Abstract
Altered metabolism is one of the key hallmarks of cancer. The development of sensitive, reproducible and robust bioanalytical tools such as Nuclear Magnetic Resonance Spectroscopy and Mass Spectrometry techniques offers numerous opportunities for cancer metabolism research, and provides additional and exciting avenues in cancer diagnosis, prognosis and for the development of more effective and personalized treatments. In this chapter, we introduce the current state of the art of metabolomics and metabolic phenotyping approaches in cancer research and clinical diagnostics.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Metabolomics
- Cancer metabolism
- NMR spectroscopy
- Mass spectrometry
- Diagnostics
- Metabolic imaging
- Biofluids
- Tissues
1 Metabolic Alterations in Cancer
It is well known that cancer cells undergo metabolic reprogramming in order to sustain the anabolic requirements of tumorigenesis and cellular proliferation. This can be achieved by mutations in genes regulating oncogenic signaling pathways ultimately interfering with the expression of key metabolic enzymes (Pavlova and Thompson 2016). Generally speaking, cancer cells tend to display enhanced uptake of glucose, amino acids such as glutamine and other nutrients, increased reliance on glycolysis for ATP production, TCA cycle intermediates for biosynthesis and NADPH production (Pavlova and Thompson 2016). These metabolic changes also bring about alterations in metabolite-driven gene regulation and metabolic interactions with the tumor microenvironment (Pavlova and Thompson 2016), which in turn will have implications in tumor progression and invasiveness.
The most well known metabolic change in cancer occurs in central metabolism with the increased use of aerobic glucose metabolism in which cellular glucose import is increased to generate ATP and lactic acid, known as the Warburg effect (Sanderson and Locasale 2018). The enhanced glucose consumption was the basis of the positron-emission tomography (PET) imaging in which a glucose analogue, 18F-fluoro-2-deoxyglucose (FDG), is used to detect tumor activity, enabling cancer diagnosis, staging and treatment follow-up (Zhu et al. 2011). Another important metabolic alteration is the increased glutamine uptake, which was shown to be implicated in important pathways such as the synthesis of NADPH and as a source of nitrogen in the biosynthesis of non-essential amino acids and nucleotides (Pavlova and Thompson 2016). Glutamine can also play an important role in the cellular import of essential amino acids such as leucine, isoleucine, valine, methionine, tyrosine, tryptophan, and phenylalanine by acting as an anti-porter through the LAT1 membrane transporter (Pavlova and Thompson 2016). Similarly to PET-FDG, 18F-fluoroglutamine, a compound analogous to glutamine has been tested in diagnostic PET imaging (Dunphy et al. 2018), and is particularly useful as an alternative to FDG in tissues where glucose utilization is physiologically high, such as in brain tissue. The described applications are some examples of techniques aimed at a few metabolic alterations that motivated the development of diagnostic imaging techniques which are nowadays in current clinical practice. By looking simultaneously to all possible metabolites in a tissue or body fluid, metabolomics, metabolic profiling and phenotyping techniques aim at exploring other tissue- or tumor-specific metabolic alterations, ultimately contributing to the knowledge of disease mechanisms and to the development of diagnostic tools. The current chapter aims at introducing the reader to the field of metabolomics and metabolic phenotyping and illustrate some of the most important applications in the study of cancer metabolism and diagnostic.
2 Metabolomics and Metabolic Phenotyping
Metabolic phenotyping/metabolomics aims to take a holistic view of a biological sample and is broadly defined as the comprehensive measurement and fingerprinting of low molecular weight compounds in biological samples to understand their roles in cellular functions and diseases. Whilst the terminology of metabolomics/metabonomics were introduced in the late 1990s, the concept of utilizing the distinctive color, odor or taste of human urine for clinical applications were documented as early as the sixteenth century. However, it was not until the twentieth century when molecular entities were effectively elucidated from biological samples. Separation of metabolites in urine samples achieved by 2D paper chromatography was reported in 1956 (Dalgliesh 1956) and the technique was successfully applied to identify metabolites associated with cystinuria, argininosuccinic aciduria and Hartnup disease. Development of gas chromatography meant that by the 1970s up to around 250 volatile components could be detected in urine and breath samples. NMR spectroscopy was first applied in 1967 to identify a urinary metabolite associated with an inborn error of metabolism (Tanaka and Isselbacher 1967), and the profiling of multiple chemicals in urine or blood samples by NMR were first reported by Nicholson et al. in 1984. The advent of information technology and the explosion of computational infrastructures in the early 1990s meant statistical techniques were now being developed in earnest to address the data analytics challenges – helping to analyze and visualize the multivariate datasets. Wishart et al. have made great progress in defining the composition of the human serum metabolome, and later the human urine metabolome, and in setting up the Human Metabolome Database (HMDB) containing background information and spectral data of a large collection of compounds (Wishart et al. 2013). More recently, we see a number of well-resourced specialist metabolic phenotyping centers being setup to support large-scale, high-throughput metabolomics serving researchers across the biomedical research communities.
The first studies that look for a large set of metabolites in the context of cancer can be dated to the 1980s, but the technical developments and recognizing the importance of metabolic alterations in cancer, lead to an increased interest in metabolomics applied to cancer research since 2004 (Fig. 19.1). In the first years the number of publications using MS or NMR approaches were very similar. However since 2012 the number of papers with a MS approach outperforms the studies using NMR (Fig. 19.1). The preference for MS in metabolomics studies results from its higher sensitivity, smaller sample size demands and relatively lower operational costs. The technology developments in MS instrumentation, software databases and tools introduced in the last decade, has also permitted an increase in resolution and ease of analysis (Amberg et al. 2017; Emwas 2015; Bingol 2018). Nevertheless, NMR spectroscopy continues to have an important role in cancer metabolomics and more importantly, the combination of NMR and MS approaches provides additional metabolite coverage (Psychogios et al. 2011; Wishart et al. 2013).
Number of papers using metabolomic approaches in cancer research by year (search was performed on March 2019 in Scopus limited to original papers that mention ((“nuclear magnetic resonance” OR NMR) AND Cancer AND (metabolomics OR metabonomics OR “metabolic profile”)) or ((“Mass spectrometry” OR MS) AND Cancer AND (metabolomics OR metabonomics OR “metabolic profile”)) in the title, abstract or keyword)
Important information about metabolic pathways and fluxes can be drawn from metabolic studies using stable isotope tracers, which can be considered as one of the next-generation applications of metabolomics. In this field, NMR methodologies have unique advantages, since it allows the determination of the position of the isotopomers from isotopically enriched metabolites, the identification and structure elucidation of unknown metabolites as well as the analysis of metabolic pathways dynamics in vivo and in situ in cell culture, tissues and whole organisms (Fan et al. 2012; Fan and Lane 2016).
Metabolomics studies are usually divided into two categories: targeted, where only selected metabolites are analyzed, e.g. from one single metabolic pathway; and untargeted studies which do not focus on any particular set of metabolites and all signals from either NMR or MS are analyzed.
2.1 Nuclear Magnetic Resonance (NMR) Spectroscopy
Nuclear Magnetic Resonance Spectroscopy (NMR) is a powerful and versatile analytical technique. It is used in diverse fields from the structural elucidation of macromolecules and small molecules to the quantification of metabolites present in a sample. NMR was discovered in the mid 1940s, by two different groups (Purcell et al. 1946; Bloch et al. 1946) and from the beginning it was used to characterize molecules, with the first commercial spectrometer developed in 1952 (Marion 2013). The basic principle of NMR involves the atomic nucleus. A nucleus with a non-zero nuclear spin (an odd atomic number), when placed in an external magnetic field can absorb and re-emit radiofrequency with a frequency characteristic of the magnetic field acting on the nucleus. The magnetic field of each nucleus in the sample depends on the action of the external magnetic field and the weak magnetic fields each nucleus in its vicinity (Marion 2013). As a result, each nucleus in a molecule has a resonance at a characteristic frequency, which allows the identification and structural characterization of a molecule.
NMR spectroscopy is extremely useful for studying biological systems, since one of the most sensitive nuclei, the hydrogen isotope (1H or proton), has a natural abundance of almost 100%. The protons with similar molecular environment are called equivalent and produce signals in the 1H-NMR spectrum at specific frequencies (Fig. 19.2). The signal intensity is directly proportional to the number of protons that originate the signal, and also to the concentration of the molecule in the sample. The proximity of other nuclei inside the molecule also produces signal splitting (multiplicity) which varies according to the number of nuclei in the vicinity. These characteristics make NMR spectroscopy a very popular and powerful technique for metabolomics. Moreover, NMR is highly reproducible even between different spectrometers (at the same magnetic field strength and similar hardware configurations) and/or operators (Dona et al. 2014). It is also a very versatile technique, making it possible to analyze intact tissues or biofluids, in most of the cases, with minimal sample preparation (Fig. 19.2). The sample is not consumed in the analysis, thus it can be reanalyzed for as long as it remains stable. The application of different NMR techniques enables the identification of unknown compounds and its structure elucidation (Graça et al. 2019). NMR versatility to study biological systems is extended beyond proton to other magnetic nuclei present in organic molecules (e.g. 31P, 13C or 15N) (Gowda and Raftery 2017). An example is the use of 31P-NMR in prostate cancer, which can be used to measure the changes in phospholipid contents in the prostate tissues induced by the carcinoma (Cornel et al. 1993; Komoroski et al. 2011). The major drawback of NMR is the low sensitivity when compared to MS techniques, as it detects compounds with concentrations of >50 μM while MS compounds with concentrations >10–100 nM (Emwas et al. 2019). Recent advances to improve NMR sensitivity included developments of novel pulse sequences, new probes, spectrometers with higher magnetic fields strengths and by applying enhanced signal polarization techniques such as dynamic nuclear polarization (DNP) (Ardenkjaer-Larsen et al. 2015).
Example of NMR spectra of different biofluids used in cancer NMR metabolomics: (a) Ascitic fluid from an ovarian tumor patient; (b) Cerebrospinal fluid (CSF) from a non-Hodgkin lymphoma patient; (c) Serum from an ovarian tumor patient; and (d) Urine from a paraganglioma patient. All spectra were acquired in an 800 MHz spectrometer at 298 K, except the serum which was acquired in a 600 MHz spectrometer at 310 K.A noesygppr1d pulse program was used for the CSF and urine, while in serum and ascitic fluid a cpmgpr1d pulse program was used to suppress the signals from macromolecules (proteins and lipoproteins). Some of the metabolites detected are indicated: 1-formate, 2-histidine, 3-phenylalanine, 4-tyrosine, 5-urea, 6-glucose, 7-ascorbate, 8-lactate, 9-creatinine 10-myo-inositol, 11-creatine, 12-phosphocreatine, 13-glycine, 14-choline, 15-phosphocholine, 16-dimethylamine, 17-citrate, 18-glutamate, 19-pyruvate, 20-acetoacetate, 21-acetone, 22-acetate, 23-glutamine, 24-alanine, 25-3-hydroxybutyrate, 26-valine, 27-leucine, 28-isoleucine
Another challenge in NMR metabolomics is spectral resolution. Biological samples contain hundreds to thousands of metabolites which produce hundreds of NMR signals leading to significant signal overlap (Fig. 19.2), which makes metabolite identification and concentration determination difficult tasks. These challenges have been tackled through the development of spectral deconvolution software such as Chenomx (Chenomx Inc., Edmonton, Canada) and BATMAN (Hao et al. 2012) and comprehensive spectral databases, such as HMDB (Wishart et al. 2013). Two-dimensional NMR experiments (2D NMR), where signals are dispersed into more than one frequency dimension, constitute also an essential tool for metabolite identification in such complex samples (Emwas 2015; Graça et al. 2019). Despite their advantages, the use of 2D NMR as a profiling platform in metabolomics has been hindered by the long experimental times of 1 to several hours. For this reason, 1D NMR experiments are still routinely used and reported in vast majority of the studies. Nevertheless, new fast 2D techniques which reduce significantly the 2D spectral acquisition time, have been introduced in biofluids NMR metabolomics with very promising results. These include ultrafast (UF) NMR and non-uniform sampling (NUS) (Guennec et al. 2014; Marchand et al. 2017).
2.2 Mass Spectrometry Methods
Mass spectrometry (MS) is a very popular and powerful analytical technique which has found particular use for the elucidation of molecular structures and quantitative analysis of small molecules such as metabolites. MS was introduced in the beginning of the twentieth century and gained widespread popularity in the late 1950s. The technique is based on the detection of charged molecules in the gas phase (Glish and Vachet 2003). Because, not all compounds are easily ionizable and volatile, the technique was limited to gaseous samples for several years since its introduction. It was not until the late 1980s that new ionization techniques such as electrospray ionization (ESI) and matrix-assisted desorption-ionization (MALDI) enabled the direct ionization of molecules from the liquid and solid samples, respectively, into the gas phase (Glish and Vachet 2003). While ESI enables the generation of ions from liquid samples by generation of charged micro-particles after passing the sample through a charged needle, in MALDI, the sample is mixed with a light absorbing compound (matrix) which, when excited with laser light promotes de ionization and displacement of ions from the sample (Glish and Vachet 2003; Bodzon-Kulakowska and Suder 2016). Other important group of ionization techniques are the ambient-pressure ionization techniques which enable, for instance, the withdrawal of ions directly from solid samples into the mass spectrometer (Glish and Vachet 2003; Hänel et al. 2019). Among these, desorption-electrospray ionization (DESI) technique has special importance in mass spectrometry imaging, as it will be described in more detail in Sect. 19.3.7.3. In DESI, ions are withdrawn from the sample by a jet of gas and charged micro droplets usually oriented at an angle close to 45° to sample surface, then injected into the ESI MS source (Hänel et al. 2019). Another ionization technique with important applications in imaging is secondary ion MS (SIMS), in which ions are extracted from the sample surface (secondary ions) after collision with primary ions from an inorganic ion beam (Bodzon-Kulakowska and Suder 2016).
Ionization occurs at the inlet of the mass spectrometer, known as the source. After ionization, the ions are transmitted to the mass analyzer, which is composed of a series of charged metal plates under vacuum, where the ions are separated according to their mass-to-charge ratio (m/z), before hitting the detector (Glish and Vachet 2003). A mass spectrum, a representation of the ion abundance as a function of each ion m/z is then produced (Fig. 19.3a).
Mass spectra: (a) Leucine enkephalin peptide acquired by electrospray ionization in negative ion mode (ESI−); (b) human blood serum lipid extract acquired in ESI−, (c) LC-MS chromatogram of human blood serum lipid extract acquired in positive ion mode (ESI+), where mass spectra are acquired continuously during chromatographic separation; RT retention time
In comparison to NMR spectroscopy, MS is more sensitive, also requiring lower amounts of sample. On the other hand, the sample is consumed during analysis because the ions are lost after reaching the detector.
The MS spectrum of a pure compound can be very simple as the one shown in Fig. 19.3a. However, in applications such as metabolomics analysis, where complex mixtures are analyzed, the spectra can become quite convoluted and difficult to interpret. As an example of such complexity an MS spectrum of negatively charged molecules from a human blood serum lipid extract is shown in Fig. 19.3b. Apart from the complexity stemming from the peak overlap, in practice the most abundant ions can suppress the ionization of other ions. To resolve such problems, MS is usually coupled with online compound physical separation such as gaseous- or liquid-chromatography, the techniques being termed gas- or liquid- chromatography – MS (GC-MS and LC-MS respectively) or capillary electrophoresis (CE-MS). GC-MS is commonly applied to volatile samples, whereas LC-MS and CE-MS are usually employed to analyze liquid samples and solid sample extracts (Emwas 2015). An example of a LC-MS chromatogram from a human blood serum lipid extract sample is shown in Fig. 19.3c. It is clear that the resolution and number of observed peaks (ions) increased quite dramatically in comparison to the direct MS analysis (Fig. 19.3c).
Another important feature to MS-based techniques, is that multiple ions can be generated from a single compound. As an example, in ESI one organic compound can ionize by capturing or releasing a proton or by forming adducts with other ions already present in the sample (e.g. [M + H]+, [M + Na]+, [M-H]−, [M + Formate]−, where M represents the organic compound). After ionization, some molecules also break into charged fragments. Moreover, due to the sensitivity of the technique, several forms of each compound containing one or more naturally occurring isotopes such as 13C (1% abundance), isotopologues, can also be detected increasing the complexity of the spectrum. These factors will lead to higher number of peaks than detected molecules in MS datasets, which is something that needs to be accounted for when interpreting the data. Metabolite identification from MS spectra is therefore a non-trivial task. Often, the analyst will need to perform searches with the measured m/z values on publicly available databases and, eventually, run additional ion fragmentation experiments to get more insight into the molecule identity (Emwas 2015).
2.3 Statistical Data Analysis
Metabolomics experiments generate large quantities of data composed of thousands of variables if simultaneous measurements are collected as in untargeted metabolomics experiments. In most cases, metabolomics datasets need additional processing, such as spectral baseline correction, peak alignment, normalization and variable scaling before statistical analysis can be performed. These operations are required to correct for sample dilution, sample preparation and/or analytical bias and to scale the relevant contributions of each variable (Emwas 2015). Adequate statistical analysis tools are then employed to extract meaningful information from the data. Both univariate and multivariate statistical approaches can be utilized for these purposes. However, care should be taken when using univariate analysis tests for untargeted metabolomics data. In those cases, multiple tests are usually performed simultaneously and the risk of false-discovery results is high. In those cases suitable multiple correction strategies should be used (Broadhurst and Kell 2006).
Multivariate analysis (MVA) are the most commonly used methods in untargeted metabolomics. They have the double advantage of generating interpretable statistical summaries of the data, which are necessary to pursue biological and physiological interpretations, and also enabling the development of predictive models of the disease under investigation. MVA methods can be divided into unsupervised methods, where no a priori sample classification or patient information is taken into account in the analysis; and supervised methods, where the information regarding patient diagnostic is included in the analysis (Trygg et al. 2007). Examples of unsupervised analysis methods are Principal Component Analysis (PCA) and Hierarchical Clustering, which are used to investigate similarities between samples and trends in the data. Supervised methods such as Partial Least Squares – Discriminant Analysis (PLS-DA) and related variants (e.g. Orthogonal PLS-DA), Random Forests, Support Vector Machines and other machine learning approaches are commonly used to develop classification models and look for metabolites that correlate with the disease studied (Trygg et al. 2007; Gromski et al. 2015). Supervised methods are obtained in two-stages: (1) model training, where samples of known class are used to generate classification models; and (2) model validation, where subsets of training data or an external sample set (cross-validation and test-set validation, respectively) are used to test the model classification performance (Trygg et al. 2007; Gromski et al. 2015).
3 Applications to Cancer Diagnostics
Metabolomics studies in cancer diagnosis usually involve the comparison of matched groups of patients (at one or more stages of cancer) versus healthy control or benign cases. One or several types of biological material are obtained and analyzed, whose selection is based on the affected organ(s). A remarkable collection of studies on the application of metabolomics to study cancer is available in the literature. It is beyond the scope of this chapter to provide a systematic review of all studies performed to date for all cancer types. Instead applications of metabolomics and metabolic phenotyping to diagnosis, prognosis and treatment monitoring of major cancer types are illustrated with studies from 2008 to 2019 period, organized by sample type.
3.1 Blood Serum and Plasma
Blood serum and plasma are the most studied biological fluids in cancer metabolomics, as they reflect metabolite levels entering systemic circulation and directly provide an accessible snapshot of the physiological condition of an individual without the need of tissue biopsies. Blood metabolites could be valuable biomarkers for early disease detection. For example, the median survival interval of patients with pancreatic cancer is currently less than 12 months and one study has identified elevated blood branched-chain amino acids as an early risk factor in human pancreatic adenocarcinoma development (Mayers et al. 2014). Using data from targeted LC-MS methods comparing plasma samples from 450 patients to their matched controls collected before the onset of the disease, the same study found that elevated blood branched-chain amino acids were associated with a twofold increase in future risk in developing pancreatic cancer (Mayers et al. 2014). Multiple reports indicated that alteration of circulating amino acids including tryptophan, glutamine, glutamate, phenylalanine and branched chain amino acids and lysophosphatidylcholine (C18:0, C18:2) could potentially serve as useful diagnostic biomarkers for pancreatic tumors (Sakai et al. 2016; Fukutake et al. 2015; Akita et al. 2016) which may be linked to pancreatic adenocarcinoma-associated cachexia, insulin resistance or hyperglycemia.
Tumors located in different organ sites could have distinctive footprint on the blood metabolome. For example elevated levels of circulating ketone bodies (including 3-hydroxybutyric acid), sugars and free fatty acids, and lower levels of glycolytic and TCA metabolites have previously been reported in ovarian cancer patients (N = 158) which may be the consequence of increased activity of fatty acid oxidation in specific tumor organ sites (Hilvo et al. 2016). Furthermore, it has been shown that metabolomics performed on plasma and serum samples could be applied to monitor treatment response in patients. A number of studies have demonstrated that pharmacodynamic response to inhibitors targeting oncogenic signaling could be successfully monitored in patient’s plasma samples. One such study was able to show that changes in phosphatidylcholines and sphingomyelins levels were observed in responder patients with advanced melanoma treated with a mitogen-activated protein kinase (MEK) inhibitor, and that pre-treatment levels of a panel of lipids were predictive of inhibitor treatment response (Ang et al. 2017). Furthermore, it has been shown in a separate study that time and dose-dependent response to Phosphoinositide 3-kinases (PI3K) inhibitor could be observed in patients enrolled in a phase I dose-escalation trial (Ang et al. 2016), demonstrating plasma metabolomics could be a valuable resource for translating and validating preclinical findings in patients. Also, plasma metabolomics have been applied to predict future cancer risk. A Danish study analyzed plasma samples from 838 women by 1H-NMR, where half of the women had developed breast cancer between the time of enrolment in the study and the follow-up date. The inclusion of the NMR data in the risk predictive model increased its sensitivity and specificity to above 80%, and glycerol, ethanol and formate, were amongst the metabolites contributing to the prediction model (Bro et al. 2015).
3.2 Urine
Urine is a noninvasive, accessible and concentration and volume-rich biofluid for clinicians to collect, and many urinary metabolomics studies have focused on tumors located in the urinary tract. For example, it has been reported that the levels of metabolites involved in glycolysis and fatty acid oxidation are altered in patients with bladder tumors (N = 138) compared to control subjects (N = 121) and this may be related to changes to carnitine transferase and pyruvate dehydrogenase complex expression in the patient group (Jin et al. 2014). One study has identified dopamine 4-sulfate, aspartyl-histidine, and tyrosyl-methionine to be discriminatory between non-muscle invasive bladder cancer patients (N = 167) and healthy controls (N = 117), with higher levels of tryptophan metabolites in urine found patients with higher grade tumor (Cheng et al. 2018). Kidney cancer has also been investigated, and in particular urinary levels of acylcarnitines have been found to discriminate patients with low- and high- grade tumors (Ganti et al. 2012).
In addition, urine has been applied to study tumors that are remote from the urinary tract, and has been successful in differentiating patients with malignancies ranging from prostate, lung and gastrointestinal cancers such as gastric cancer, from their healthy controls (Dinges et al. 2019). For example, with NMR metabolomics urinary 2-hydroxyisobutyrate, 3-indoxylsulfate, and alanine, were identified as discriminatory between patients with gastric cancer (N = 43), healthy individuals (N = 40) and nonmalignant gastric conditions (N = 40) with a classification accuracy of 95% as indicated through the area under the receiver operating characteristic curve (Chan et al. 2016). In addition, some reports have indicated that tumors of distinct organ systems could have unique urine metabolic signatures (Woo et al. 2009; Slupsky et al. 2010), which would be an important consideration if urine metabolomics were to be utilized for cancer diagnostics in clinics.
Urine metabolomics has also been applied to examine the treatment effects of chemotherapy. For example, one study used 2D 1H-1H J-resolved NMR data to follow the effects of cisplatin in patients with non-small-cell lung cancer (N = 5) and show that cisplatin alters urinary amino acids levels (Doskocz et al. 2015).
3.3 Cerebrospinal Fluid
Cerebrospinal fluid is traditionally the fluid of choice to study neurological conditions. However, it has been shown to be an important source of biomarkers of malignant cell invasion to the leptomeninges, which is a relatively rare condition of late stage solid and hematologic cancers. In this context, two separate studies inspected CSF metabolic composition by 1H-NMR spectroscopy in leptomeningeal invasion from lung cancer and B-cell non-Hodgkin lymphoma and found metabolite alterations related to the presence of malignant cells in CSF (An et al. 2015; Graça et al. 2017). An et al. compared CSF samples from controls affected by neurologic conditions (N = 41) with samples from patients diagnosed with leptomeningeal carcinomatosis from lung adenocarcinoma (N = 26). Changes in the levels of myo-inositol, creatine, lactate, alanine and citrate were the most discriminatory CSF metabolites between the two groups of patients (An et al. 2015). These authors also reported a good correlation between the metabolic profile and the grading of radiological leptomeningeal enhancement accessed by magnetic resonance imaging (MRI), suggesting the potential utility of CSF metabolic profile in grading of leptomeningeal carcinomatosis (An et al. 2015). Graça et al. compared the CSF metabolic profiles of B-cell non-Hodgkin lymphoma patients with positive (N = 5) and negative (N = 13) diagnosis of leptomeningeal invasion. Among the most significant metabolite alterations glycine, alanine, pyruvate, acetylcarnitine, carnitine, phenylalanine as well as protein signals seemed to be increased in the positively diagnosed patients (Graça et al. 2017). The authors also found that leptomeningeal invasion chemotherapy treatment produced sharp decreases in the levels of those metabolites in a group of follow-up positively diagnosed patients (Graça et al. 2017).
3.4 Ascitic Fluid
Malignant ascites is the abnormal buildup of tumor-cell containing fluid in the abdomen, the ascitic fluid (Sangisetty and Miner 2012). The presence of malignant ascites is generally signal of an advanced stage of the disease and poor prognostic in ovarian, uterine, colorectal and pancreatic cancers (Garrison et al. 1986). Because ascitic fluid can also accumulate in other diseases, such as cirrhosis, it is important to devise a quick method to determine the causes of ascite origin in cases. Some studies investigated the origin of ascitic fluid using metabolomics, by comparing ovarian carcinoma patients with cirrhotic patients showing promising results (Bala et al. 2008; Shender et al. 2014). Important differences were observed in the levels of fatty acids, cholesterol, ceramide, glycerol-3-phosphate, glucose, and glucose-3-phosphate between ovarian cancer patients (N = 10) and cirrhotic patient (N = 5) in a study using GC-MS (Shender et al. 2014). In a study using 1H-NMR 3-hydroxybutyric acid, lactate, citrate, and tyrosine were the metabolites that discriminated between ovarian cancer (N = 15) and cirrhotic patients (N = 47) (Bala et al. 2008). 1H-NMR metabolomics was also applied to identify the metabolic differences induced by chemotherapy in ovarian serous carcinoma effusions, indicating that the ascitic fluid levels of glucose and lipids increase while the levels of lactate and β-hydroxybutyrate decrease after chemotherapy (N = 35) when compared with ascitic fluid before chemotherapy (N = 44) (Vettukattil et al. 2013).
Animal models have also been used to investigate the development of ascites and ascitic fluid in cancer. A metabolomics study of two murine xenograft ovarian carcinoma models, one with a mouse ID8-vascular endothelial growth factor (VEGF)-Defb29 cell line (N = 8) and the human OVCAR3 cell line (N = 5), was carried out to characterize the malignant ascites metabolic features (Bharti et al. 2017). Despite the two cell lines lead to different metabolic profiles, some metabolites were common to both xenograft models: β-hydroxybutyric acid, maleic acid and citrate (Bharti et al. 2017).
3.5 Exhaled Breath Analysis
The analysis of exhaled breath is an established non-invasive technique for specific applications such as alcoholemia and Helicobacter Pylori testing (measurement of 13C urea). Its application to cancer diagnostic focuses on the measurement of endogenous volatile organic compounds (VOCs), which can be defined as carbon-containing volatile compounds at room temperature (Hanna et al. 2019). Due to their physico-chemical properties, VOCs are well detected and measured using MS-related techniques, notably GC-MS but also direct-MS measurements (Hanna et al. 2019).
Exhaled breath VOC analysis can provide means of early diagnosis and patient stratification, particularly in population groups at higher risk for cancer development, e.g. smokers or individuals exposed to volatile and particulate contaminants; but also for patients presenting non-specific symptoms associated with cancer. Therefore, it can help clinicians decide on more invasive diagnostic or imaging procedures. The non-invasiveness of exhaled breath analysis can also lead to more patient enrolment (Hanna et al. 2019).
Applications of exhaled breath analysis in cancer seem particularly suitable in early diagnosis of cancer from the respiratory and digestive systems such as lung (Fu et al. 2014; Li et al. 2015), gastroesophageal (Kumar et al. 2015), oral cavity (Bouza et al. 2017) and laryngeal cancers (Garcia et al. 2014) since the affected organs have direct contact with breath. Nevertheless, some authors have also explored the application to cancers from distant organs such as liver, breast, prostate or ovarian (Qin et al. 2010; Barash et al. 2015; Peng et al. 2010; Amal et al. 2015).
The most popular application is by far the discrimination of groups of lung cancer patients from control subjects. Two representative studies have reported that VOCs analyses provided sensitivity values close or above 90% and specificity values above 80% for discrimination between controls and lung cancer patients (Fu et al. 2014; Li et al. 2015). Levels of carbonyl compounds levels were found elevated in patients with lung tumors (N = 85) (Li et al. 2015), whereas the concentrations of 2-butanone, 2-hydroxyacetaldehyde, 3-hydroxy-2-butanone, and 4-hydroxyhexenal in the exhaled breath of lung cancer patients (N = 97) were found significantly higher than in the exhaled breath of healthy smoker and non-smoker controls (N = 88) (Fu et al. 2014). Another interesting application is related to esophagogastric cancer. In a representative study, Kumar et al. identified 12 VOCs (pentanoic acid, hexanoic acid, phenol, methyl phenol, ethyl phenol, butanal, pentanal, hexanal, heptanal, octanal, nonanal, and decanal) increased in exhaled breath from esophageal (N = 48) and gastric adenocarcinoma (N = 33) when compared to non-cancer controls (N = 129), which provides specificity and sensitivity values for patient discrimination above 80% (Kumar et al. 2015).
Nanoarray-based sensor technology developments are also making it possible to measure breath VOCs. This technology has several advantages over GC-MS, particularly regarding operational costs and portability. It has been tested in the analysis of breath analysis from patients with several types of cancers, such as ovarian cancer (Amal et al. 2015), gastric cancers (Amal et al. 2016) as well as lung, breast, colorectal and prostate cancers (Peng et al. 2010), with discrimination performances similar to those of GC-MS.
Regardless of the application of exhaled breath analysis in cancer, additional analytical bias assessment and the introduction of standardized sampling procedures are key elements in the development and transitioning of the technique and its results to clinical applications (Hanna et al. 2019).
3.6 Other Noninvasive Biological Matrices: Saliva, Sputum, and Feces
In addition to the biological fluids/ matrices described above, metabolomics investigations have also been performed in numerous other matrix types. For example, saliva obtained from oral, breast and pancreatic cancer patients has successfully been analyzed (Sugimoto et al. 2010). Oncogenic MYC has been reported to regulate polyamine biosynthesis leading to accumulation in cancer cells, and Asai et al. have used CE-MS for detecting polyamines, and found spermine, N-acetylspermidine, and N-acetylspermine levels in saliva successfully discriminate patients with pancreatic cancer (N = 39) from controls (N = 26) (Asai et al. 2018). Similarly, the levels of several polyamines in saliva have also been found elevated in relapsed breast cancer patients (N = 22) in another study using targeted LC-MS (Tsutsui et al. 2013).
Sputum consists of mucus produced in the respiratory tract and is potentially relevant for the diagnosis of lung cancer. There are currently very limited cancer metabolomics literature available on sputum, however, one study has successfully utilized flow infusion MS and GC–MS for distinguishing their 34 lung cancer patients and 33 healthy controls (Cameron et al. 2016).
Feces, rather like urine is readily available and information rich, as it contains undigested food passed from the gastrointestinal tract (GI) and metabolite compositions reflect dietary habits, mammalian– gut microbial interactions, as well as health status of the GI tract. Compared to health controls, colorectal cancer patients may have altered levels of acetate, butyrate, propionate, isovalerate, isobutyrate, valerate, and bile acids in their feces (Lin et al. 2016; Le Gall et al. 2018).
3.7 Biopsy and Cytology Material
Tissue biopsies and cytology aspirates are obtained from tumors and their metastases to confirm the diagnosis, molecular typing and staging which are performed at cyto- and histo-pathological analysis. The metabolomic analysis of such materials offers complementary metabolic information for further disease characterization and phenotyping. Moreover, it can be used as a diagnostic tool on its own. As mentioned in previous sections, both NMR and MS techniques are suitable for analysis of tissues and cells, either by analysis of extracts or intact material.
3.7.1 Analysis of Cells and Tissue Extracts
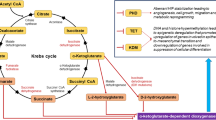
Cell and tissue extractions break up cellular structures and releases metabolites for in-depth or targeted biochemical analysis, for instance with focus on lipids or in hydrophilic metabolites. However, extractions may have reproducibility issues. For this reason, extraction procedures must ensure an effective arrest of cellular metabolism and minimize metabolite loss. Nevertheless, the analysis of cell and tissue extracts have been a valuable resource in in vitro tumor metabolism studies. One such studies is the study of isocitrate hydrogenase mutation in specific types of tumors. Isocitrate dehydrogenase 1 and 2 (IDH1/2) are enzymes important for energy metabolism, redox control and DNA methylation. Mutations in the genes encoding for these enzymes are frequent, including in majority of gliomas (Yan et al. 2009) and cartilage tumours (Pansuriya et al. 2011), and can be found in a significant portion of acute myeloid leukaemia (Molenaar et al. 2015). In a landmark paper, Dang et al. has shown that tumors harboring IDH1/2 mutations gain the ability to convert α-ketoglutarate to 2-hydroxyglutarate, leading to accumulation in 2-hydroxyglutarate in tumor cells. Comparing to the wild type gliomas, 2-hydroxyglutarate level in IDH mutant human tumors increased by 100-fold (Dang et al. 2009). The gain-of-function mutations are phenotypically specific and, in fact, 2-hydroxyglutarate could be detected directly in vivo in patients with glioma using magnetic resonance spectroscopy (MRS) acquired in MRI instruments (Choi et al. 2012). The conversion of α-ketoglutarate to 2-hydroxyglutarate could be measured in real time in vivo by using the same methodology with increased sensitivity through substrate dynamic nuclear polarization (DNP-MRS) (Chaumeil et al. 2013).
3.7.2 Analysis of Intact Tissues
A specific NMR technique, high-resolution magic-angle spinning (HRMAS), allows the analysis of micro-grams of tissue biopsies with similar resolution of liquid NMR (Emwas 2015). HRMAS can be used for the analysis of human and animal tumor tissues ex vivo, however, freezing delay time should be minimized as it could adversely bias analysis. Significant metabolite changes have been observed in samples frozen after 30 min of resection, and some metabolites are affected by prolonged experiment time due to sample spinning and degradation (Haukaas et al. 2016). Nevertheless, HRMAS can be useful in identifying diagnostic markers if experiments were designed and samples were handled with care. This has been illustrated for some types of cancer such as prostate or colorectal. Indeed, using tissue samples, spermine, spermidine, choline, kynurenine, sarcosine, citrate have been proposed as potential candidates as markers of diagnosis or staging in prostate tumors (de Vogel et al. 2014; Sreekumar et al. 2009; McDunn et al. 2013; Liu et al. 2015; Giskeodegard et al. 2013). Increased levels of lactate, taurine, and isoglutamine and decreased levels of lipids/triglycerides have been found in colorectal cancer (N = 88) relative to healthy mucosa (N = 83) (Mirnezami et al. 2014).
Surgical evaluation of tumor margins is routinely performed during tumor-extracting surgery, in which the surgeon decides on the extent of malignant tissue to extract while trying to maintain healthy tissue intact. This delicate process is usually assisted by a trained histopathologist who analyses the frozen surgically extracted tissues by light microscopy. The whole process needs to be done quickly while the patient is under anesthesia (Ifa and Eberlin 2016; Hänel et al. 2019).
Although HRMAS NMR could be applied in the analysis of tumor margins (Bathen et al. 2013; Paul et al. 2018), there is a great advantage in using MS techniques due to their higher sensitivity and smaller sample amounts requirement. Ambient-ionization MS techniques seem the most useful as they allow the acquisition of MS spectra in real-time and are easily operated by non-specialists, which gives the technique great advantage in surgical tumor diagnostics (Ifa and Eberlin 2016; Hänel et al. 2019).
Several ambient-ionization MS techniques have been introduced in cancer tissue analysis such as DESI, rapid evaporative MS (REIMS), “MasSpec pen” and picosecond infrared laser (PIRL) with some promising results towards intact ex vivo sample analysis (Hänel et al. 2019). While all of them focus on the analysis of lipid content, each one has specific characteristics regarding the amount of sample consumed, cross-contamination, preanalytical issues, surface scanning and transferability towards clinical diagnostic application (Hänel et al. 2019). REIMS is the most popular of ambient-ionization MS techniques because it is also applicable in vivo (Balog et al. 2013).
The most well known setup of REIMS, known as “iKnife” or “intelligent scalpel”, has been used intra-surgically. It consists of a hand-held device connected to an electrosurgical instrument which transfers the aerosols produced by cutting through the tissue directly into the MS spectrometer. The MS spectrum produced contains a signature of the lipidome profile of the tissue being cut (Fig. 19.4). MS spectra, collected in real-time, are immediately tested in a multivariate discriminant model (trained on real benign and malignant tissue spectra from samples classified via histopathology) giving a classification of the tissue cut by the surgeon (Balog et al. 2013). The iKnife has been tested on hundreds of patients with several types of tumor such as liver, lung, colorectal, breast, gynecologic, glioma, glioblastoma as well as in metastasis from lung and colon cancer to the brain enabling classification of sampled tissues with high sensitivity (90–98%) and specificity (94–100%) values (Balog et al. 2013; St John et al. 2017; Phelps et al. 2018). A version of the iKnife procedure was also introduced in the endoscopic analysis of colon polyps (Balog et al. 2015). The major disadvantages of REIMS compared to the above mentioned methods are sample consumption, possible cross-contamination and analyte degradation during tissue cutting due to the high temperatures generated (Hänel et al. 2019).
Rapid evaporative mass spectrometry “iKnife” analysis of intact tissue applied to ovarian cancer. Electrical current, produced from the generator, is applied to the tissue and the resultant charged particles are extracted through the custom-designed hand-piece and drawn into the REIMS atmospheric inlet and analyzed in a Xevo G2-XS mass spectrometer to produce tissue-specific mass spectra, which are then subjected to multivariate statistical analysis using Principal Component – Linear Discriminant Analysis (PC-LDA). Within 1–2 s, real-time tissue diagnosis is displayed on a screen for the surgeon to see. Adapted with permission from Phelps et al. (2018) under Creative Commons Attribution 4.0 License
The PIRL method is a promising method for in vivo applications and it has some advantages over REIMS. PIRL uses infrared laser to cut through tissue which enable MS spectra to be obtained from smaller areas of tissue and even single cells and avoid damaging adjacent tissue (Hänel et al. 2019). It also has the potential to achieve better spatial resolution in vivo compared to REIMS (Hänel et al. 2019).
3.7.3 MS Imaging of Intact Tissue
Perhaps one of the most interesting applications of MS is imaging (MSI). In MSI, samples are prepared into fine slices or smears, much like in histologic preparations. Then the sample surface is scanned in small areas (10–200 μm) corresponding to image pixels, and ions are withdrawn and analyzed in the mass spectrometer (Bodzon-Kulakowska and Suder 2016). One MS spectrum is acquired from every small area (pixel) of the sample. An image can be then generated by mapping the intensity of any selected ion into the optical image of the tissue (Fig. 19.5).
Negative-ion-mode DESI-MS images of a breast tissue sample from an invasive ductal carcinoma patient. Upper panel shows images of specific fatty acid (FA) and phosphatidylinositols (PI) ions highlighting their distribution in the tissue slice. Bottom panel shows the Hematoxylin and Eosin staining optical imaging; expansions of the sectioned tissue shows the delimited stromal and tumoral cells areas and abundance of PI(36:1) and PI(38:4) ions. Lipid species are described by the numbers of fatty acid chain carbons and double bonds. Adapted with permission from Porcari, et al. (2018). Copyright 2018 American Chemical Society
In order to get into the fine molecular imaging detail, MSI spectrometers are equipped with ionization techniques such as MALDI, DESI or SIMS and very high resolution detectors such as time-of-flight (TOF), orbitrap or ion-cyclotron resonance, to ensure both high image and MS resolutions. Although SIMS provides higher sensitivity and resolution than MALDI and DESI, the latter two being “soft” ionization methods find more wide-spread application in tumor tissue MSI (Bodzon-Kulakowska and Suder 2016).
Due to the fine molecular detail provided, MSI has an enormous potential both as a diagnostic and research tool in cancer and can be viewed as a form of augmented histology. Indeed changes in metabolites such as lipids as those illustrated in Fig. 19.5 can be mapped into tumor tissue sections, providing finer detail about tumor heterogeneity and help in the diagnosis of invasive ductal carcinoma (Porcari et al. 2018). MSI also permits the in situ study of metabolic pathways that may be altered due to reprogramming. For instance, Sun et al. effectively mapped several metabolites from tumor-associated metabolic pathways, including proline biosynthesis, glutamine metabolism, uridine metabolism, histidine metabolism, fatty acid biosynthesis, and polyamine biosynthesis in tissues from 256 esophageal cancer patients, thus helping to uncover abnormal expression of enzymes pyrroline-5-carboxylate reductase 2 (PYCR2) and uridine phosphorylase 1 (UPase1) in esophageal squamous cell carcinoma (Sun et al. 2019).
Finally, MSI has found an increasing applicability in pharmaceutical research and drug development in oncology, particularly in drug biodistribution, pharmacodynamic biomarker research and in toxicology assessment studies (Goodwin and Webborn 2015).
4 Final Remarks and Future Prospects
Metabolomics and metabolic phenotyping are established tools in the study of cancer metabolism. They have benefited from technological developments in both NMR and MS analytical instrumentation coupled with state-of-the art data analysis, particularly in the last decade. Both analytical platforms seem well suited for the development of diagnostic methods in cancer. However, the higher investment and operational costs of NMR hinder its wide-spread adoption. One exception is in vivo NMR spectroscopy (MRS), which can be performed in diagnostic MRI instruments and, in fact, it is an approved diagnostic tool to investigate certain types of brain tumors (Horská and Barker 2010). However, in comparison with ex vivo NMR, in vivo MRS has limited resolution and sensitivity which are factors that may have limited the translation of ex vivo discoveries to in vivo diagnostic MRS. The introduction of hyperpolarized substrates using DNP techniques, showed very promising results in preclinical studies and may form the basis for future metabolic imaging applications using NMR (Julià-Sapé et al. 2019). On the other hand, MS-based techniques have been present in clinical chemistry laboratories as diagnostic tool for several decades, especially are used in drug monitoring, newborn screening and in the diagnosis of metabolic diseases (Hänel et al. 2019), which make them ideally suited to metabolomics/metabolic profiling based diagnostic applications. Major advances in MS-based approaches such as intra-operative MS and MS imaging are opening the door for real clinical applications. Nevertheless, there is still a long road ahead until the development of truly diagnostic metabolomics approaches in cancer comes to fruition in the clinics, particularly if less-invasive and early diagnosis applications are to be considered.
The number of published studies in metabolomics/ metabolic phenotyping applications in oncology is already vast, covering a wide range of malignancies at different stages of the disease, across numerous types of biological samples and diverse patient/subject background and of varying sample size. As the amount of scientific literature grows, putting all the information into context in order to draw meaningful conclusions useful for diagnostic application becomes a challenge. This is in part due to the varying study designs, different reporting details of patient data, diverging sample preparation and acquisition protocols as well as insufficient reporting of analytical bias, which makes knowledge integration (for instance through meta-analysis) a difficult task. Therefore, standardized reporting of study design, sampling, experimental protocols, metadata and rigorous metabolite identification and analytical bias reporting would facilitate knowledge integration and would also help promote replication studies which are needed for biomarker validation.
References
Akita H, Ritchie SA, Takemasa I et al (2016) Serum metabolite profiling for the detection of pancreatic cancer: results of a large independent validation study. Pancreas 45:1418–1423
Amal H, Shi DY, Ionescu R et al (2015) Assessment of ovarian cancer conditions from exhaled breath. Int J Cancer 136:E614–E622. https://doi.org/10.1002/ijc.29166
Amal H, Leja M, Funka K et al (2016) Detection of precancerous gastric lesions and gastric cancer through exhaled breath. Gut 65:400–407
Amberg A, Riefke B, Schlotterbeck G et al (2017) NMR and MS methods for metabolomics. Methods Mol Biol 1641:229–258. https://doi.org/10.1007/978-1-4939-7172-5_13
An YJ, Cho HR, Kim TM et al (2015) An NMR metabolomics approach for the diagnosis of leptomeningeal carcinomatosis in lung adenocarcinoma cancer patients. Int J Cancer 136:162–171
Ang JE, Pandher R, Ang JC et al (2016) Plasma metabolomic changes following PI3K inhibition as pharmacodynamic biomarkers: preclinical discovery to phase I trial evaluation. Mol Cancer Ther 15:1412–1424
Ang JE, Pal A, Asad YJ et al (2017) Modulation of plasma metabolite biomarkers of the MAPK pathway with MEK inhibitor RO4987655: pharmacodynamic and predictive potential in metastatic melanoma. Mol Cancer Ther 16:2315–2323
Ardenkjaer-Larsen J-H, Boebinger GS, Comment A et al (2015) Facing and overcoming sensitivity challenges in biomolecular NMR spectroscopy. Angew Chem Int Ed Engl 54:9162–9185
Asai Y, Itoi Y, Sugimoto M et al (2018) Elevated polyamines in saliva of pancreatic cancer. Cancers 10:E43. https://doi.org/10.3390/cancers10020043
Bala L, Sharma A, Yellapa RK et al (2008) 1H NMR spectroscopy of ascitic fluid: discrimination between malignant and benign ascites and comparison of the results with conventional methods. NMR Biomed 21:606–614
Balog J, Sasi-Szabó L, Kinross J et al (2013) Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med 5:194ra93. https://doi.org/10.1126/scitranslmed.3005623
Balog J, Kumar S, Alexander J et al (2015) In vivo endoscopic tissue identification by rapid evaporative ionization mass spectrometry (REIMS). Angew Chem 54:11059–11062
Barash O, Zhang W, Halpern JM et al (2015) Differentiation between genetic mutations of breast cancer by breath volatolomics. Oncotarget 6:44864–44876. https://doi.org/10.18632/oncotarget.6269
Bathen TF, Geurts B, Sitter B et al (2013) Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS One 8:e61578. https://doi.org/10.1371/journal.pone.0061578
Bharti SK, Wildes F, Hung C-F et al (2017) Metabolomic characterization of experimental ovarian cancer ascitic fluid. Metabolomics 113. https://doi.org/10.1007/s11306-017-1254-3
Bingol K (2018) Recent advances in targeted and untargeted metabolomics by NMR and MS/NMR methods. High-Throughput 7:E9. https://doi.org/10.3390/ht7020009
Bloch F, Hansen WW, Packard M (1946) The nuclear induction experiment. Phys Rev 70:474–485. https://doi.org/10.1103/PhysRev.70.474
Bodzon-Kulakowska A, Suder P (2016) Imaging mass spectrometry: instrumentation, applications, and combination with other visualization techniques. Mass Spectrom Rev 35:147–169
Bouza M, Gonzalez-Soto J, Pereiro R et al (2017) Exhaled breath and oral cavity VOCs as potential biomarkers in oral cancer patients. J Breath Res 11:016015. https://doi.org/10.1088/1752-7163/aa5e76
Bro R, Kamstrup-Nielsen MH, Engelsen SB et al (2015) Forecasting individual breast cancer risk using plasma metabolomics and biocontours. Metabolomics 11:1376–1380
Broadhurst DI, Kell DB (2006) Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2:171–196
Cameron SJS, Lewis KE, Beckmann M et al (2016) The metabolomic detection of lung cancer biomarkers in sputum. Lung Cancer 94:88–95
Chan AW, Mercier P, Schiller D et al (2016) 1H-NMR urinary metabolomic profiling for diagnosis of gastric cancer. Br J Cancer 114:59–62
Chaumeil MM, Larson PEZ, Yoshihara HAI et al (2013) Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun 4:2429
Cheng XM, Liu XY, Liu X et al (2018) Metabolomics of non-muscle invasive bladder cancer: biomarkers for early detection of bladder cancer. Front Oncol 8:494. https://doi.org/10.3389/fonc.2018.00494
Choi C, Ganji SK, DeBerardinis RJ et al (2012) 2-hydroxyglutarate detection by magnetic resonance spectroscopy in subjects with IDH-mutated gliomas. Nat Med 18:624–629
Cornel EB, Smits GA, Oosterhof GO et al (1993) Characterization of human prostate cancer, benign prostatic hyperplasia and normal prostate by in vitro 1H and 31P magnetic resonance spectroscopy. J Urol 150:2019–2024
Dalgliesh CE (1956) Two-dimensional paper chromatography of urinary indoles and related substances. Biochem J 64:481–485
Dang L, White DW, Gross S et al (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744
de Vogel S, Ulvik A, Meyer K et al (2014) Sarcosine and other metabolites along the choline oxidation pathway in relation to prostate cancer-a large nested case-control study within the JANUS cohort in Norway. Int J Cancer 134:197–206
Dinges SS, Hohm A, Vandergrift LA et al (2019) Cancer metabolomic markers in urine: evidence, techniques and recommendations. Nat Rev Urol 16:339–362
Dona AC, Jiménez B, Schäfer H et al (2014) Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal Chem 86:9887–9894
Doskocz M, Marchewka Z, Jeż M et al (2015) Preliminary study on J-resolved NMR method usability for toxic kidney’s injury assessment. Adv Clin Exp Med 24:629–635
Dunphy MPS, Harding JJ, Venneti S (2018) In vivo PET assay of tumor glutamine flux and metabolism: in-human trial of 18F-(2S,4R)-4-fluoroglutamine. Radiology 287:667–675
Emwas AH (2015) The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol 1277:161–193
Emwas AH, Roy R, McKay RT et al (2019) NMR spectroscopy for metabolomics research. Meta 9. https://doi.org/10.3390/metabo9070123
Fan TW-M, Lane AN (2016) Applications of NMR spectroscopy to systems biochemistry. Prog Nucl Magn Reson Spectrosc 92-93:18–53
Fan TW-M, Lorkiewicz PK, Sellers K et al (2012) Stable isotope-resolved metabolomics and applications for drug development. Pharmacol Ther 133:366–391
Fu XA, Li MX, Knipp RJ et al (2014) Noninvasive detection of lung cancer using exhaled breath. Cancer Med 3:174–181
Fukutake N, Ueno M, Hiraoka N et al (2015) A novel multivariate index for pancreatic cancer detection based on the plasma free amino acid profile. PLoS One 10:e0132223. https://doi.org/10.1371/journal.pone.0132223
Ganti S, Taylor SL, Kim K et al (2012) Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer 130:2791–2800
Garcia RA, Morales V, Martin S et al (2014) Volatile organic compounds analysis in breath air in healthy volunteers and patients suffering epidermoid laryngeal carcinomas. Chromatographia 77:501–509
Garrison RN, Kaelin LD, Galloway RH et al (1986) Malignant ascites. Clinical and experimental observations. Ann Surg 203:644–651
Giskeodegard GF, Bertilsson H, Selnaes KM et al (2013) Spermine and citrate as metabolic biomarkers for assessing prostate cancer aggressiveness. PLoS One 8:e62375. https://doi.org/10.1371/journal.pone.0062375
Glish GL, Vachet RW (2003) The basics of mass spectrometry in the twenty-first century. Nat Rev Drug Dis 2:140–150
Goodwin RJA, Webborn PJH (2015) Future directions of imaging MS in pharmaceutical R&D. Bioanalysis 7(20):2667–2673
Graça G, Desterro J, Sousa J et al (2017) Identification of putative biomarkers for leptomeningeal invasion in B-cell non-Hodgkin lymphoma by NMR metabolomics. Metabolomics 13:136
Graça G, Serrano-Contreras JI, Chekmeneva E (2019) Nuclear magnetic resonance spectroscopy: pulse sequences for chemical analysis. In: Worsfold P, Poole C, Townshend A, Miró M (eds) Encyclopedia of analytical science, vol 7, 3rd edn. Elsevier, Amsterdam, pp 354–365
Gromski PS, Muhamadali H, Ellis DI et al (2015) A tutorial review: metabolomics and partial least squares-discriminant analysis - a marriage of convenience or a shotgun wedding. Anal Chim Acta 879:10–23
Guennec AL, Giraudeau P, Caldarelli S (2014) Evaluation of fast 2D NMR for metabolomics. Anal Chem 86:5946–5954
Hänel L, Kwiatkowski M, Heikaus L et al (2019) Mass spectrometry-based intraoperative tumor diagnostics. Future Sci OA 5:FSO373. https://doi.org/10.4155/fsoa-2018-0087
Hanna GB, Boshier PR, Markar SR et al (2019) Accuracy and methodological challenges of volatile organic compound-based exhaled breath tests for cancer diagnosis: a systematic review and meta-analysis. JAMA Oncol 5:e182815. https://doi.org/10.1001/jamaoncol.2018.2815
Hao J, Astle W, De Iorio M et al (2012) BATMAN – an R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a Bayesian model. Bioinformatics 28:2088–2090
Haukaas TH, Moestue SA, Vettukattil R et al (2016) Impact of freezing delay time on tissue samples for metabolomic studies. Front Oncol 6:17. https://doi.org/10.3389/fonc.2016.00017
Hilvo M, de Santiago I, Gopalacharyulu P et al (2016) Accumulated metabolites of hydroxybutyric acid serve as diagnostic and prognostic biomarkers of ovarian high-grade serous carcinomas. Cancer Res 76:796–804
Horská A, Barker PB (2010) Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin N Am 20(3):293–310
Ifa DR, Eberlin LS (2016) Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin Chem 62:111–123
Jin X, Yun SJ, Jeong P et al (2014) Diagnosis of bladder cancer and prediction of survival by urinary metabolomics. Oncotarget 5:1635–1645
Julià-Sapé M, Candiota AP, Arús C (2019) Cancer metabolism in a snapshot: MRS(I). NMR Biomed 11:e4054. https://doi.org/10.1002/nbm.4054
Komoroski RA, Holder JC, Pappas AA et al (2011) 31P NMR of phospholipid metabolites in prostate cancer and benign prostatic hyperplasia. Magn Reson Med 65:911–913
Kumar S, Huang J, Abbassi-Ghadi N et al (2015) Mass spectrometric analysis of exhaled breath for the identification of volatile organic compound biomarkers in esophageal and gastric adenocarcinoma. Ann Surg 262:981–990
Le Gall G, Guttula K, Kellingray L et al (2018) Metabolite quantification of faecal extracts from colorectal cancer patients and healthy controls. Oncotarget 9:33278–33289
Li MX, Yang DK, Brock G et al (2015) Breath carbonyl compounds as biomarkers of lung cancer. Lung Cancer 90:92–97
Lin Y, Ma CC, Liu CK et al (2016) NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget 7:29454–29464
Liu W, Bai XF, Liu YJ et al (2015) Topologically inferring pathway activity toward precise cancer classification via integrating genomic and metabolomic data: prostate cancer as a case. Sci Rep 5:13192
Marchand J, Martineau E, Guitton Y et al (2017) Multidimensional NMR approaches towards highly resolved, sensitive and high-throughput quantitative metabolomics. Curr Opin Biotechnol 43:49–55
Marion D (2013) An introduction to biological NMR spectroscopy. Mol Cell Proteomics 12:3006–3025
Mayers JR, Wu C, Clish CB et al (2014) Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 20:1193–1198
McDunn JE, Li Z, Adam KP et al (2013) Metabolomic signatures of aggressive prostate cancer. Prostate 73:1547–1560
Mirnezami R, Jiménez B, Li JV et al (2014) Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann Surg 259:1138–1149
Molenaar RJ, Thota S, Nagata Y et al (2015) Clinical and biological implications of ancestral and non-ancestral IDH1 and IDH2 mutations in myeloid neoplasms. Leukemia 29:2134–2142
Nagana Gowda GA, Raftery D (2017) Recent advances in NMR-based metabolomics. Anal Chem 89:490–510
Nicholson JK, O'Flynn MP, Sadler PJ et al (1984) Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. Biochem J 217:365–375
Pansuriya TC, van Eijk R, d'Adamo P et al (2011) Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet 43:1256–1261
Paul A, Kumar S, Raj A et al (2018) Alteration in lipid composition differentiates breast cancer tissues: a 1H HRMAS NMR metabolomic study. Metabolomics 14:119
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Peng G, Hakim M, Broza YY et al (2010) Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br J Cancer 103:542–551
Phelps DL, Balog J, Gildea LF et al (2018) The surgical intelligent knife distinguishes normal, borderline and malignant gynaecological tissues using rapid evaporative ionisation mass spectrometry (REIMS). Br J Cancer 118:1349–1358
Porcari AM, Zhang J, Garza KY et al (2018) Multi-center study using desorption-electrospray-ionization-mass-spectrometry imaging for breast cancer diagnosis. Anal Chem 90:11324–11332
Psychogios N, Hau DD, Peng J et al (2011) The human serum metabolome. PLoS One 6:e16957. https://doi.org/10.1371/journal.pone.0016957
Purcell EM, Torrey HC, Pound RV (1946) Resonance absorption by nuclear magnetic moments in a solid. Phys Rev 69:37–38
Qin T, Liu H, Song Q et al (2010) The screening of volatile markers for hepatocellular carcinoma. Cancer Epidemiol Biomark Prev 19:2247–2253
Sakai A, Suzuki M, Kobayashi T et al (2016) Pancreatic cancer screening using a multiplatform human serum metabolomics system. Biomark Med 10:577–586
Sanderson SM, Locasale JW (2018) Revisiting the Warburg effect: some tumors hold their breath. Cell Metab 28:669–670
Sangisetty SL, Miner TJ (2012) Malignant ascites: a review of prognostic factors, pathophysiology and therapeutic measures. World J Gastrointest Surg 4:87–95
Shender VO, Pavlyukov MS, Ziganshin RH et al (2014) Proteome-metabolome profiling of ovarian cancer ascites reveals novel components involved in intercellular communication. Mol Cell Proteomics 13:3558–3571
Slupsky CM, Steed H, Wells TH et al (2010) Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res 16:5835–5841
Sreekumar A, Poisson LM, Rajendiran TM et al (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457:910–914
St John ER, Balog J, Mckenzie JS et al (2017) Rapid evaporative ionisation mass spectrometry of electrosurgical vapours for the identification of breast pathology: towards an intelligent knife for breast cancer surgery. Breast Cancer Res 19:59. https://doi.org/10.1186/s13058-017-0845-2
Sugimoto M, Wong DT, Hirayama A et al (2010) Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6:78–95
Sun C, Lia T, Song X et al (2019) Spatially resolved metabolomics to discover tumor associated metabolic alterations. PNAS 116:52–57
Tanaka K, Isselbacher KJ (1967) The isolation and identification of N-isovalerylglycine from urine of patients with isovaleric acidemia. J Biol Chem 242:2966–2972
Trygg J, Holmes E, Lundstedt T (2007) Chemometrics in metabonomics. J Proteome Res 6:469–479
Tsutsui H, Mochizuki T, Inoue K et al (2013) High-throughput LC-MS/MS based simultaneous determination of polyamines including N-acetylated forms in human saliva and the diagnostic approach to breast cancer patients. Anal Chem 85:11835–11842
Vettukattil R, Hetland TE, Flørenes VA et al (2013) Proton magnetic resonance metabolomic characterization of ovarian serous carcinoma effusions: chemotherapy-related effects and comparison with malignant mesothelioma and breast carcinoma. Hum Pathol 44:1859–1866
Wishart DS, Jewison T, Guo AC et al (2013) HMDB 3.0 — the human metabolome database in 2013. Nucleic Acids Res 41:D801–D807. https://doi.org/10.1093/nar/gks1065
Woo HM, Kim KM, Choi MH et al (2009) Mass spectrometry based metabolomic approaches in urinary biomarker study of women's cancers. Clin Chim Acta 400:63–69
Yan H, Parsons DW, Jin GL et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Zhu A, Lee D, Shim H (2011) Metabolic PET imaging in cancer detection and therapy response. Semin Oncol 38:55–69
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Graça, G., Lau, CH.E., Gonçalves, L.G. (2020). Exploring Cancer Metabolism: Applications of Metabolomics and Metabolic Phenotyping in Cancer Research and Diagnostics. In: Serpa, J. (eds) Tumor Microenvironment . Advances in Experimental Medicine and Biology, vol 1219. Springer, Cham. https://doi.org/10.1007/978-3-030-34025-4_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-34025-4_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34024-7
Online ISBN: 978-3-030-34025-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)