sci U S A 114(35):9463–9468. https://doi.org/10.1073/pnas.1701829114
Ahmadiantehrani S, London SE (2017b) A reliable and flexible gene manipulation strategy in posthatch zebra finch brain. Sci Rep 7:43244. https://doi.org/10.1038/srep43244
Ahmadiantehrani S, Gores EO, London SE (2018) A complex mTOR response in habituation paradigms for a social signal in adult songbirds. Learn Mem 25(6):273–282. https://doi.org/10.1101/lm.046417.117
Alberini CM (2009) Transcription factors in synaptic plasticity and learning and memory A2 - squire, Larry R. In: Encyclopedia of neuroscience. Academic, Oxford, pp 1081–1092. https://doi.org/10.1016/B978-008045046-9.00828-7
Allis CD, Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat Rev Genet 17(8):487–500. https://doi.org/10.1038/nrg.2016.59
Arnold AP (1975) The effects of castration and androgen replacement on song, courtship, and aggression in zebra finches (Poephila guttata). J Exp Zool 191(3):309–326. https://doi.org/10.1002/jez.1401910302
Arnold AP, Chen X (2009) What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol 30(1):1–9. https://doi.org/10.1016/j.yfrne.2008.11.001
Arnold AP, Schlinger BA (1993) Sexual differentiation of brain and behavior: the zebra finch is not just a flying rat. Brain Behav Evol 42(4–5):231–241. https://doi.org/10.1159/000114157
Arnold AP, Xu J, Grisham W, Chen X, Kim YH, Itoh Y (2004) Minireview: sex chromosomes and brain sexual differentiation. Endocrinology 145(3):1057–1062. https://doi.org/10.1210/en.2003-1491
Aronov D, Andalman AS, Fee MS (2008) A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science (New York, NY) 320(5876):630–634. https://doi.org/10.1126/science.1155140
Barbu S, Nardy A, Chevrot J-P, Guellaï B, Glas L, Juhel J, Lemasson A (2015) Sex differences in language across early childhood: family socioeconomic status does not impact boys and girls equally. Front Psychol 6:1874–1874. https://doi.org/10.3389/fpsyg.2015.01874
Bolhuis JJ, Okanoya K, Scharff C (2010) Twitter evolution: converging mechanisms in birdsong and human speech. Nat Rev Neurosci 11(11):747–759. https://doi.org/10.1038/nrn2931
Boyden ES (2011) A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biol Rep 3:11. https://doi.org/10.3410/B3-11
Bozon B, Davis S, Laroche S (2002) Regulated transciption of the immediate-early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus 12(5):570–577. https://doi.org/10.1002/hipo.10100
Braaten RF, Petzoldt M, Colbath A (2006) Song perception during the sensitive period of song learning in zebra finches (Taeniopygia guttata). J Comp Psychol 120(2):79–88. https://doi.org/10.1037/0735-7036.120.2.79
Braaten RF, Miner SS, Cybenko AK (2008) Song recognition memory in juvenile zebra finches: effects of varying the number of presentations of heterospecific and conspecific songs. Behav Proc 77(2):177–183. https://doi.org/10.1016/j.beproc.2007.10.010
Burkett ZD, Day NF, Kimball TH, Aamodt CM, Heston JB, Hilliard AT, Xiao X, White SA (2018) FoxP2 isoforms delineate spatiotemporal transcriptional networks for vocal learning in the zebra finch. eLife 7:e30649. https://doi.org/10.7554/eLife.30649
Carruth LL, Reisert I, Arnold AP (2002) Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci 5(10):933–934. https://doi.org/10.1038/nn922
Cheng HY, Clayton DF (2004) Activation and habituation of extracellular signal-regulated kinase phosphorylation in zebra finch auditory forebrain during song presentation. J Neurosci 24(34):7503–7513. https://doi.org/10.1523/JNEUROSCI.1405-04.2004
Clayton DF (2000) The genomic action potential. Neurobiol Learn Mem 74(3):185–216. https://doi.org/10.1006/nlme.2000.3967
Clayton DF, Balakrishnan CN, London SE (2009) Integrating genomes, brain and behavior in the study of songbirds. Curr Biol 19(18):R865–R873. https://doi.org/10.1016/j.cub.2009.07.006
Cohen NJ, Vallance DD, Barwick M, Im N, Menna R, Horodezky NB, Isaacson L (2000) The Interface between ADHD and Language Impairment: An Examination of Language, Achievement, and Cognitive Processing. J Child Psychol Psychiatry and Allied Disc 41 (3):353–362. doi:undefined
Condro MC, White SA (2014) Distribution of language-related Cntnap2 protein in neural circuits critical for vocal learning. J Comp Neurol 522(1):169–185. https://doi.org/10.1002/cne.23394
Dewulf V, Bottjer SW (2005) Neurogenesis within the juvenile zebra finch telencephalic ventricular zone: a map of proliferative activity. J Comp Neurol 481(1):70–83. https://doi.org/10.1002/cne.20352
Dong S, Clayton DF (2008) Partial dissociation of molecular and behavioral measures of song habituation in adult zebra finches. Genes Brain Behav 7(7):802–809. https://doi.org/10.1111/j.1601-183X.2008.00423.x
Dong S, Clayton DF (2009) Habituation in songbirds. Neurobiol Learn Mem 92(2):183–188. https://doi.org/10.1016/j.nlm.2008.09.009
Doupe AJ, Kuhl PK (1999) Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci 22:567–631. https://doi.org/10.1146/annurev.neuro.22.1.567
Dugas-Ford J, Rowell JJ, Ragsdale CW (2012) Cell-type homologies and the origins of the neocortex. Proc Natl Acad Sci U S A 109(42):16974–16979. https://doi.org/10.1073/pnas.1204773109
Eales LA (1985) Song learning in zebra finches - some effects of song model availability on what is learnt and when. An Behav 33 (Nov):1293–1300. doi:Doi https://doi.org/10.1016/S0003-3472(85)80189-5
Eales LA (1987) Song learning in female-raised zebra finches - Another look at the sensitive phase. An Behav 35:1356–1365. doi:Doi https://doi.org/10.1016/S0003-3472(87)80008-8
Enard W, Przeworski M, Fisher SE, Lai CSL, Wiebe V, Kitano T, Monaco AP, Pääbo S (2002) Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418:869–872. https://doi.org/10.1038/nature01025
Etchell A, Adhikari A, Weinberg LS, Choo AL, Garnett EO, Chow HM, Chang SE (2018) A systematic literature review of sex differences in childhood language and brain development. Neuropsychologia 114:19–31. https://doi.org/10.1016/j.neuropsychologia.2018.04.011
Fisher SE, Scharff C (2009) FOXP2 as a molecular window into speech and language. Trends Genetics 25(4):166–177. https://doi.org/10.1016/j.tig.2009.03.002
Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18(2):168–170. https://doi.org/10.1038/ng0298-168
Fu Y, Dominissini D, Rechavi G, He C (2014a) Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genetics 15:293–306. https://doi.org/10.1038/nrg3724
Fu L, Shi Z, Luo G, Tu W, Wang X, Fang Z, Li X (2014b) Multiple microRNAs regulate human FOXP2 gene expression by targeting sequences in its 3′ untranslated region. Mol Brain 7:71. https://doi.org/10.1186/s13041-014-0071-0
Gahr M, Metzdorf R (1999) The sexually dimorphic expression of androgen receptors in the song nucleus Hyperstriatalis Ventrale pars Caudale of the zebra finch develops independently of gonadal steroids. J Neurosci 19(7):2628–2636. https://doi.org/10.1523/jneurosci.19-07-02628.1999
Gebert LFR, MacRae IJ (2018) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20:21–37. https://doi.org/10.1038/s41580-018-0045-7
Gervain J, Vines BW, Chen LM, Seo RJ, Hensch TK, Werker JF, Young AH (2013) Valproate reopens critical-period learning of absolute pitch. Front Syst Neurosci 7:102. https://doi.org/10.3389/fnsys.2013.00102
Gilad Y, Oshlack A, Smyth GP, Speed TP, White K (2006) Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440:242–245. https://doi.org/10.1038/nature04559
Gobes SMH, Jennings RB, Maeda RK (2017) The sensitive period for auditory-vocal learning in the zebra finch: consequences of limited-model availability and multiple-tutor paradigms on song imitation. Behav Process 163:5–12. https://doi.org/10.1016/j.beproc.2017.07.007
Gunaratne PH, Lin YC, Benham AL, Drnevich J, et al. (2011) Song exposure regulates known and novel microRNAs in the zebra finch auditory forebrain. BMC genomics 12, doi.org/10.1186/1471-2164-12-277
Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C (2004) FoxP2 expression in avian vocal learners and non-learners. J Neurosci 24(13):3164–3175. https://doi.org/10.1523/JNEUROSCI.4369-03.2004
Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C (2007) Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus area X. PLoS Biol 5(12):e321. https://doi.org/10.1371/journal.pbio.0050321
Halladay AK, Bishop S, Constantino JN, Daniels AM, Koenig K, Palmer K, Messinger D, Pelphrey K, Sanders SJ, Singer AT, Taylor JL, Szatmari P (2015) Sex and gender differences in autism spectrum disorder: summarizing evidence gaps and identifying emerging areas of priority. Mol Autism 6(1):36. https://doi.org/10.1186/s13229-015-0019-y
Hammock EA, Young LJ (2005) Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308:1630–1634. https://doi.org/10.1126/science.1111427
Heston JB, White SA (2015) Behavior-linked foxP2 regulation enables zebra finch vocal learning. J Neurosci 35(7):2885–2894. https://doi.org/10.1523/JNEUROSCI.3715-14.2015
Heston JB, Jt S, Day NF, Coleman MJ, White SA (2018) Bidirectional scaling of vocal variability by an avian cortico-basal ganglia circuit. Physiol Rep 6(8):e13638–e13638. https://doi.org/10.14814/phy2.13638
Hisey E, Kearney MG, Mooney R (2018) A common neural circuit mechanism for internally guided and externally reinforced forms of motor learning. Nat Neurosci 21(4):589–597
Hoeffer CA, Klann E (2010) mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci 33(2):67–75. https://doi.org/10.1016/j.tins.2009.11.003
Hollins SL, Cairns MJ (2016) MicroRNA: small RNA mediators of the brains genomic response to environmental stress. Prog Neurobiol 143:61–81. https://doi.org/10.1016/j.pneurobio.2016.06.005
Horita H, Wada K, Rivas MV, Hara E, Jarvis ED (2010) The dusp1 immediate early gene is regulated by natural stimuli predominantly insensory input neurons. J Comp Neurol 518(14):2873–2901. https://doi.org/10.1002/cne.22370
Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP (2007) Dosage compensation is less effective in birds than in mammals. J Biol 6(1):2. https://doi.org/10.1186/jbiol53
Jarvis ED, Nottebohm F (1997) Motor-driven gene expression. Proc Natl Acad Sci U S 94(8):4097–4102. https://doi.org/10.1073/pnas.94.8.4097
Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F (1998) For whom the bird sings: context-dependent gene expression. Neuron 21(4):775–788. https://doi.org/10.1016/S0896-6273(00)80594-2
Jin H, Clayton DF (1997) Localized changes in immediate-early gene regulation during sensory and motor learning in zebra finches. Neuron 19(5):1049–1059. https://doi.org/10.1016/S0896-6273(00)80396-7
Karten HJ (2013) Neocortical evolution: neuronal circuits arise independently of lamination. Curr Biol 23(1):R12–R15. https://doi.org/10.1016/j.cub.2012.11.013
Kelly TK, Ahmadiantehrani S, Blattler A, London SE (2018) Epigenetic regulation of transcriptional plasticity associated with developmental song learning. Proc Royal Soc B: Biol Sci 285(1878):20180160. https://doi.org/10.1098/rspb.2018.0160
Kim D, Sung YM, Park J, Kim S, Kim J, Park J, Ha H, Bae JY, Kim S, Baek D (2016) General rules for functional microRNA targeting. Nat Genetics 48:1517–1526. https://doi.org/10.1038/ng.3694
Kim YH, Perlman WR, Arnold AP (2004) Expression of androgen receptor mRNA in zebra finch song system: developmental regulation by estrogen. J Comp Neurol 469(4):535–547. https://doi.org/10.1002/cne.11033
Kimpo RR, Doupe AJ (1997) FOS is induced by singing in distinct neuronal populations in a motor network. Neuron 18(2):315–325. https://doi.org/10.1016/S0896-6273(00)80271-8
Knudsen EI (2004) Sensitive periods in the development of the brain and behavior. J Cog Neurosci 16(8):1412–1425. https://doi.org/10.1162/0898929042304796
Kreitzer AC (2009) Physiology and pharmacology of striatal neurons. Ann Rev Neurosci 32(1):127–147. https://doi.org/10.1146/annurev.neuro.051508.135422
Kruse AA, Stripling R, Clayton DF (2004) Context-specific habituation of the zenk gene response to song in adult zebra finches. Neurobiol Learn Mem 82(2):99–108. https://doi.org/10.1016/j.nlm.2004.05.001
Kvande MN, Belsky J, Wichstrøm L (2018) Selection for special education services: the role of gender and socio-economic status. European J Sp Needs Edu 33(4):510–524. https://doi.org/10.1080/08856257.2017.1373493
Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413(6855):519–523. https://doi.org/10.1038/35097076
Lin D, Hong P, Zhang S, Xu W, Jamal M, Yan K, Lei Y, Li L, Ruan Y, Fu ZF, Li G, Cao G (2018) Digestion-ligation-only hi-C is an efficient and cost-effective method for chromosome conformation capture. Nat Genetics 50(5):754–763. https://doi.org/10.1038/s41588-018-0111-2
Lin LC, Vanier DR, London SE (2014) Social information embedded in vocalizations induces neurogenomic and behavioral responses. PLoS One 9(11):e112905. https://doi.org/10.1371/journal.pone.0112905
Liu W, Kohn J, Szwed SK, Pariser E, Sepe S, Haripal B, Oshimori N, Marsala M, Miyanohara A, Lee R (2015) Human mutant huntingtin disrupts vocal learning in transgenic songbirds. Nat Neurosci 18(11):1617–1622. https://doi.org/10.1038/nn.4133
London SE (2016) Influences of non-canonical neurosteroid signaling on developing neural circuits. Curr Opin Neurobiol 40:103–110. https://doi.org/10.1016/j.conb.2016.06.018
London SE (2017) Developmental song learning as a model to understand neural mechanisms that limit and promote the ability to learn. Behav Process 163:13–23. https://doi.org/10.1016/j.beproc.2017.11.008
London SE, Clayton DF (2008) Functional identification of sensory mechanisms required for developmental song learning. Nat Neurosci 11(5):579–586. https://doi.org/10.1038/nn.2103
London SE, Schlinger BA (2007) Steroidogenic enzymes along the ventricular proliferative zone in the developing songbird brain. J Comp Neurol 502(4):507–521. https://doi.org/10.1002/cne.21335
London SE, Monks DA, Wade J, Schlinger BA (2006) Widespread capacity for steroid synthesis in the avian brain and song system. Endocrinology 147(12):5975–5987. https://doi.org/10.1210/en.2006-0154
London SE, Dong S, Replogle K, Clayton DF (2009a) Developmental shifts in gene expression in the auditory forebrain during the sensitive period for song learning. Dev Neurobiol 69(7):437–450. https://doi.org/10.1002/dneu.20719
London SE, Remage-Healey L, Schlinger BA (2009b) Neurosteroid production in the songbird brain: a re-evaluation of core principles. Front Neuroendocrinol 30(3):302–314. https://doi.org/10.1016/j.yfrne.2009.05.001
Luo GZ, Hafner M, Shi Z, Brown M, Feng GH, Tuschl T, Wang XJ, Li X (2012) Genome-wide annotation and analysis of zebra finch microRNA repertoire reveal sex-biased expression. BMC Genomics 13(1):727. https://doi.org/10.1186/1471-2164-13-727
MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha-Khadem F, McKenzie F, Smith RL, Monaco AP, Fisher SE (2005) Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet 76(6):1074–1080. https://doi.org/10.1086/430841
Marty V, Cavaillé J (2019) Imprinted small noncoding RNA genes in brain function and behaviour. Curr Opin Behav Sci 25:8–14. https://doi.org/10.1016/j.cobeha.2018.05.009
McQuown SC, Wood MA (2011) HDAC3 and the molecular brake pad hypothesis. Neurobiol Learn Mem 96(1):27–34. https://doi.org/10.1016/j.nlm.2011.04.002
McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA (2011) HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci 31(2):764–774. https://doi.org/10.1523/JNEUROSCI.5052-10.2011
Mello CV, Velho TAF, Pinaud R (2004) Song-induced gene expression: a window on wong auditory processing and perception. Ann N Y Acad Sci 1016(1):263–281. https://doi.org/10.1196/annals.1298.021
Minatohara K, Akiyoshi M, Okuno H (2016) Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front Mol Neurosci 8:78–78. https://doi.org/10.3389/fnmol.2015.00078
Mooney R, Konishi M (1991) Two distinct inputs to an avian song nucleus activate different glutamate receptor subtypes on individual neurons. Proc Natl Acad Sci U S A 88(10):4075–4079
Mori C, Wada K (2015) Audition-independent vocal crystallization associated with intrinsic developmental gene expression dynamics. J Neurosci 35(3):878–889. https://doi.org/10.1523/JNEUROSCI.1804-14.2015
Murugan M, Harward S, Scharff C, Mooney R (2013) Diminished FoxP2 levels affect dopaminergic modulation of corticostriatal signaling important to song variability. Neuron 80(6):1464–1476. https://doi.org/10.1016/j.neuron.2013.09.021
Nguyen TC, Zaleta-Rivera K, Huang X, Dai X, Zhong S (2018) RNA, action through interactions. Trends Genetics 34(11):867–882. https://doi.org/10.1016/j.tig.2018.08.001
Nottebohm F, Arnold AP (1976) Sexual dimorphism in vocal control areas of the songbird brain. Science 194(4261):211–213. https://doi.org/10.1126/science.959852
Nudel R, Newbury DF (2013) FOXP2. Wiley Interdiscip Rev Cogn Sci 4(5):547–560. https://doi.org/10.1002/wcs.1247
O’Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol 9:402–402. https://doi.org/10.3389/fendo.2018.00402
O’Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP, Goldberg SL, Kraatz BP, Luo Z-X, Meng J, Ni X, Novacek MJ, Perini FA, Randall ZS, Rougier GW, Sargis EJ, Silcox MT, Simmons NB, Spaulding M, Velazco PM, Weksler M, Wible JR, Cirranello AL (2013) The placental mammal ancestor and the post–K-Pg radiation of placentals. Science 339(6120):662–667. https://doi.org/10.1126/science.1229237
Panaitof SC, Abrahams BS, Dong H, Geschwind DH, White SA (2010) Language-related Cntnap2 gene is differentially expressed in sexually dimorphic song nuclei essential for vocal learning in songbirds. J Comp Neurol 518(11):1995–2018. https://doi.org/10.1002/cne.22318
Person AL, Gale SD, Farries MA, Perkel DJ (2008) Organization of the songbird basal ganglia, including area X. J Comp Neurol 508(5):840–866. https://doi.org/10.1002/cne.21699
Petkov CI, Jarvis ED (2012) Birds, primates, and spoken language origins: behavioral phenotypes and neurobiological substrates. Front Evol Neurosci 4:12. https://doi.org/10.3389/fnevo.2012.00012
Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, Mountcastle J, Moseley MA, Thompson JW, Soderblom EJ, Iriki A, Kato M, Gilbert MTP, Zhang G, Bakken T, Bongaarts A, Bernard A, Lein E, Mello CV, Hartemink AJ, Jarvis ED (2014) Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346(6215):1256846–1256846. https://doi.org/10.1126/science.1256846
Phan ML, Gergues MM, Mahidadia S, Jimenez-Castillo J, Vicario DS, Bieszczad KM (2017) HDAC3 inhibitor RGFP966 modulates neuronal memory for vocal communication signals in a songbird model. Front Sys Neurosci 11:65–65. https://doi.org/10.3389/fnsys.2017.00065
Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR (2016) A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 534:S7–S8. https://doi.org/10.1038/nature19417
Rankin CH, Abrams T, Barry RJ, Bhatnagar S, Clayton DF, Colombo J, Coppola G, Geyer MA, Glanzman DL, Marsland S, McSweeney FK, Wilson DA, Wu CF, Thompson RF (2009) Habituation revisited: an updated and revised description of the behavioral characteristics of habituation. Neurobiol Learn Mem 92(2):135–138. https://doi.org/10.1016/j.nlm.2008.09.012
Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, Gong G, Hasselquist D, Hernandez AG, Kim R, Lewin HA, Liu L, Lovell PV, Mello CV, Naurin S, Rodriguez-Zas S, Thimmapuram J, Wade J, Clayton DF (2008) The songbird Neurogenomics (SoNG) initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics 9:131. https://doi.org/10.1186/1471-2164-9-131
Roberts TF, Gobes SMH, Murugan M, Ölveczky BP, Mooney R (2012) Motor circuits are required to encode a sensory model for imitative learning. Nat Neurosci 15(10):1454–1459. https://doi.org/10.1038/nn.3206
Roper A, Zann R (2006) The onset of song learning and song tutor selection in fledgling zebra finches. Ethology 112(5):458–470. https://doi.org/10.1111/J.1439-0310.2005.01169.X
Roth BL (2016) DREADDs for neuroscientists. Neuron 89(4):683–694. https://doi.org/10.1016/j.neuron.2016.01.040
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech 32:347–355. https://doi.org/10.1038/nbt.2842
Scharff C, Adam I (2013) Neurogenetics of birdsong. Curr Opin Neurobiol 23(1):29–36. https://doi.org/10.1016/j.conb.2012.10.001
Scharff C, Haesler S (2005) An evolutionary perspective on FoxP2: strictly for the birds? Curr Opin Neurobiol 15(6):694–703. https://doi.org/10.1016/j.conb.2005.10.004
Schlinger BA, Arnold AP (1992) Circulating estrogens in a male songbird originate in the brain. Proc Natl Acad Sci U S A 89(16):7650–7653. https://doi.org/10.1073/pnas.89.16.7650
Shi Z, Luo G, Fu L, Fang Z, Wang X, Li X (2013) miR-9 and miR-140-5p target FoxP2 and are regulated as a function of the social context of singing behavior in zebra finches. J Neurosci 33(42):16510–16521. https://doi.org/10.1523/jneurosci.0838-13.2013
Shukla GC, Singh J, Barik S (2011) MicroRNAs: processing, maturation. target recognition and regulatory functions Mol Cell Pharmacol 3(3):83–92
Smith KS, Bucci DJ, Luikart BW, Mahler SV (2016) DREADDS: use and application in behavioral neuroscience. Behav Neurosci 130(2):137–155. https://doi.org/10.1037/bne0000135
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403(6765):41–45
Takesian AE, Hensch TK (2013) Balancing plasticity/stability across brain development. Prog Brain Res 207:3–34. https://doi.org/10.1016/B978-0-444-63327-9.00001-1
Tanaka M, Sun F, Li Y, Mooney R (2018) A mesocortical dopamine circuit enables the cultural transmission of vocal behaviour. Nature 563(7729):117–120. https://doi.org/10.1038/s41586-018-0636-7
Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA (2004) Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci 24(13):3152–3163. https://doi.org/10.1523/JNEUROSCI.5589-03.2004
The ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. https://doi.org/10.1038/nature11247
Thompson RF, Spencer WA (1966) Habituation: a model phenomenon for the study of neuronal substrates of behavior. Psych Rev 73(1):16–43. https://doi.org/10.1037/h0022681
Tischmeyer W, Grimm R (1999) Activation of immediate early genes and memory formation. Cell Mol Life Sci CMLS 55(4):564–574. https://doi.org/10.1007/s000180050315
Urban DJ, Roth BL (2015) DREADDs (designer receptors exclusively activated by designer drugs): Chemogenetic tools with therapeutic utility. Ann Rev Pharm Toxicol 55(1):399–417. https://doi.org/10.1146/annurev-pharmtox-010814-124803
Van Hedger SC, Heald SLM, Koch R, Nusbaum HC (2015) Auditory working memory predicts individual differences in absolute pitch learning. Cognition 140:95–110. https://doi.org/10.1016/j.cognition.2015.03.012
Vargha-Khadem F, Watkins KE, Price CJ, Ashburner J et al (1998) Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci U S A 95(21):12695–12700. https://doi.org/10.1073/pnas.95.21.12695
Vates GE, Broome BM, Mello CV, Nottebohm F (1996) Auditory pathways of caudal telencephalon and their relation to the song system of adult male zebra finches. J Comp Neurol 366(4):613–642. https://doi.org/10.1002/(SICI)1096-9861(19960318)366:4. <613::AID-CNE5>3.0.CO;2-7
Venter JC, Adams MD, Myers EW, Li PW et al (2001) The sequence of the human genome. Science 291(5507):1304–1351. https://doi.org/10.1126/science.1058040
Vignal C, Andru J, Mathevon N (2005) Social context modulates behavioural and brain immediate early gene responses to sound in male songbird. Eur J Neurosci 22(4):949–955. https://doi.org/10.1111/j.1460-9568.2005.04254.x
Wade J, Arnold AP (2004) Sexual differentiation of the zebra finch song system. Ann N Y Acad Sci 1016:540–559. https://doi.org/10.1196/annals.1298.015
Walder DJ, Seidman LJ, Cullen N, Su J, Tsuang MT, Goldstein JM (2006) Sex differences in language dysfunction in schizophrenia. AmJ psychiatry 163(3):470–477. https://doi.org/10.1176/appi.ajp.163.3.470
Wallentin M (2009) Putative sex differences in verbal abilities and language cortex: a critical review. Brain Lang 108:175–183. https://doi.org/10.1016/j.bandl.2008.07.001
Wang W, Kwon EJ, Tsai L-H (2012) MicroRNAs in learning, memory, and neurological diseases. Learn Mem 19(9):359–368. https://doi.org/10.1101/lm.026492.112
Warren WC, Clayton DF, Ellegren H, Arnold AP et al (2010) The genome of a songbird. Nature 464(7289):757–762. https://doi.org/10.1038/nature08819
Werker JF, Hensch TK (2015) Critical periods in speech perception: new directions. Annu Rev Psychol 66:173–196. https://doi.org/10.1146/annurev-psych-010814-015104
Whitney O, Soderstrom K, Johnson F (2000) Post-transcriptional regulation of zenk expression associated with zebra finch vocal development. Brain Res Mol Brain Res 80(2):279–290. https://doi.org/10.1016/S0169-328X(00)00178-9
Whitney O, Pfenning AR, Howard JT, Blatti CA et al (2014) Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science 346(6215):1256780. https://doi.org/10.1126/science.1256780
Woolley SC, Doupe AJ (2008) Social context–induced song variation affects female behavior and gene expression. PLoS Biol 6(3):e62. https://doi.org/10.1371/journal.pbio.0060062
Xiao L, Chattree G, Oscos FG, Cao M, Wanat MJ, Roberts TF (2018) A Basal Ganglia circuit sufficient to guide birdsong learning. Neuron 98(1):208–221. e205. https://doi.org/10.1016/j.neuron.2018.02.020
Zann RA (1996) The Zebra finch: a synthesis of Field and laboratory studies. Oxford University Press, New York
Zhao BS, Roundtree IA, He C (2016) Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol18:31. doi:https://doi.org/10.1038/nrm.2016.132
Compliance with Ethics Requirements
Sarah E. London declares that she has no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
London, S.E. (2020). Linking Features of Genomic Function to Fundamental Features of Learned Vocal Communication. In: Sakata, J., Woolley, S., Fay, R., Popper, A. (eds) The Neuroethology of Birdsong. Springer Handbook of Auditory Research, vol 71. Springer, Cham. https://doi.org/10.1007/978-3-030-34683-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-34683-6_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34682-9
Online ISBN: 978-3-030-34683-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)