Abstract
The year 2018 marks the 10-year anniversary of the discovery of the diadenylate cyclase enzyme and its capacity to synthesize the broadly conserved second messenger cyclic di-AMP. Since this discovery, our understanding of the physiological processes controlled by this dinucleotide has advanced rapidly, with the discovery of both cyclic di-AMP responsive riboswitch gene control elements and protein binding partners. Additionally, cyclic di-AMP has been implicated as a cross-kingdom signal between bacteria and eukaryotic hosts. While the physiological processes modulated by these signaling partners are as diverse as the bacteria that produce cyclic di-AMP, a key theme that has emerged is the regulation of cellular metabolism. In this chapter, we will focus on the biological impacts of metabolic regulation imposed by cyclic di-AMP at both the transcriptional/translational and posttranslational levels, as well as the molecular mechanism of this regulation. We will highlight the regulation of central carbon metabolism through pyruvate carboxylase, the regulation of cell wall metabolism through the ydaO riboswitch, and the impact on host cell inflammatory response through competitive inhibition of the host binding protein RECON.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Cyclic dinucleotide
- Cyclic di-AMP
- Bacterial signaling
- Host immune response
- Central metabolism
- Pyruvate carboxylase
- RECON
- Riboswitch
- Listeria monocytogenes
- Lactococcus lactis
- Milk acidification
1 Introduction
The biosynthesis of cyclic di-AMP (cdA) was first discovered 10 years ago [1]. Since then, the diadenylate cyclase responsible for cyclic di-AMP synthesis has been described among many phyla of bacteria (Fig. 10.1a) and this second messenger has been found to have crucial roles in many bacterial processes, including central metabolism, cell wall metabolism, DNA repair, potassium homeostasis, osmotic regulation, sporulation, stress response, antibiotic resistance, biofilm formation, and virulence [2,3,4,5,6]. Moreover, cyclic di-AMP is essential for many of those bacteria that produce it, while high levels of this compound can be toxic [7]. The capacity of cyclic di-AMP to mediate these pleiotropic effects is predicated upon the presence of both nucleic acid and protein effectors that mediate transcriptional and posttranslational changes in protein function.
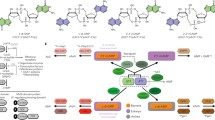
Cyclic di-AMP in bacteria and host–microbe interactions. (a) Phylogenetic tree depicting Archaea, Eukaryota, and Bacterial phyla. Those organisms with diadenylate cyclase (DAC) enzymes that can produce cyclic di-AMP are in bold black lines. DAC enzymes have been found to be largely essential among organisms in the Firmicutes and Bacteroidales phyla but dispensable in Actinobacteria and Cyanobacteria. (b) Cyclic di-AMP is sensed during infection by a variety of organisms. Activation of STING results in IRF-3 phosphorylation and modest NF-kB phosphorylation, resulting in the induction of Type I IFN responses. Inhibition of RECON by cyclic di-AMP results in augmented NF-κB activation
In addition to the signaling role within bacteria, cyclic di-AMP has also emerged as a key signal between bacteria and their eukaryotic hosts (Fig. 10.1b). The diadenylate cyclase responsible for cyclic di-AMP production is broadly conserved among most major phyla of bacteria and essential among many genera [2], but notably absent among eukaryotes. Due to its small molecule chemical nature and essentiality among many pathogens [8,9,10,11,12,13], cyclic di-AMP is difficult for organisms to evolve away from or to chemically alter to mask immune detection. Based on these characteristics, cyclic di-AMP is an ideal signature of bacterial presence within eukaryotic hosts that do not produce this molecule. Indeed, the role of cyclic di-AMP as a mediator of host inflammation to bacterial infection has garnered significant interest in the last decade, with some host sensors for bacterial cyclic di-AMP having been identified, and the mechanism of cyclic di-AMP signaling in host cells beginning to be understood [14,15,16]. STING-dependent detection of cyclic di-AMP results in the induction of Type I interferon during infection by a variety of organisms, including L. monocytogenes, C. trachomatis, M. tuberculosis (MTB), S. aureus, and S. agalactiae (GBS) [17,18,19,20,21]. GBS and MTB have been reported to utilize specific phosphodiesterases to degrade cyclic di-AMP to evade STING-mediated immune sensing [20, 21]. More recently, a second host cyclic di-AMP sensor named RECON was identified as an enzyme that, upon cyclic di-AMP binding, promotes NF-κB dependent inflammatory gene expression [14].
In this chapter, we will focus on the involvement of cyclic di-AMP in regulating metabolic processes, especially its regulation of the central metabolic enzyme pyruvate carboxylase (PC), the host metabolic enzyme RECON, and the riboswitch ydaO involved in cell wall metabolism.
2 Regulation of Pyruvate Carboxylase (PC) by Cyclic di-AMP
2.1 Identification of PC as a Direct Target of Cyclic di-AMP
PC catalyzes the carboxylation of pyruvate to produce oxaloacetate and is a central metabolic enzyme in most organisms [22, 23]. It has an anaplerotic role to replace the intermediates in the TCA cycle, and it is also crucial for gluconeogenesis, glyceroneogenesis, neurotransmitter release, and other cellular processes. PC deficiency in humans is linked to lactic acidemia, psychomotor retardation, and other symptoms, while PC overexpression has been observed in some cancers [24, 25].
The PC enzyme of the human pathogen Listeria monocytogenes (LmPC) was first identified as a direct target of cyclic di-AMP by a chemical proteomics approach [26]. Cyclic di-AMP was covalently immobilized on a resin and incubated with L. monocytogenes extract. Bound proteins were visualized by SDS gel and identified by mass spectrometry. LmPC was one of 12 proteins that were identified with statistical significance by this approach. The direct interaction between LmPC and cyclic di-AMP was confirmed using a radioactivity-based binding assay, and the K d of the complex was determined as 8 μM. Kinetic studies showed that cyclic di-AMP reduced the apparent k cat of the PC reaction while having only a small effect on the apparent K m, suggesting that cyclic di-AMP does not compete with the pyruvate substrate and is an allosteric inhibitor of LmPC. The kinetic studies also showed that LmPC is selective for cyclic di-AMP, while cyclic di-GMP and cGAMP had no effect on the catalysis.
With the elucidation of the cyclic di-AMP binding site in LmPC (see Sect. 10.2.2), residues that are important for recognizing cyclic di-AMP were identified. Sequence analysis then identified a few other PC enzymes that could also bind cyclic di-AMP. Among these, Enterococcus faecalis PC (EfPC) [26] and Lactococcus lactis PC (LlPC) [27] have been confirmed to be direct targets of cyclic di-AMP. Like LmPC, LlPC is selective for cyclic di-AMP, while cyclic di-GMP has very little effect on the catalysis.
In contrast, most other PCs, including human PC, are not targets of cyclic di-AMP binding and regulation.
2.2 Molecular Mechanism of PC Regulation by Cyclic di-AMP
PC is a biotin-dependent enzyme and contains two separate active sites [22, 23]. Biotin carboxylase (BC) catalyzes the carboxylation of biotin coupled with the hydrolysis of ATP to ADP, and bicarbonate is the CO2 donor. Carboxyltransferase (CT) catalyzes the transfer of CO2 from carboxybiotin to the pyruvate acceptor to produce oxaloacetate. Biotin is linked covalently to the biotin carboxyl carrier protein (BCCP). Most PC enzymes are ~120 kDa single-chain, multi-domain proteins, with BC, CT, and BCCP domains. They function only as tetramers, and most of them are activated by acetyl-CoA. They also contain a PT domain that mediates PC tetramerization as well as allosteric regulation by acetyl-CoA. A large amount of structural information is available for these enzymes, showing structural conservation of the domains but extensive variability in the relative organizations of the domains and the architectures of the holoenzymes, despite their strong sequence conservation [23, 28].
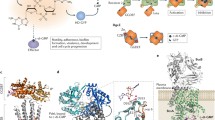
The overall structure of the 500 kDa PC tetramer is in the shape of a diamond, with BC and CT dimers located at alternate corners (Fig. 10.2a). Cyclic di-AMP assumes a folded, U-shaped conformation and is bound to a pocket at the dimer interface of CT in LmPC [26]. The twofold symmetry axis of cyclic di-AMP is aligned with that of the CT dimer, and therefore each LmPC tetramer binds only two molecules of cyclic di-AMP. Three important interactions are observed between cyclic di-AMP and LmPC (Fig. 10.2b): (1) π-stacking between the adenine base and the side chain of Tyr722; (2) direct hydrogen bond between the phosphate and the side chain of Tyr749; and (3) van der Waals interactions between the ribose and Ala752-Ala753 (two small side chains). Mutations of these residues can severely reduce the binding, for example, changing Tyr722 to Thr, its equivalent in human PC (Fig. 10.2c), essentially abolished binding to cyclic di-AMP. This binding site is not well conserved among PC enzymes (Fig. 10.2d), indicating that only a small subset of them are likely targets of cyclic di-AMP.
Molecular basis for the regulation of PC by cyclic di-AMP. (a) Schematic drawing of the structure of LmPC tetramer in complex with cyclic di-AMP. The domains of monomer 1 are colored according to the diagram at the bottom of the panel. Cyclic di-AMP is shown as a sphere model and labeled cdA (carbon atoms in black). The metal ion in the active site of CT is shown as a gray sphere. The BC and CT active sites are indicated with the asterisks in brown. (b) Comparison of the binding mode of cyclic di-AMP (black) in LlPC (green and yellow) with that in LmPC (gray). The 7° rotation for the adenine base of cyclic di-AMP in the two structures is indicated with the red arrow. (c) Alignment of residues in the cyclic di-AMP binding site of LmPC (highlighted in red) with equivalent residues in selected bacterial PCs and human PC. (d) Molecular surface of the cyclic di-AMP binding site in LmPC, colored based on sequence conservation (purple: conserved; cyan: not conserved) using the program ConSURF [44]. The structure figures were produced with PyMOL (www.pymol.org)
The overall interactions between LlPC and cyclic di-AMP are similar to those for LmPC [27]. Cyclic di-AMP assumes a slightly more open conformation in LlPC, and a conformational change for Tyr715 (equivalent to Tyr722 of LmPC) is observed to maintain the π-stacking interactions (Fig. 10.2b). Tyr749 of LmPC is replaced by Ile742 in LlPC (Fig. 10.2c), but the hydrogen bond to the phosphate group is maintained through Ser745 (Ala752 in LmPC). In both structures, the adenine base does not appear to be specifically recognized through hydrogen-bonding interactions, and the molecular basis of how these enzymes are selective for cyclic di-AMP over cyclic di-GMP is still not understood.
The binding site for cyclic di-AMP is located far away from the CT and BC active sites (Fig. 10.2a), consistent with kinetic data showing that cyclic di-AMP is an allosteric inhibitor. The exact molecular mechanism for how binding of cyclic di-AMP in this pocket can inhibit PC is not yet fully understood. There are large structural differences between free LmPC and the cyclic di-AMP complex. In addition, the conformations of the four monomers of the tetramer are essentially identical in the cyclic di-AMP complex, while substantial differences among them are observed without cyclic di-AMP. This led to the hypothesis that PC needs to undergo significant conformational changes during catalysis, and cyclic di-AMP inhibits the enzyme by “freezing” it into a single state [26, 28].
2.3 Biological Impacts of PC Regulation by Cyclic di-AMP
L. monocytogenes is an intracellular pathogen and is often associated with food poisoning outbreaks. It does not have a complete TCA cycle, and the oxaloacetate product of LmPC is crucial for glutamate/glutamine (Glx) biosynthesis [26]. LmPC is essential for L. monocytogenes growth [29], although the exact mechanism is not known. A strain with reduced levels of cyclic di-AMP, and hence higher LmPC activity, showed greatly enhanced Glx biosynthesis, while aspartate levels were not affected [26]. This metabolic imbalance led to defects in L. monocytogenes growth in mouse immortalized bone marrow-derived macrophages and fibroblasts, as well as in liver and spleen tissues in a mouse model of acute listeriosis. On the other hand, the deletion of citrate synthase, just downstream of PC and the first committed step for Glx biosynthesis, restored Glx levels as well as intracellular growth. Activation of the host pyroptosis pathway is partly responsible for the reduced L. monocytogenes growth at lower levels of cyclic di-AMP.
The L. monocytogenes strain with reduced cyclic di-AMP also accumulates higher levels of citrate, giving rise to defects in growth in rich media and to sensitivity to the β-lactam antibiotic cefuroxime [30]. Mutations in the acetyl-CoA binding site of LmPC can suppress the growth defects and restore resistance to the cefuroxime. Therefore, regulation of LmPC by cyclic di-AMP is crucial for bacterial growth in rich media and antibiotic resistance.
L. lactis is an industrially important bacterium and is used for milk acidification. Like L. monocytogenes, L. lactis does not have a complete TCA cycle. However, in contrast to L. monocytogenes, L. lactis does not have a functional glutamate dehydrogenase and cannot synthesize glutamate from oxaloacetate de novo. The oxaloacetate product of LlPC is instead essential for the biosynthesis of aspartate, which is responsible for milk acidification. A L. lactis strain lacking PC had a significantly slower rate of acidification [27]. Cyclic di-AMP regulates LlPC and thereby the milk acidification property of L. lactis. A strain with elevated cyclic di-AMP had greatly reduced levels of aspartate, which could be restored only with a mutant LlPC (Y715T) that is insensitive to cyclic di-AMP, suggesting that the effect of cyclic di-AMP on aspartate levels in L. lactis is mediated primarily through LlPC.
3 Regulation of Host Metabolic Enzyme RECON by Cyclic di-AMP
3.1 Identification of RECON as a Direct Target for Cyclic di-AMP
Like the identification of PC, a similar chemical proteomics approach was used to identify host proteins that can bind cyclic di-AMP [14]. The oxidoreductase AKR1C13 (aldo-keto reductase family 1, member C13) is a highly abundant cyclic di-AMP binding protein in mouse liver extract, and the protein is renamed RECON (reductase controlling NK-κB). RECON has a high affinity for cyclic di-AMP, with a K d of 87 nM, while cyclic di-GMP, 2′,3′-cGAMP, NAD+, and other nucleotides show no competition for binding at 200–400 μM concentration. Cyclic di-AMP inhibits the oxidoreductase activity of RECON, while cyclic di-GMP and host-synthesized 2′,3′-cGAMP have no effects. In comparison, 3′,3′-cGAMP of bacterial origin competes with cyclic di-AMP for binding and also inhibits the catalytic activity of RECON. Overall, the host protein RECON is a direct target of cyclic di-AMP, with high affinity and selectivity for binding this bacterial second messenger.
3.2 Molecular Mechanism of RECON Binding by Cyclic di-AMP
In the complex with RECON, cyclic di-AMP assumes a nearly fully extended conformation and occupies the binding sites for NAD+ and the substrate of this enzyme [14] (Fig. 10.3a–c). One of the AMP moieties of cyclic di-AMP (AMP1) overlaps closely with the AMP portion of NAD+ (PDB entry 3LN3) (Fig. 10.3c), and the adenine base is recognized by hydrogen bonds to RECON (Fig. 10.3b). The other AMP moiety (AMP2) has essentially no overlap with NAD+, but its adenine base is located close to the redox-active C4 atom of the nicotinamide ring of NAD (Fig. 10.3c). Therefore, AMP2 likely has steric clashes with the expected substrate of the enzyme [31]. The structure of the complex illuminates the molecular basis for the selectivity of RECON for cyclic di-AMP and the inhibitory activity of the compound. AMP2 interacts with unique features in RECON and does not have overlap with NAD+ in the binding site, possibly explaining why cyclic di-AMP does not interact strongly with all NAD+-binding proteins.
Molecular basis for the regulation of RECON by cyclic di-AMP. (a) Overall structure of RECON (green) in complex with cyclic di-AMP (magenta). (b) Detailed interactions between cyclic di-AMP (magenta) and RECON (green). Hydrogen bonding interactions are indicated with dashed lines in red. W: solvent water. (c) Overlay of the binding modes of cyclic di-AMP (magenta) and NAD+ (cyan) to RECON. The position of the progesterone substrate in AKR1C1 is also shown (orange) [31]
3.3 Biological Impacts of RECON Binding by Cyclic di-AMP
The inflammatory activity of cyclic di-AMP was first identified due to its capacity to engage the host receptor STING in murine macrophages, resulting in the production of Type I IFN and other IRF-3 regulated genes. Within many host cells, two receptors for cyclic di-AMP are present, RECON and STING. The presence of RECON, which has over tenfold higher affinity for cyclic di-AMP than STING, results in sequestration of cyclic di-AMP secreted by L. monocytogenes during infection, and thereby negatively regulates the expression of STING-dependent inflammatory genes including interferon-β, CCL5 (RANTES), CXCL10 (IP-10), CXCL11 (I-TAC), interleukin-1β, and Nos2 [14].
While RECON presence negatively regulates STING activation in infected macrophages, it also represses NK-κB activation in infected hepatocytes, which are devoid of STING [14]. The expression of RECON itself does not change significantly during infection, and the catalytic activity of RECON is essential for its regulatory activity on cyclic di-AMP signaling. His117 is the general acid-base for the oxidoreductase activity of RECON. The H117A mutant of RECON is catalytically inactive but maintains an ability to bind cyclic di-AMP (with a K d of 288 nM). However, this mutant cannot complement the loss of wild-type RECON in infected hepatocytes in terms of NK-κB activation. RECON-deficient hepatocytes have elevated inflammatory responses upon L. monocytogenes infection, including NO production, and demonstrate enhanced intercellular spread of the bacteria [32]. The catalytic activity of RECON is also required for this effect.
Together these observations reveal that RECON enzyme activity is crucial for its capacity to sense cyclic di-AMP and augment inflammatory gene expression. This strongly supports a model in which accumulation of a substrate(s) of RECON, upon inhibition by cyclic di-AMP, mediates the NF-κB activation upon bacterial infection (Fig. 10.4). Aldoketoreductases like RECON are well known for their capacity to metabolize several lipophilic aldehyde and alcohol-containing metabolic intermediates, including steroid hormones, isoprenoids, retinoids, and oxidized lipids [33], which have pleiotropic effects on inflammation and cellular homeostasis in eukaryotes. Identification of the metabolic intermediate(s) targeted by RECON will not only reveal the mechanisms of inflammatory gene induction but may also provide evidence of broader impacts on host cell processes mediated by the RECON-cyclic di-AMP signaling axis.
Activation of NF-κB by RECON. Cyclic di-AMP potently inhibits the enzymatic activity of RECON, which is required for suppression of NF-κB. Evidence supports that inflammatory metabolite(s) downstream of TLR stimulation is/are enzymatically detoxified by RECON and cyclic di-AMP inhibition promotes antibacterial responses through blockade of this metabolic function
4 Regulation of the ydaO Riboswitch by Cyclic di-AMP
4.1 Identification of the ydaO Riboswitch as a Direct Target for Cyclic di-AMP
The ydaO riboswitch is widely distributed in Gram-positive bacteria and regulates cell wall metabolism, osmotic stress, sporulation, amino acid transporters, and other processes [34, 35]. Yeast extract contains a ligand that can bind to this riboswitch and cause changes in the pattern of its spontaneous cleavage. While the yeast extract-associated ligand was identified to be AMP [36], a thorough characterization of nucleotides containing the AMP moiety identified the endogenous bacterial derived ligand as cyclic di-AMP as the most potent and biologically relevant ligand of this gene control element [37]. The ydaO riboswitch has high affinity for cyclic di-AMP, with K d of 0.1 nM or lower under optimal assay conditions. The stoichiometry between ydaO and cyclic di-AMP is 1:1. Under conditions similar to bacterial cytosol, K d is ~10 nM. ydaO is highly selective for cyclic di-AMP, while cyclic di-GMP, cyclic di-IMP, AMP, ADP, and other compounds show much weaker binding. The hydrolysis product of cyclic di-AMP, pApA, has a K d of ~300 nM. Cyclic di-AMP binding to the riboswitch causes transcription termination in in vitro assays, and reduced levels of the second messenger lead to increased expression of a reporter under ydaO control in Bacillus subtilis, consistent with the riboswitch being a negative regulator.
The B. subtilis ydaO riboswitch has a weak affinity for ATP (K d of 0.6 mM) [36], but the binding is lost under physiological conditions and ATP has no effect on transcription termination regulated by ydaO [37].
4.2 Molecular Mechanism of ydaO Regulation by Cyclic di-AMP
The structure of the sensing domain of ydaO riboswitch has pseudo twofold symmetry, thereby creating two pockets to bind two cyclic di-AMP molecules (Fig. 10.5a) [38,39,40]. This 1:2 stoichiometry is confirmed by isothermal titration calorimetry experiments for Thermoanaerobacter tengcongensis but not B. subtilis ydaO, and it is not clear why there is a discrepancy with the results from in-line probing studies [37]. Cyclic di-AMP assumes a partially extended conformation and has extensive interactions with ydaO, including π-stacking interactions for the adenine base, and hydrogen-bonding interactions for the adenine base, ribose 3′-hydroxyl, and phosphate of cyclic di-AMP (Fig. 10.5b). Moreover, the interactions with the two AMP moieties of cyclic di-AMP are mostly equivalent, indicating a pseudo twofold symmetry within each binding site as well.
Molecular basis for the regulation of the ydaO riboswitch by cyclic di-AMP. (a) Overall structure of the Thermoanaerobacter tengcongensis ydaO riboswitch in complex with two cyclic di-AMP molecules. The riboswitch is shown as a cartoon and a semitransparent surface (orange), and cyclic di-AMP as sticks (green), labeled cdA. The pseudo twofold symmetry axis in the structure is indicated with the oval. (b) Detailed interactions between cyclic di-AMP and ydaO in one of the binding sites. Hydrogen-bonding interactions are indicated with the dashed lines (red). Equivalent interactions are observed in the other binding site
Specifically, the N1, N6, and N7 atoms of the adenine base have interactions with the hydroxyls of two different riboses (Fig. 10.5b), explaining the selectivity of this riboswitch for cyclic di-AMP over the other nucleotides. The 3′-hydroxyl of cyclic di-AMP is hydrogen-bonded to the carbonyl group of a cytosine base, consistent with the weaker affinity of deoxy cyclic di-AMP (K d ~ 20 nM) [37]. The phosphate group of cyclic di-AMP is hydrogen bonded to a guanine base. Mutations that disrupt the structure of the riboswitch or the π-stacking interactions with the adenine bases reduce the affinity for cyclic di-AMP. More extensive mutations in one binding site could reduce or abolish binding in the other site [39], suggesting communications between the two sites and/or the mutations have disrupted the overall structure of the riboswitch. Small angle X-ray scattering studies indicate that the riboswitch undergoes an extensive conformation change upon cyclic di-AMP binding, with the free riboswitch in a partially unfolded state [40]. This folding transition upon ligand binding has also been observed in other riboswitches.
4.3 Biological Impacts of ydaO Riboswitch Regulation by Cyclic di-AMP
Bioinformatic studies identified the ydaO riboswitch as a broadly conserved genetic element primarily found within the Gram-positive bacteria [35]. Analysis of the location of the ydaO motif within the transcriptional and translational unit of genes revealed the presence of transcriptional terminators and ribosome binding sites, supporting both transcriptional and translational mediated mechanisms of gene regulation. Those genes associated with the ydaO riboswitch are broadly involved in cell wall metabolism, amino acid transport, and osmolyte regulation, among others. These associations point to mechanisms by which cyclic di-AMP broadly shapes peptidoglycan synthesis and turnover, as well as cellular responses to osmotic stress.
Among the Actinobacteria, cyclic di-AMP-mediated control of muralytic enzymes involved in cellular resuscitation from dormancy suggests a role in promoting cellular growth [41, 42]. While originally identified bioinformatically, the validation and characterization of the ydaO riboswitch were first conducted in the model organism B. subtilis. Here, the ydaO riboswitch is associated with the potassium transporter ktrAB and the gene ydaO, a gene of unknown function that was recently revealed to also function as a potassium importer [12]. The role of cyclic di-AMP as a regulator of potassium in response to osmotic stress encountered by bacteria has emerged as a conserved physiological function of this second messenger [43]. However, the means by which cyclic di-AMP levels are controlled within cells are not yet clear, and early work with the ydaO riboswitch found that disruption of key genes involved in cellular respiration, including the NADH dehydrogenase and MenH involved in menaquinone biosynthesis, strongly promoted ydaO controlled transcription [12]. These observations support an intriguing possibility that not only do cyclic di-AMP levels modulate cellular metabolism but that central metabolic changes also regulate cyclic di-AMP levels to coordinate transcriptional changes that contribute to growth, including genes involved in cell wall metabolism and osmolyte/potassium accumulation, which are key requirements involved in controlling cellular turgor that drives growth of Gram-positive organisms.
5 Conclusions
Since its discovery 10 years ago, much has been learned about the crucial, pleiotropic effects of cyclic di-AMP in bacteria, such as metabolism, DNA repair, stress response, biofilm formation, and others, as well as the cellular receptors that mediate this myriad of functions of cyclic di-AMP. Moreover, this dinucleotide has an important role in host immune response to bacteria, and the molecular and functional mechanisms of this communication have begun to be elucidated. Overall, these studies demonstrate the biological significance of cyclic di-AMP and suggest that many new discoveries remain to be made in this exciting field.
References
Witte G, Hartung S, Buttner K, Hopfner K-P (2008) Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombinantion intermediates. Mol Cell 30:167–178
Romling U (2008) Great times for small molecules: c-di-AMP, a second messenger candidate in bacteria and archaea. Sci Signal 1:pe39
Corrigan RM, Grundling A (2013) Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524
Commichau FM, Dickmanns A, Gundlach J, Ficner R, Stulke J (2015) A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol Microbiol 97:189–204
Pham TH, Liang ZX, Marcellin E, Turner MS (2016) Replenishing the cyclic-di-AMP pool: regulation of diadenylate cyclase activity in bacteria. Curr Genet 62:731–738
Krasteva PV, Sondermann H (2017) Versatile modes of cellular regulation via cyclic dinucleotides. Nat Chem Biol 13:350–359
Huynh TN, Woodward JJ (2016) Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol 30:22–29
Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA 3rd, Smith HO, Venter JC (2006) Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A 103:425–430
Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289
Whiteley AT, Pollock AJ, Portnoy DA (2015) The PAMP c-di-AMP is essential for Listeria growth in macrophages and rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 17:788–798
Fahmi T, Port GC, Cho KH (2017) c-di-AMP: an essential molecule in the signaling pathways that regulate the viability and virulence of Gram-positive bacteria. Genes 8:E197
Gundlach J, Herzberg C, Kaever V, Gunka K, Hoffmann T, Weiss M, Gibhardt J, Thurmer A, Hertel D, Daniel R, Bremer E, Commichau FM, Stulke J (2017) Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci Signal 10:eaal3011
Blotz C, Treffon K, Kaever V, Schwede F, Hammer E, Stulke J (2017) Identification of the components involved in cyclic di-AMP signaling in Mycoplasma pneumoniae. Front Microbiol 8:1328
McFarland AP, Luo S, Ahmed-Qadri F, Zuck M, Thayer EF, Goo YA, Hybiske K, Tong L, Woodward JJ (2017) Sensing of bacterial cyclic dinucleotides by the oxidoreductase RECON promotes NF-kB activation and shapes a proinflammatory antibacterial state. Immunity 46:433–445
Devaux L, Kaminski PA, Trieu-Cuot P, Firon A (2018) Cyclic di-AMP in host-pathogen interactions. Curr Opin Microbiol 41:21–28
Xia P, Wang S, Xiong Z, Zhu X, Ye B, Du Y, Meng S, Qu Y, Liu J, Gao G, Tian Y, Fan Z (2018) The ER membrane adaptor ERAdP senses the bacterial second messenger c-di-AMP and initiates anti-bacterial immunity. Nat Immunol 19:141–150
Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH (2013) STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio 4:e00018–00013
Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee JH, Bishai WR (2015) A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat Med 21:401–406
Gries CM, Bruger EL, Moormeier DE, Scherr TD, Waters CM, Kielian T (2016) Cyclic di-AMP released from Staphylococcus aureus biofilm induces a macrophage type I interferon response. Infect Immun 84:3564–3474
Andrade WA, Firon A, Schmidt T, Hornung V, Fitzgerald KA, Kurt-Jones EA, Trieu-Cuot P, Golenbock DT, Kaminski PA (2016) Group B Streptococcus degrades cyclic-di-AMP to modulate STING-dependent type I interferon production. Cell Host Microbe 20:49–59
Dey RJ, Dey B, Zheng Y, Cheung LS, Zhou J, Sayre D, Kumar P, Guo H, Lamichhane G, Sintim HO, Bishai WR (2017) Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat Chem Biol 13:210–217
Jitrapakdee S, St. Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV (2008) Structure, mechanism and regulation of pyruvate carboxylase. Biochem J 413:369–387
Tong L (2013) Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70:863–891
Sellers K, Fox MP, Bousamra M 2nd, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, Lane AN, Fan TW (2015) Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Investig 125:687–698
Phannasil P, Thuwajit C, Warnnissorn M, Wallace JC, MacDonald MJ, Jitrapakdee S (2015) Pyruvate carboxylase is up-regulated in breast cancer and essential to support growth and invasion of MDA-MB-231 cells. PLoS One 10:e0129848
Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer J-D, Tong L, Woodward JJ (2014) The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158:1389–1401
Choi PH, Vu TMN, Pham HT, Woodward JJ, Turner MS, Tong L (2017) Structural and functional studies of pyruvate carboxylase regulation by cyclic-di-AMP in lactic acid bacteria. Proc Natl Acad Sci U S A 114:E7226–E7235
Tong L (2017) Striking diversity in holoenzyme architecture and extensive conformational variability in biotin-dependent carboxylases. Adv Protein Chem Struct Biol 109:161–194
Schar J, Stoll R, Schauer K, Loeffler DIM, Eylert E, Joseph B, Eisenreich W, Fuchs TM, Goebel W (2010) Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J Bacteriol 192:1774–1784
Whiteley AT, Garelis NE, Peterson BN, Choi PH, Tong L, Woodward JJ, Portnoy DA (2017) c-di-AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Mol Microbiol 104(2):212–233
Couture JF, Legrand P, Cantin L, Luu-The V, Labrie F, Breton R (2003) Human 20a-hydroxysteroid dehydrogenase: crystallographic and site-directed mutagenesis studies lead to the identification of an alternative binding site for C21-steroids. J Mol Biol 331:593–604
McFarland AP, Burke TP, Carletti AA, Glover RC, Tabakh H, Welch MD, Woodward JJ (2018) RECON-dependent inflammation in hepatocytes enhances Listeria monocytogenes cell-to-cell spread. mBio 9:e00526–00518
Rizner TL (2012) Enzymes of the AKR1B and AKR1C subfamilies and uterine diseases. Front Pharmacol 3:34
Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR (2004) New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A 101:6421–6426
Block KF, Hammond MC, Breaker RR (2010) Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol 192:3983–3989
Watson PY, Fedor MJ (2012) The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol 8:963–965
Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR (2013) Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9:834–839
Ren A, Patel DJ (2014) c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol 10:780–786
Gao A, Serganov A (2014) Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol 10:787–792
Jones CP, Ferre-D’Amare AR (2014) Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. EMBO J 33:2692–2703
St-Onge RJ, Haiser HJ, Yousef MR, Sherwood E, Tschowri N, Al-Bassam M, Elliot MA (2015) Nucleotide second messenger-mediated regulation of a muralytic enzyme in Streptomyces. Mol Microbiol 96:779–795
Schwenk S, Moores A, Nobeli I, McHugh TD, Arnvig KB (2018) Cell-wall synthesis and ribosome maturation are co-regulated by an RNA switch in Mycobacterium tuberculosis. Nucleic Acids Res 46:5837–5849
Gundlach J, Commichau FM, Stulke J (2018) Perspective of ions and messengers: an intricate link between potassium, glutamate, and cyclic di-AMP. Curr Genet 64:191–195
Armon A, Graur D, Ben-Tal N (2001) ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol 307:447–463
Acknowledgment
The research on cyclic di-AMP signaling in the authors’ laboratories is supported by NIH grant R01AI116669.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Tong, L., Woodward, J.J. (2020). Metabolic Regulation by Cyclic di-AMP Signaling. In: Chou, SH., Guiliani, N., Lee, V., Römling, U. (eds) Microbial Cyclic Di-Nucleotide Signaling. Springer, Cham. https://doi.org/10.1007/978-3-030-33308-9_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-33308-9_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-33307-2
Online ISBN: 978-3-030-33308-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)