Abstract
Epigenetics refers to mitotically/meiotically heritable mechanisms that regulate gene transcription without a need for changes in the DNA code. Covalent modifications of DNA, in the form of methylation, and histone post-translational modifications, in the form of acetylation and methylation, constitute the epigenetic code of a cell. Both DNA and histone modifications are highly dynamic and often work in unison to define the epigenetic state of a cell. Most epigenetic mechanisms regulate gene transcription by affecting localized/genome-wide transitions between heterochromatin and euchromatin states, thereby altering the accessibility of the transcriptional machinery and in turn, reduce/increase transcriptional output. Altered chromatin structure is associated with cancer progression, and epigenetic plasticity primarily governs the resistance of cancer cells to therapeutic agents. In this chapter, we specifically focus on regulators of histone methylation and acetylation, the two well-studied chromatin post-translational modifications, in the context of prostate cancer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Introduction
Prostate Cancer (PCa) is a disease characterized by genetic and epigenetic alterations. Accumulation of somatic genomic alterations such as mutations and chromosomal rearrangements and global changes to the chromatin landscape contribute to prostate cancer initiation, progression, and therapy resistance. Specifically, genomic alterations in genes that encode chromatin regulators and chromatin-remodeling factors are enriched in advanced, metastatic castration resistant prostate cancer (mCRPC). Epigenetic reprogramming of chromatin alters the accessible regions of the genome resulting in differential transcriptional output during prostate oncogenesis and progression.
Within the cell nucleus, the linear genomic DNA is organized into a highly compact form called chromatin , which helps fit the entire 2-m-long genomic DNA into a cell nucleus measuring only ~10 μm in diameter. This compaction is reversible and is mediated by interactions between the negatively charged DNA and a set of four proteins called histones. The fundamental unit of chromatin is a nucleosome that contains 145–147 bp of DNA wrapped about 1.65 times around a globular octamer complex formed from homo-dimers of the core histone proteins H2A, H2B, H3, and H4. This chromatin conformation resembles a “beads on a string” structure [1, 2]. A fifth histone protein H1 binds to the DNA between two adjacent nucleosomes and acts as a nucleosome-linker and further promotes compaction and stabilization of the chromatin into a 30 nm fiber representing a higher order chromatin structure [3, 4]. This higher order chromatin structure can be further classified into “heterochromatin” that represents a highly condensed “transcriptionally inactive” state not accessible to the transcriptional machinery, and “euchromatin” that represents an open “transcriptionally active” state containing most of the active genes and accessible to the transcription machinery.
Recent advances in chromosome conformation capture technologies (e.g., 4C, HiC, and its derivatives) [5, 6] to study the three dimensional (3D) organization of chromatin have shown that the chromatin architecture is complex and is organized in a less-random fashion, resulting in a higher order 3D organization of genome with heterochromatic and euchromatic compartments [7]. At the sub-chromosomal level, the gene regulatory regions are classified into promoters that are located near the Transcription Start Site (TSS) of genes, and enhancers that are typically located at a considerable distance from the TSS [8]. Upon activation by tissue-specific transcription factors, these enhancers modulate gene expression by physically interacting with their target gene promoters through chromatin looping. This looping, which facilitates the physical interaction between distal enhancers and gene promoters, is further propagated by proteins that promote this contact, thereby constituting gene-modulatory landscape [9]. Based on such high levels of internal interactions, chromatin structure is sub-divided into many domains known as topologically associating domain (TAD) . They are the chromatin domains with high levels of internal interactions and are separated from each other by regions of low interaction called boundary elements. TADs constitute fundamental units of the 3D organization of the genome, promoting enhancer-promoter interactions [10]. Characterization of these gene regulatory units like promoters, enhancers and their 3D interactions has opened avenues for understanding mechanisms of transcriptional control of genes. Chromosomal architecture is primarily mediated by the CCCTC-binding factor (CTCF)/cohesin complex. CTCF is an 11-zinc-finger transcription factor that is enriched at boundary elements of TADs and loop domains and is essential for the recruitment of cohesin to chromatin [11, 12]. This suggests that the microscopic structures of chromatin configurations are much more complex, and that the dynamic, yet controlled modulations of these chromatin conformations are essential for timely, tissue-specific, and coordinated gene expression. Efficient regulation of gene expression and cellular processes are mediated through modulating chromatin structure by three mechanisms involving the DNA and histones: (1) Covalent modifications of DNA, (2) Post-translational modifications of histone tails by histone modifying enzymes, and (3) Disruption of histone-DNA contacts by ATP dependent chromatin remodeling proteins. These epigenetic regulatory mechanisms work independently or in unison to modulate chromatin architecture, which in turn regulates gene expression, thereby governing cellular function.
DNA Methylation as an Epigenetic Code for Prostate Tumor Development
Methylation patterns of cytosine residues within CpG dinucleotide sequences play an important role in key cellular process such as DNA repair, recombination and replication, and regulation of gene expression [13, 14]. DNA methylation based regulatory mechanisms are highly dynamic where in nearly 60–80% of CpG sites in the mammalian genome display altered methylation patterns. Cytosine methylation is catalyzed by DNA methyltransferases DNMT1, DNMT3A, and DNMT3B that transfer a methyl group from S-adenosylmethionine to the 5′ carbon of the cytosine ring to form 5-methylcytosine (5-mC). CpG dinucleotide sequences are enriched at active gene promoters and hypermethylation of these regions leads to the preferential binding of methyl CpG binding domain (MBD) proteins, such as MBD1, MBD2, MBD3, MBD4, Kaiso, and MECP2 [15]. These MBD proteins contain transcriptional repression domains, which in association with histone deacetylases (HDACs), repress target gene transcription. In prostate cancer development, aberrant hypermethylation is commonly observed in the promoter regions of genes associated with tumor-suppressor activity (e.g. APC, RARβ, RASSF1A, p16), DNA-repair (e.g. GSTP1, MGMT, GSTM1), cell cycle control (e.g. CCNA1, CDKN2A, CCND2, H1C1, SFN), apoptosis (e.g. PYCARD, DAPK, SLC5A8, SLC18A2, TNFRSF10C, RUNX3), and maintenance of cell-cell contacts (e.g. CDH1, CD44), whose repression enables the growth and stabilization of neoplastic phenotypes [13, 16, 17]. Additionally, DNA hypermethylation represses the transcription of microRNAs leading to upregulation of their oncogenic targets that can drive tumorigenesis [18]. DNA demethylation proceeds primarily through oxidative reactions catalyzed by the TET family oxygenases that convert 5-methylcytosine → 5-hydroxymethylcytosine → 5-formylcytosine → 5-carboxylcytosine, which is then converted to cytosine through the action of thymine DNA glycosylase [19]. DNA hypomethylation, which activates gene transcription primarily at the promoter regions of oncogenes, is observed more frequently in mCRPC compared to PCa [13]. Genes regulated by aberrant DNA hypomethylation in prostate cancer include MYC, RAS, uPA, PLAU, HPSE, CYP1B1, WNT5A, S100P, and CRIP1. More details on the role of DNA methylation in Pca and their potential for use as diagnostic and prognostic biomarkers may be found in several excellent reviews on this topic [13,14,15,16,17,18, 20].
Histone Post-translational Modifications
Amino acid residues in the unstructured N-terminal tails of histone proteins H3 and H4, and both N- and C-termini tails of H2A and H2B, display a variety of covalent, reversible, post-translational modifications (PTMs) that define the epigenetic code. PTMs are strongly associated with specific amino acid residues. Some of the well-studied PTMs include methylation (occurring on K and R residues), acetylation (K, S, T), phosphorylation (S, T, Y, H), ubiquitination (K), sumoylation (K), ADP ribosylation (K, E), succinylation (K), 2-hydroxyisobutyrylation (K) formylation (K), malonylation (K), propionylation (K), butyrylation (K), crotonylation (K), hydroxylation (Y), citrullination, a.k.a. deamination (R), O-GlcNAcylation (S, T), and proline isomerization [21,22,23,24,25]. The highly dynamic histone PTMs regulate cell function by altering DNA-histone interactions, nucleosomal assembly, and global/local higher order structures of chromatin, all of which in turn directly control the accessibility of genes to transcription factors. The major cellular processes regulated by the histone PTM epigenetic code include gene transcription, gene-repair, metabolism, replication, and chromatin condensation [23].
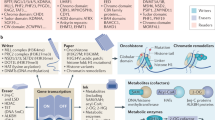
The vast number and combinations of histone modifications define highly complex epigenetic regulatory states of chromatin. Unlike the CpG specific DNA methylation discussed earlier, histones can be methylated at any of the lysine or arginine residues. Hence, the position and state of methylations, and in effect that for all PTMs, represents integral part of the epigenetic code. For example, tri-methylations H3K9me3 and H3K27me3 are primarily linked to compact and closed chromatin conformations (heterochromatin) that repress gene transcription [26] (see Fig. 1a), while H3K4me1/me2/me3, and H3K36me3 are associated with relaxed chromatin conformations (euchromatin) that activate gene transcription [11] (see Fig. 1b).
Effect of state-dependent methylation on transcriptional activity. Tri-methylation (me3) of histone H3K9 or K27 leads to closed chromatin conformations, i.e., heterochromatin, resulting in transcriptional repression (a), while mono/di/tri-methylation of H3K4 and H3K36 leads to open and relaxed chromatin conformation, i.e., euchromatin, resulting in transcriptional activation (b)
In PCa, histone methylation and acetylation are often associated with progression and metastasis. Compared to non-malignant phenotypes, the levels of H3K4me1, H3K9me2, H3K9me3 are often reduced in primary prostate cancer and increased in mCRPC. Acetylation of H3K9, H318, and H4K12 also often display a similar trend [3, 17, 27]. Similarly, acetylation of the histone variant H2A.Z at active promoter sites is often associated with oncogene activation in PCa [28].
Factors governing epigenetic regulation can be broadly classified into three distinct groups namely readers—proteins that recognize a PTM, writers—enzymes that catalyze the addition of a PTM, and erasers—enzymes that catalyze the removal of a PTM. Each histone PTM has a specific set of reader, writer, and eraser proteins. In this chapter, we will specifically focus on the well-studied enzymatic machineries governing histone acetylation and methylation in PCa. Histone methylation and demethylation are mediated by histone methyltransferases (HMTs) and demethylases (HDMs), while acetylation and deacetylation are mediated by histone acetyltransferases (HATs), histone deacetylases (HDACs) and sirtuins . A significant number of these epigenetic readers, writers and erases are dysregulated and mutated in PCa.
Histone Methylation in Prostate Cancer
Histone methylation occurs primarily on the side chains of all basic amino acid residues, i.e. lysine (K), arginine (R) and histidine (H). Lysines can be mono (me1), di (me2), or tri (me3) methylated on their ε-amine group, Arginines can be mono (me1), symmetrically dimethylated (me2s), or asymmetrically dimethylated (me2a) on their guanidinyl group, whereas histidines have been reported to be monomethylated, although this is a rare form of methylation. Interestingly, unlike acetylation, histone methylation does not alter the charge of the histone protein [29]. Rather, the position and the state of methylation (me1/me2/me3) define different regulatory states. Histone methylation plays a central role in (1) transcriptional activation/silencing via chromosomal looping and chromatin remodeling, (2) recruitment of cell specific transcription factors via interactions with initiation and elongation factors, as well as (3) RNA splicing [30, 31]. Histone methylation dynamics regulates a variety of cell functions including cell-cycle regulation, DNA damage and stress response, development and differentiation. Thus, aberrations in histone methylation patterns and the enzymes regulating histone methylation play major roles in cancer development and growth.
Histone lysine methyltransferases (HMTs) catalyze the transfer of a methyl group from S-adenosylmethionine (SAM) to a lysine ε-amino group on the N-terminal tails of histones. Since the discovery of the first HMT, SUV39H1 that catalyzes H3K9me3, a variety of methyltransferases that target lysine on H3 and H4 histones have been identified [21]. A characteristic feature of all lysine methylating HMTs is that they contain a SET domain, which harbors the enzymatic activity, except for DOT1L, which lacks a SET domain. HMTs are further classified into the SUV39, EZH, SET1, SET2, PRDM, and SMYD sub-families based on the sequence homology in and around the SET domain [32]. Some of the most extensively studied histone methylation sites include H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20. The enzymatic machinery that regulates the methylation of these residues is shown in Fig. 2. Generally, H3K4, H3K36, and H3K79 methylations mark sites of active transcription, while methylations of H3K9, H3K27, and H4K20 are associated with sites of silenced transcription [31]. Arginine methylation is catalyzed by arginine methyltransferases, which are divided into two subclasses—Class I and Class II enzymes. Together, these two types of arginine methyltransferases constitute a relatively large protein family with a total of 11 members that are referred to as PRMTs.
The highly stable methyl bonds on methylated lysines are removed through an oxidative mechanism catalyzed by Histone lysine demethylases (KDMs). Demethylases are classified into two major families: (1) those with an amine oxidase domain that use Flavin adenine dinucleotide (FAD) as a cofactor, and (2) those containing a Jumonji C (JmJC)-domain that use iron and α-ketoglutarate as cofactors to catalyze their oxidative reactions [31, 33, 35,36,37]. The first type belongs to the KDM1 family of KMTs, with two members LSD1/KDM1A and LSD2/KDM1B, that only catalyze demethylation of mono- and di-methyl groups. The JmJC family of KDMs contain 28 members that are further subdivided into the KDM2, KDM3, KDM4, KDM5, JARID2, KDM6, KDM7, and KDM8 subfamilies. Each subfamily is characterized by the similarities in their DNA binding, DNA recognition domains, and specific histone and non-histone substrates. The JmJC KDMs demethylate all mono-, di-, and tri-methylated histone lysines.
The following section will focus on the H3 and H4 lysine methylations/demethylations displayed in Fig. 2. It should be noted that in addition to these well studied marks, histone methylation has also been detected in all basic residues in all the four nucleosomal histones [29] and their specific roles in cell function remain to be elucidated.
H3K4 Methylation
A genome-wide study of H3K4 methylation at 44 loci in the human genome selected by the ENCODE consortium showed that H3K4me1 is a mark for active or poised enhancers, H3K4me3 is a mark for promoter regions of genes poised for or undergoing active transcription, while H3K4me2 marks both enhancer and promoter regions [38]. These findings were also verified in genome-wide studies of a panel of five human cell lines [39]. Though these marks are evolutionarily conserved, their exact role in active transcription of a specific gene depends on a number of factors such as the transcription frequency, elongation rate, and COMPASS activity [40]. H3K4me3 is highly enriched at TSSs, but totally depleted in the elongation regions [41] and CpG islands [42]. A majority of the highly transcribed genes have a gradient of H3K4me3 > H3K4me2 > H3K4me1 along the gene body from the 5′ end to the 3′ end, but the exact methylation patterns are gene-specific [40]. While observations on the function of a H3K4 mark holds true in most of the cases, its exact function is determined by the activity of the reader protein that recognizes the mark. For example, both H3K4me2 and H3K4me3 marks bound by plant homeodomain (PHD)-domain containing ING2 (inhibitor of growth family member 2) proteins are associated with transcriptional repression. Also, H3K4 methylation levels correlate with DNA damage signaling [43].
A large body of evidence points to the importance of H3K4 methylation in PCa tumorigenesis and metastasis. Tissue microarray analysis of prostate tumor tissues revealed that patients with reduced H3K4me2 levels were at an increased risk for relapse [27, 44], and these results were verified through analysis of patient microarray data [45]. Studies using CRPC cell lines and tissues found that H3K4me1 and H3K4me2 marks were selectively enriched at androgen receptor (AR)-regulated enhancers of cell cycle genes such as UBE2C and CDK1 that promote CRPC growth [46]. Further, an increase in H3K4me3 marks correlated with activation of cell growth and survival genes FGFR1 and BCL2 [47]. A genome-wide ChIP-seq study using LNCaP cells showed that H3K4me2 precisely marked the nucleosomes flanking AR binding motifs at distal enhancers regulating the transcription of key AR target genes such as TMPRSS2, and PSA [48]. Alteration in H3K4 levels are also a result of dysregulation/mutations in the enzymatic machinery that regulates methylation. For example, the tumor suppressor gene PTEN is frequently deleted in primary PCa, and is lost to a greater extent in the mCRPC tumors [49], leading to deregulated PI3K signaling. This is turn affects the subcellular localization of the KDM5A demethylase [50] leading to genome-wide alteration of H3K4 levels. Reduced H3K4me2 levels in primary PCa was shown to be associated with increased risk for recurrence and metastasis [29]. Whole exome sequencing studies identified AR interactions with proteins of the KMT2 complex [51], specifically with Menin and ASH2L [52]. In the following section we will briefly describe the major methylation writers, and erasers that govern the dynamics of H3K4 methylation and their known role in the development of PCa.
H3K4 Methylation Writers
In mammalian cells, methylation of H3K4 is catalyzed by the KMT2/MLL/COMPASS family of methyltransferases; MLL1/KMT2A, MLL2/KMT2D, MLL3/KMT2C, MLL4/KMT2B, MLL5/KMT2E, SETD1A/KMT2F, andSETD1B/KMT2G; SMYD family methyltransferases; SMYD1, SMYD2, and SMYD3; SETD7, and PRDM9 (see Fig. 2) [29, 31, 53].
KMT2/MLL/COMPASS family Methyltransferases : COMPASS (Complex proteins associated with Set1) proteins in mammalian cells are a family of six methyltransferases. All proteins in this family of methyltransferases contain a SET1 domain in complex with four common subunits—namely, WDR5, ASH2L, RbBP5, and DPY30—and other protein specific subunits chosen from CXXC1, WDR82, HCF1, HCF2, Menin, PTIP, PA1, and NCOA6 [31, 54]. The 130–140 amino acid long SET1 (Suppressor of variegation, Enhancer of Zeste, Trithorax) domain is conserved across all KMT2 family methyltransferases and is responsible for catalyzing lysine methylation activity [54,55,56]. An analysis of 12 different cancers in the cancer genome atlas (TCGA) dataset revealed that KMT2 family methyltransferases are frequently dysregulated and mutated in nearly all cancers, and prominently in bladder, lung, and endometrial cancers [57, 58].
KMT2A and KMT2B (MLL1 and MLL4) writers : MLL1/KMT2A is the founding member of the KMT2 family of methyltransferases, which was originally observed in the 11q23 chromosomal translocation known to be the key driver of acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). This family of methyltransferases carries the unique COMPASS protein subunits Menin, and HCF1 or HCF2. MLL1 and MLL4 both contain a N-terminal CXXC domain and a C-terminal SET domain. MLL1, which also contains an AT-hook, stably binds to mitotic chromatin at both enhancer and promoter regions through multivalent interactions of its AT-hook and CXCC domains with AT-rich and unmethylated CpG dinucleotides, respectively [58]. It remains bound to the chromatin during DNA replication and mitosis to ensure the propagation of a cell’s transcription state to the daughter cells. MLL1 and MLL4 catalyze high levels of mono-, di-, and low levels of tri- methylation of H3K4, and MLL1 is localized to regions of RNA polymerase II activity, specifically at the 5′ end of actively transcribed genes [59]. MLL1 is also associated with microRNAs involved in cancer and hematopoiesis. Loss of MLL1 affects embryogenesis, transcriptional elongation and cancer development. Both MLL1 and MLL4 harbor cleavage sites for threonine aspartase, taspase1, hence their activity is also regulated by Taspase1. Furthermore, MLL1 is regulated by histone H2B ubiquitination. While the role of 11q23 translocations impacting MLL1 is well established in ALL and AML, analyses of the Catalogue of Somatic Mutations in Cancer (COSMIC) database showed that greater than 60% of lung, breast, bladder, endometrium, and large intestine cancer patients harbored one or more mutations in MLL1, but fewer in MLL4.
KMT2C and KMT2D (MLL3 and MLL2) writers : MLL2 and MLL3 primarily catalyze mono-methylation of H3K4 at enhancer regions [60, 61]. The unique COMPASS protein subunits in MLL2 and MLL3 are PTIP, PA1, NCOA6, and UTX. The stable binding of MLL2 and MLL3 to chromatin is mediated by their high mobility group I (HMG-I) and LXXLL binding motifs that are common in most transcription factors and coactivators [58]. Both MLL2 and MLL3 contain seven PHD domains via which they bind to arginine residues H3R3me0 and H3R3me2a within intergenic regions [62]. MLL2 is a known co-activator of the estrogen receptor (ER)-, and is required for ER-α transcriptional activity as well as proliferation of ER-α positive breast cancer cell lines, such as MCF7 [63]. MLL3 mono-methylation is essential for IgG class switching, adipogenesis, and nuclear receptor co-activators [54]. Loss of MLL3 results in developmental defects, reduction in white adipose tissue, embryogenesis and growth [58]. Analysis of the COSMIC database showed that MLL2 and MLL3 were mutated in >60% of lung, large intestine, breast, endometrium, and bladder cancers, and ~25% of non-Hodgkin lymphoma, medulloblastoma, and primitive neuroendodermal cancers. A recent clinical study on 46 Chinese PCa patients revealed that MLL2 is mutated in 63% of the samples and drives PCa progression by activating LIFR and KLF4 [64]. Whole exome sequencing studies of 50 mCRPC samples identified recurrent MLL2 mutations in 8.6% of patients [51].
KMT2F and KMT2G (SETD1A and SETD1B) writers : SETD1A and SETD1B catalyze mono-, di-, and tri-methylation of H3K4. In most cells, these enzymes catalyze the bulk of H3K4me3 methylations. They are the smallest subgroup of the KMT2 family and contain a SET domain that catalyzes methylation, an RNA recognition motif (RRM) at the N-terminal, a N-SET domain at the C-terminal, a WD-repeat 82 domain that interacts with RNA Pol II, and a CXXC finger domain that preferentially binds to CpG dinucleotides. Genome-wide ChIP-seq analysis of SETD1A and SETD1B binding patterns reveal that they preferentially bind to gene promoter regions justifying their strong preference for H3K4 tri-methylation [58, 65]. SETD1A is recruited in a transcription factor dependent manner and is essential for development, cell proliferation, and induced pluripotency [58]. Analysis of lung, large intestine, endometrium, liver and skin datasets in the COSMIC database found that ~62% of the chosen cancers harbored SETD1A mutations and ~60% harbored mutations in the SETD1B enzyme [58]. However, a similar analysis for the prostate adenocarcinoma dataset showed no evidence of SETD1A or SETD1B mutations (<0.5%) or dysregulation in PCa, consistent with other reports [54].
SMYD family methylation writers : SET and MYND domain-containing proteins (SMYD) are another family of five histone and non-histone substrate methyltransferases, of which only SMYD1, and SMYD3 are known to catalyze mono-, di-, and tri-methylation of H3K4 [66,67,68] (also see Fig. 1). SMYD2, on the other hand catalyzes H3K36 methylation [68]. SMYD3 monomethylates H4K5 [69] and trimethylates H4K20 [70]. SMYD family KMTs contains a bi-lobal architecture with the SET and MYND domains on the N-terminal lobe and a TPR-like domain on the C-terminal lobe. The interface between the N- and C-terminal lobes act as cavernous binding sites for protein substrates, which bind a wide variety of proteins including p53 (protein data bank ID: 3TG5), and ER-α (protein data bank ID: 406F) [67]. SMYD family proteins are key regulators of development and function of skeletal and cardiac muscles, and specifically SMYD1 is essential for thick filament organization of myosin [71].
SMYD3 is overexpressed in a broad range of cancers, including prostate, breast, and many colorectal and hepatocellular carcinomas. It regulates transcriptional activity of nearly 80 genes by forming a complex with RNA pol II and binding to their promoter regions. A well-studied example is the SMYD3 regulation of homeobox gene NKX2.8 that is commonly upregulated in hepatocellular carcinoma. The role of SMYD3 in oncogenesis was demonstrated by its siRNA-mediated knockdown, which suppressed proliferation in HCC, while its introduction into 3T3 cells enhanced cell growth [72]. IHC staining in paired normal prostate and prostate tumor samples showed that SMYD3 was highly upregulated in tumors, and was primarily localized to the cytoplasm (in 92% of samples) compared to the nucleus (32% of samples) [73]. These findings suggest that the role of SMYD3 in cellular function extends beyond its methyltransferase activity. AR signaling was downregulated upon siRNA and shRNA mediated knockdown of SMYD3 leading to increased apoptosis and S phase accumulation, and decreased proliferation, colony formation, transwell cell migration, and invasion [73]. Interestingly, MMP-9, which is transcriptionally regulated by AR and associated with invasiveness in PCa cells, was upregulated by SMYD3 activity. This points to a cross-talk between the SMYD3 and AR regulatory networks [74, 75].
Currently, much research is focused on developing strategies to effectively inhibit SMYD3 [76]. For instance, one group synthesized a small molecule inhibitor BCl-121, that interrupts the SMYD3-substrate interaction at the histone peptide binding channel and thus prevents SMYD3 chromatin localization. Treatment with a high dose of 100 μM was able to induce S-phase arrest and thereby inhibit growth of cancer cells that specifically overexpress SMYD3. Noteworthy, this growth inhibitory effect of BCl-121 was observed in the LNCaP and DU145 prostate cancer cell lines, where proliferation was reduced by approximately 70% [77]. Epizyme has developed sulfonamides and sulfamides that inhibit the enzyme, including the orally bioavailable EPZ031686 [78]. GSK designed a small molecule, GSK2807, based on the crystal structure of SMYD3/MEKK2/SAH that can compete with SAM to link SMYD3 with its substrate [79]. H3K4 methylation is also catalyzed by other KMTs such as SETD7, and RMTs such as PRDM9. For detailed reviews on this topic see reference [67].
H3K4 Methylation Erasers
H3K4 demethylation is primarily catalyzed by KDM1A, KDM1B, KDM5A, KDM5B, KDM5C, KDM5D, and MAPJD KMTs [31, 33].
KDM1 family H3K4 methylation erasers : The KDM1 family consists of KDM1A/LSD1 and KDM1B/LSD2, both of which demethylate H3K4me1 and H3K4me2 residues (see Fig. 2) and is one of the well-studied lysine demethylases. LSD1 is a known activator of AR, and its overexpression is essential for the maintenance of malignant phenotypes in mCRPC, and also in other cancers including bladder, colorectal, AML, neuroblastoma, and estrogen-receptor-negative breast cancer [36, 66, 80]. LSD1 dysregulation is an oncogenic driver of PCa due to its ability to: (1) control phenotypic plasticity of PCa cells [81, 82], (2) promote AR-independent survival in mCRPC cells in a non-canonical, demethylase-independent manner [83], (3) regulate expression of the AR-V7 splice variant [84], (4) activate mCRPC gene networks independent of its demethylase activity [83], and (5) control metastasis of CRPC [85]. It also demethylates p53 and represses p53 function, inhibits differentiation of neuroblastoma and leukemia, and interacts with EWS/FLI1 in osteosarcoma [35]. LSD2 is a homolog of LSD1, but little is known about its role in cancer.
KDM5 family H3K4 methylation erasers : The KDM5/JARID1 family of lysine methyltransferases, KDM5A/JARID1A, KDM5B/JARID1B, KDM5C/JARID1C, and KDM5D/JARID1D, catalyze demethylation of di-, and tri-methylated H3K4. The JARID1 family of proteins is unique among all JmJc family KDMs in that they contain a DNA binding ARID domain and a methylation binding PHD domain intersecting their JmJN and JmJC domains [86]. The KDM5 clusters are primarily associated with the removal of the active H3K4me2 and H3K36me2 marks that leads to gene silencing and are important for neuronal and hematopoietic development, as well as drug resistance [87]. KDM5A, KDM5B, and KDM5C function as oncogenic drivers [88] and are overexpressed in prostate cancer [33, 89], while KDM5D functions as a putative tumor suppressor and is down-regulated/deleted in prostate cancer [88]. KDM5A, originally identified as a retinoblastoma-binding protein (RBP) [90], is an integral part of the Notch/RBP-J repressor complex mediated gene silencing machinery that regulates Notch-signaling pathways. KDM5A plays an important role in homeostasis and carcinogenesis [35, 91] and is implicated in dysregulated Notch signaling with concomitant increase in Notch family proteins such as Jagged2, Notch3, and Hes6. KDM5A has been observed in highly invasive, high grade prostate cancer [92,93,94]. KDM5A overexpression has been linked to chemoresistance in both prostate and lung cancer cell lines [33, 80]. KDM5B, also known to demethylate H3K4me1 [29], is associated with normal development since it protects developmental genes from aberrant H3K4me3 modifications [36]. Overexpression of KDM5B attenuated transcription of genes related to melanoma progression [33] and promoted the aggressiveness of non-small cell lung cancer [95], while its knockdown inhibited tumor growth [33]. KDM5B is also regulated by AKT levels where it was recently shown that AKT inhibition in PTEN knockout mice reduced expression of KDM5B [96]. Analysis of Oncomine data showed that KDM5C expression was elevated in mCRPC and PCa, and could be used as a biomarker to predict the metastatic potential of primary tumors [88]. Furthermore, KDM5B has been shown to regulate a number of genes including BRCA1, CAV1, and HOXA5 and as a result is one of most altered KDMs in a number of cancers [31, 97]. The KDM5C gene, found on the X chromosome, is overexpressed in prostate cancer and is a prognostic marker for tumor relapse post radical prostatectomy [98]. Moreover, BRD4 mediated transcription of KDM5C led to mCRPC cell sensitivity to BET inhibitors [99]. Its inactivation triggered genomic instability in sporadic renal cancer [100]. KDM5C, independent of its enzymatic activity, functions as an oncogene by repressing the TGFβ-dependent transcription factor SMAD3 which in turn promotes PCa tumorigenesis [88]. KDM5D, a homolog of KDM5C found on the Y chromosome, is a putative tumor suppressor that is downregulated in prostate cancer. A recent analysis of the Oncomine datasets showed that prostate tumors with high Gleason score expressed lower levels of KDM5C, its loss lead to docetaxel resistance, and could be used as a marker for prostate cancer invasion and metastasis [101]. Analysis of whole exome sequencing data of metastatic prostate tumors showed that KDM5D was deleted in 25% of cases and perturbed nearly three times more frequently than other KDMs and KMTs [101]. An earlier study on prostate tumor samples obtained through radical prostatectomy found that 52% of prostate epithelial cells had a deletion of the genomic region containing KDM5D [102].
NO66 (nucleolar protein 66) is another JmJC-domain containing demethylase of H3K4me1 and H3K4me3. It is an oncogenic driver in prostate cancer that regulates genes associated with survival, invasion and metastasis [103].
Epigenetic Therapies Targeting H3K4 Methylation
Due to the high prevalence of mutations in many cancer types, there have been many efforts to target the MLL complex as a therapeutic target [104]. One group developed MM-401, the first small molecule inhibitor to block the KMT2A/WDR5 interaction, which was shown to ablate KMT2A HMTase activity while not affecting other KMT2 family members [105]. Recent studies have focused on peptidomimetic and small molecules that interrupt KMT2A/WDR5 binding, as well as small molecules that target the KMT2A/menin interaction [106,107,108]. Recently it was discovered that prostate cancer can specifically be targeted by using a small-molecule that inhibits this MLL1/menin interaction [52]. Additionally, a number of compounds have been developed to target the activity of KDM1A/LSD1 (see reference [109] for the complete list). In Table 1 we provide a list of drugs in development and in clinical trials for epigenetic targeting of H3K4 methylation writers and erasers.
H3K9 Methylation
H3K9me1 is enriched at active promoters thus marking active gene transcription, while both H3K9me2, and H3K9me3 are significantly enriched within silent genes thus serving as a mark for heterochromatin and gene silencing [11]. Microarray analysis of PCa tumor samples showed that patients with increased levels of PSA also displayed elevated levels of H3K9me2 and H3K9me3 [3]. Methylated H3K9 preferentially recruits Heterochromatin protein 1 (HP1), which is known to repress transcription of euchromatic genes and promote the formation of heterochromatin [33].
H3K9 Methylation Writers
Methyltransferases known to be involved in the methylation of H3K9 belong to the KMT1 (SUV39H1/KMT1A, SUV39H2/KMT1B, G9a/EHMT2/KMT1C, GLP/EHMT1/KMT1D, SETDB1/KMT1E, and SETDB2/KMT1F), and the KMT8 (PRDM2) family of methyltransferases. Both of these families of methyltransferases contain a SET domain that mediates enzymatic activity. The SET domain in the KMT1/SUV39 (Suppressor of variegation 3–9) subfamily of KMTs are flanked by two cysteine-rich domains called pre-SET and post-SET. These domains bind to three zinc ions, a crucial step required for the enzymatic activity of the SET domain [110].
KMT1A/B (SUV39H1/2) writers : The SUV39 homologs SUV39H1 and SUV39H2 were the first SET domain containing methyltransferases identified. These enzymes primarily function within highly compact and transcriptionally silent chromatin regions called pericentric heterochromatin [111]. They are known to di- or trimethylate H3K9me1 substrates and their activity is critical for the establishment and maintenance of heterochromatin structure and genome stability [112,113,114]. SUV39H1/2 function as tumor suppressors that tightly control the repression of oncogenes within heterochromatin. They are aberrantly expressed in many cancers including prostate, AML, lung, liver and colorectal cancers. SUV39H1/2 both contain a chromatin modifier domain (chromodomain) that recognizes and binds to methylated lysines. SUV39H2 is a known co-activator of AR that is overexpressed in prostate cancer. It colocalizes with the AR coregulator melanoma antigen-11 (MAGE-A11) [99] that enhances AR transcriptional activity.
KMT1C and KMT1D (G9a and GLP) writers : G9a and GLP (G9a-like protein) are also members of the SUV39 KMT subfamily that catalyze mono- and di-methylation of H3K9 in euchromatin and facultative heterochromatin regions. Unlike SUV39H1/2, which are involved in stable repression, G9a and GLP proteins play central roles in dynamic transcriptional repression [110]. They form homo- and hetero-dimers and share 80% sequence identity in their catalytic domains [115]. G9a and GLP methylate both histone and non-histone substrates. For instance, G9a- and GLP-catalyzed methylation of LaminB1 plays an important role in regulating H3K9me2-marked heterochromatin anchorage to the nuclear periphery [116]. In addition to the SUV39 specific SET domain, G9a and GLP also contain seven ankyrin repeat domains that mediate protein–protein interactions [117, 118]. Interestingly, G9a can act both as a coactivator and a corepressor of transcription. For instance, G9a association with mediator complex MED1 leads to transcriptional activation whereas G9a association with the K3K4 demethylase KDM5A leads to transcriptional repression [119]. The activity of G9a is essential for prostate differentiation. Its interactions with the homeobox gene NKX3.1 form a highly regulated transcriptional network comprised of NKX3.1, G9a, and the H3K27me3 demethylase UTY, which regulates normal prostate differentiation. Dysregulation of this network results in predisposition to prostate cancer [120]. Similarly, G9a activity promotes coactivation of Runx2-induced expression of genes involved in processes such as epithelial to mesenchymal transition to induce prostate cancer progression and metastasis [121]. G9a is overexpressed in many types of cancers, including prostate cancer [122], esophageal squamous cell carcinoma, hepatocellular carcinoma, aggressive lung cancer, brain cancer, multiple myeloma, and aggressive ovarian carcinoma. Higher expression levels of G9a correlate with poor prognosis.
H3K9 Methylation Erasers
Enzymes demethylating mono-, di-, and tri-methylation marks on H3K9 belong to the KDM/JHDM2, KDM4/JMJD2, and KDM7 subfamilies of lysine demethylases [34]. Additionally, the H3K4 demethylase, LSD1/KDM1A associates with AR and de-represses AR target genes by demethylating H3K9 without altering the status of the H3K4 methylations [123].
KDM3 methylation erasers : The KDM3 subfamily of demethylases only contain the catalytic JmJC domain and consist of three members, namely KDM3A/JHDM2A, KDM3B/JHDM2B, and KDM3C/JHDM3C. JHDM2A/B demethylate H3K9me1 and H3K9me2 [124]. However, there is no direct evidence of JHDM2C demethylating H3K9. JHDM2A is overexpressed in colorectal cancer and renal cell carcinoma [33]. In the former, higher expression of JHDM2C correlated with poor prognosis [33].
KDM4 methylation erasers : The KDM4 subfamily of KDMs consists of five members, KDM4A–E/JMJD2A–E, all of which contain the N-terminal JmJN and the catalytic JmJC domain. The KDM2A-C proteins also contain two PHD domains and two Tudor domains, that are essential for their function as histone methylation readers. These three proteins share higher than 50% sequence identity. KDM4A-D preferentially demethylate H3K9me2 and H3K9me3 while KDM4E only catalyzes the demethylation of H3K9me3 to H3K9me1 through the removal of two methyl groups [37]. Analysis of expression data for diseased and normal tissues showed that the KDM4 family is expressed in 6–9 diseased tissues, with the highest expression in the testes and spleen, making them highly relevant to prostate cancer [37]. Similarly, analysis of cancer mutation datasets from the CBio portal and TCGA showed that the KDM4 encoding genes, particularly KDM4C, harbor a variety of structural variations in prostate, lung, breast, and esophageal cancers, as well as lymphoma [37]. KDM4A interaction with activating protein transcription factors plays a major role in controlling proliferation, apoptosis and differentiation. KDM4B interacts with nuclear receptors and is particularly important in prostate and breast cancers. It is highly expressed in estrogen-positive breast cancer and regulates ER mediated transcription in an estrogen dependent manner [125]. In prostate cancer, KDM4B enhances AR stability through inhibition of AR ubiquitination [126]. Similarly, KDM4C strongly associates with AR and the H3K4 demethylase KDM1A/LSD1 and promotes H3K9me3 demethylation in an AR-ligand dependent manner, which in turn activates AR transcription [127].
Epigenetic Therapies Targeting H3K9 Methylation
There are currently no ongoing clinical trials targeting H3K9 methylation enzymes. However, several drugs are under development as shown in Table 2. Furthermore, compounds targeting LSD1 (GSK2879552, and INCB059872, see Table 1) are known to affect H3K9 levels in cells.
H3K27 Methylation
H3K27 methylation is a well-studied repressive histone PTM and is a hallmark of facultative chromatin [132]. H3K27 is either di- or tri-methylated by the enzymatic activity of the Polycomb group (PcG) protein complexes. Apart from its role as transcriptional repressor, H3K27 methylation is critical in regulating genes essential for cellular differentiation and proliferation. H3K27 methylation is dysregulated in a large number of cancers, including prostate cancer, particularly due to mutations and aberrations in its reader, writer, and eraser enzymes.
H3K27 Methylation Writers
KTM6A/B (EZH2/1) writers of H3K27: EZH1 (Enhancer of Zeste Homolog 1) and EZH2 (Enhancer of Zeste Homolog 2) are methyltransferases that catalyze the tri-methylation of H3K27. EZH1 or EZH2 constitute the catalytic subunit of the Polycomb repressor complex PRC2, that also contain the Suz12 zinc finger domain, and the EED domain that recognizes tri-methylated peptides [133]. EZH1- and EZH2-containing PRC2 complexes are found in 1:1 stoichiometry, and the latter effectively methylates H3K27. Thus, EZH2 plays a central role in governing H3K27 mediated gene repression. A number of studies have documented up-regulation of EZH2 in many tumor types, including prostate cancer, breast cancer, and lymphomas, where the expression level appears to correlate with disease progression [134, 135]. Overexpression of EZH2 in prostate cancer cell lines increases invasiveness, while EZH2 knockdown decreases proliferation, with the effect being more pronounced in AR-independent prostate cancer lines. Interestingly, EZH2 mRNA and protein levels are low in benign prostate and increase progressively from localized to metastatic tumors, suggesting that EZH2 could be a useful prognostic indicator as well as a potential therapeutic target [136]. EZH2 expression is also regulated by micro-RNA-101, which is encoded by a locus that is commonly deleted in prostate cancer. One or both alleles of the miR-101 locus is deleted in 37.5% of clinically localized prostate cancer and 66.7% of metastatic prostate tumors, suggesting that loss of micro-RNA-101 leads to EZH2 overexpression and cancer progression mediated by deregulated epigenetic mechanisms [137]. Independent of its polycomb repressor function, EZH2 also has non-canonical oncogenic roles. For instance, in LNCaP cell lines, phosphorylation of EZH2 at Ser21 by the activity of AKT kinase lead to a massive increase in AR-regulated gene transcription [138]. In prostate cancer, EZH2 manifests its oncogenic activity primarily through repression of target genes including p16INK4alpha, DAB2IP, ADRB2, WNT pathway antagonists, VASH1 and CDH1, among others; a majority of these being tumor suppressors. EZH2 activity is also regulated by long non-coding RNAs (lncRNAs), which are implicated in PCa development and progression, e.g., Metastasis-associated lung adenocarcinoma transcript-1 (MALAT1). EZH2 binds to MALAT1, and knockdown of MALAT1 impairs EZH2 recruitment to its target loci, thereby upregulating the expression of EZH2 repressed genes [139]. Furthermore, EZH2 activity affects the genome-wide three-dimensional structure of chromatin, thus making its role even more important in cancer development and progression. The gain of function mutation (EZH2 Y646X) in lymphoma completely inactivates selected topologically associated domains (TADs) that encode for tumor suppressor genes [140]. Together, these findings point to an all-encompassing role of EZH2 in cancer thus opening up new therapeutic avenues that target the canonical and non-canonical roles of EZH2 for efficacious therapeutic management and treatment of mCRPC.
H3K27 Methylation Erasers
KDM6 family of H3K27 erasers : The KDM6 family demethylases, KDM6A/UTX, KDM6B/JMJD3, and KDM6C/UTY catalyze demethylation of H3K27e2/3 substrates [31, 34, 141], as well as H3K27me1 substrates [141]. This subfamily contains the catalytic JmJC domain, and in addition KDM6A/C contains an eight TPR (tetratricopeptide) repeat (also see the uniport database) that mediates protein-protein interactions. KDM6A is dysregulated in a number of cancers including urothelial carcinoma, breast cancer and lymphoblastic leukemia [142]. Analysis of paired prostate cancer and high-grade prostate intraepithelial neoplasia found mutations in KDM6A that were specific to primary prostate cancer suggesting a role in early prostate cancer development [143]. Furthermore, KDM6A physically interacts with AR and functions as an oncogene that drives prostate tumor progression [51]. Similar functions have been observed for KDM6C, which is part of the NKX3.1-G9a-KDM6C transcriptional regulatory pathway that governs prostate differentiation [120] (also see section on “H3K9 Methylation Writer KMT1C”).
Epigenetic Therapies Targeting H3K27 Methylation
Upregulation of EZH2 expression in wide variety of cancers has suggested opportunities to develop inhibitors targeting its oncogenic activity. Multiple early phase clinical trials of EZH2 inhibitors are currently ongoing in hematological malignancies as well as solid tumors, particularly in prostate cancer. One particular trial is a Phase 1b/2 study with oral administration of CPI-1205 (ClinicalTrials.gov Identifier: NCT03480646) in combination with either enzalutamide or abiraterone/prednisone in patients with CRPC. This study is designed to determine the maximum tolerated dose (MTD) and recommended Phase II dose (RP2D) based on safety, tolerability, pharmacokinetic, and efficacy profiles of CPI-1205 in combination with either enzalutamide or abiraterone/prednisone. A phase 1 dose escalation and expanded cohort study of PF-06821497 (ClinicalTrials.gov Identifier: NCT03460977) has been implemented for the treatment of adult patients with relapsed/refractory small cell lung cancer and CRPC. Another study testing GSK-J4, which inhibits KDM6A/B activity and thereby inhibits AR deletion mutants that lack LBD (ARΔLBD), significantly reduced proliferation of CRPC cells [144]. A list of pre-clinical and clinical trials targeting H3K27 is presented in Table 3.
H3K36 Methylation
H3K36 mono-, di-, and tri-methylation are well-established histone PTM marks for active transcription and euchromatin formation, and their removal triggers transcriptional repression. H3K36me2 levels are elevated in the promoter regions of actively transcribed genes and are localized to the 5′ regions, while H3K36me3 is predominantly localized to the 3′ regions [148, 149]. H3K36 methylation has been implicated in a variety of nuclear processes including transcriptional regulation, gene dosage compensation, pre-mRNA splicing, DNA replication, recombination, and DNA damage repair [150]. H3K36 methylation levels are highly dysregulated in prostate cancer mainly due to aberrations in the levels of its methylation enzymes and its upstream-binding partners. For example, we previously reported the interactions between EZH2 and H3K36 methyltransferase NSD2/MMSET in regulating levels of H3K36 [151].
H3K36 Methylation Writers
There are at least eight known H3K36 methyltransferases in mammalian cells, of which six belong to the KMT3 subfamily of methyltransferases (NSD1/KMT3B, NSD2/KMT3G, NSD3/KMT3F, SETD2/KMT3A, SETD3/KMT3E, SMYD2/KMT3C), with the other two being SETMAR, and ASH1L/KMT2H [33, 34]. Of these, only NSD1/2 and SETD2 and SETD3 have significant methyltransferase activity and display dysregulation in cancer. NSD2 (nuclear receptor binding SET domain containing protein 2, also known as KDM3G/MMSET/WHSC1), is the best-studied methyltransferase known to mono-, and di-methylate H3K36, and has an established role in prostate cancer progression. NSD2 is overexpressed in mCRPC compared to primary prostate tumors, and its expression is strongly correlated with disease stage and poor prognosis [152]. NSD2 functions in association with a number of other upstream proteins that regulate H3K36 methylation. We showed previously that the oncogenic activity of NSD2 is regulated by EZH2, where overexpression of EZH2 lead to oncogenic phenotypes that were characterized by increased proliferation, self-renewal, and invasion, only in cells expressing NSD2. Similarly, others have found NSD2 to be a strong coactivator of the NF-kB signaling pathway and NSD2 to be a transcriptional target highly enriched in components of the NF-kB network, including IL-6, IL-8, Birc5, and VEGFA. NSD2 has also been implicated in prostate cancer through its involvement in epithelia-mesenchymal-transition, which is known to drive metastasis in different cancer types. While NSD2 knockdown in benign prostate cells did not affect the levels of H3K36me2 and H3K27me3, knockdown of NSD2 in the CRPC cell lines, DU145 and PC3, altered both H3K36me2 and H3K27me3 levels and decreased cell proliferation, soft agar colony formation, migration, and invasion [153]. Further analysis revealed that NSD2 regulates the oncogenic phenotype through its interactions with TWIST1. These results establish NSD2-mediated epigenetic regulation as a major factor in cancer development.
H3K36 Methylation Erasers
H3K36 demethylation is catalyzed by the KDM2 subfamily of demethylases, KDM2A/JHDM1A and KDM2B/JHDM1B. Both KDM2A and its paralog KDM2B contain an N-terminal JmJC domain, followed by a CXXC zinc finger domain that binds to unmethylated CpG dinucleotides, a PHD domain that recognizes methylated lysine residues, and a Fbox domain [124]. Additionally, KDM2A contains six leucine-rich repeats at its C-terminal compared to two in KDM2B. They both catalyze demethylation of mono- and di-methylated H3K36. Interestingly, the KDM2A/B demethylase genes encode various spliced isoforms in addition to the full length proteins, but these isoforms are only expressed in mammals. Though the spliced short length proteins do not have any methyltransferase activity, they play important roles in biological processes [150]. A number of studies point to the role of KDM2B’s oncogenic functions in cancer development and progression. KDM2B dysregulation is commonly seen in a majority of cancers, including, prostate, breast, pancreatic, gastric, lung, and bladder cancers, as well as ALL and AML. Its aberrant expression and activity leads to dysregulation of key cellular function such as apoptosis (inhibition of c-FLIP/c-Fos), proliferation (activation of PI3K/mTOR), inhibition of Wnt signaling (ubiquitination of β-Catenin), autophagy (inhibition of ERK) and cellular senescence (through inhibition of p53). KDM2B also interacts with EZH2 and promotes cell proliferation and metastasis, eventually leading to drug resistance [154]. Furthermore, a recent study on the cell line DU145 identified the role of KD2MB expression in modulating cell motility; KDM2B overexpression suppresses the expression of epithelial markers E-cadherin and the tight junction protein ZO-1 (thereby reducing cell-cell adhesion and increasing cell migration), and positively regulates the expression of the RhoA and RhoB GTPases (which leads to cytoskeletal rearrangement and increased cell motility) [155]. There have been significant challenges in the field to develop an inhibitor that exhibits specificity for NSD2, and several companies are actively pursuing this objective.
H3K79 Methylation
Lysine 79 on Histone H3 is located inside the globular domain, unlike K4, K9, K27, and K36 that are in the N-terminal unstructured tail. It is di- and tri-methylated by the non-SET domain containing methyltransferase KMT4/DOT1L and the H3K79me2 and H3K79me3 marks are associated with active transcription, cell cycle regulation, genome stability, and DNA damage response [156]. DOT1L methylation of H3K79 is an excellent example of crosstalk between histone PTMs. DOT1L interacts with ubiqutinated-H2BK120 and H4 to cooperatively reorient K79 buried in the H3 globular region to an accessible position [157, 158]. There are presently no known demethylases of H3K79.
H3K79 Methylation Writer
DOT1L: Disruptor of telomeric silencing 1 (DOT1) was first identified through a genetic screen for proteins whose over-expression lead to impaired telomeric silencing in yeast [159]. DOT1L transfers methyl groups from S-adenosyl-l-methionine (SAM) to mono-, di-, and tri-methylate H3K79. The structure of DOT1L is very similar to arginine methyltransferases but so far there has been no evidence for its involvement in arginine methylation. The seven-stranded beta sheets in the open α/β structure of DOT1L is characteristic of class I SAM-dependent methyltransferases [156]. DOT1L and H3K79 methylation have been implicated in diverse types of cancers including prostate, breast and lung cancer, as well as leukemia. Its role in the initiation and maintenance of mixed lineage leukemia (MLL)-rearranged leukemia has been widely studied. DOT1L interacts with MLL fusion proteins, leading to enhanced H3K79 methylation, maintenance of open chromatin, overexpression of downstream oncogenes and leukemogenesis. A DOTlL-HES6 gene fusion has been reported to drive AR-negative prostate cancer through overexpression of HES6 [160]. Loss of DOT1L has been shown to inhibit cell proliferation [161]. DOTlL was found to methylate AR and regulate its transcriptional activity through lncRNA-dependent mechanisms [162].
Epigenetic Therapies Targeting DOT1L
Therapeutic targeting of DOT1L holds significant promise due to its status as the sole H3K79 histone methyltransferase, its unique non-SET catalytic domain and its role in MLL leukaemogenesis. The first DOT1L specific small molecular inhibitor EPZ004777 displayed high specificity against DOT1L, and treatment with this inhibitor induced apoptosis in MLL-rearranged leukemia cells in vitro and also blocked leukemia progression in mice by suppressing the expression of HOXA cluster genes and Meis1 [163]. Due to its poor half-life and metabolic instability, modifications to EPZ00477 led to the synthesis of EPZ5676 [164] and SGC0946 [165] which showed improved binding affinity for DOT1L. EPZ5676 also displayed synergistic anticancer effects against MLL-rearrangement leukemia cells when used in combination with cytarabine and daunorubicin [164], and continuous intravenous treatment of rat xenografts with EPZ5676 led to dose-dependent leukemia regression [166]. Another novel DOT1L inhibitor SYC-522, synthesized by adding an additional urea group to the structure of SAH, showed increased specificity for DOT1L as well as increased anti-cancer efficacy. Treatment of MLL-rearranged leukemia cells with SYC-522 induced cell cycle arrest and cell differentiation, and treatment of primary MLL-rearranged AML cells resulted in up to 50% decrease in colony formation and promotion of monocytic differentiation [167]. The role of DOT1L in prostate cancer has not been well characterized, and therapeutic targeting of DOT1L to treat PCa and CRCPC is still a work in progress [141].
Histone Lysine Methylation Readers
Methylation reader proteins display high specificity for methylated histone lysines. These proteins act upstream of the molecular machinery associated with specific methylation marks, and their function is indispensable for epigenetic regulation of cell function. They contain one of the following methyl-lysine recognition motifs/domains, namely ADD (ATRX-DNMT3-DNMT3L), ankyrin, bromo-adjacent homology (BAH), chromo-barrel, chromodomain, double chromodomain (DCD), MBT (malignant brain tumor), PHD, tandem PHD, PWWP (Pro-Trp-Trp-Pro), Tudor domin (TTD), tandem TTD, WD40/β-propeller, zinc finger CW (zf-CW). For specific details on these domains, see references [168, 169].
Histone Acetylation in Prostate Cancer
Histone acetylation is an important epigenetic modification in which an acetyl group from acetyl coenzyme A (acetyl-CoA) is transferred to the ε-amino group of lysine residues in histones and other proteins. Acetylation, in effect, abolishes the interaction of histones with the negatively charged DNA backbone leading to open chromatin structures that facilitate the binding of RNA Pol II. In prostate cancer, acetylation of histones at gene promoter and/or enhancer regions increases AR activity and cell survival. In vitro and in vivo studies of primary and metastatic prostate cancer cells have demonstrated a significant positive correlation between the levels of prostate specific antigen (PSA) mRNA and H3 acetylation at the PSA enhancers and proximal promoter [170]. Besides its role as a mark for active transcription, acetylation has also been found to be important for genome stability, protein stability, regulation of protein-protein interactions, and even chromatin compaction [171]. Tissue microarray analysis of primary prostate cancer patients showed that acetylation of H3K18, H3K9 and H4K12, in combination with demethylation of H3K4 and H4R3 could be used as a prognostic biomarker to estimate risk for prostate tumor recurrence in patients with low-grade tumors [27]. A genome wide study of acetylation patterns at 3286 gene promoter regions in CD4+ T cells detected the presence of 17 distinct acetylation patterns [172].
Histone acetylation is catalyzed by histone acetyltransferases (HATs) which are broadly divided into five subfamilies, namely GCN5/PCAF, MYST, TAFII250, CBP/p300, and SRC. Histone deacetylation is regulated by the activity of Histone deacetylases (HDACs) which are subdivided into four subfamilies, namely class I, II, III, and IV [173]. The histone acetyl marks in turn are recognized by bromodomain-containing proteins that act upstream of the molecular machinery associated with acetylation regulated cellular signaling pathways (see Fig. 3). Dysregulation and mutations in HATs and HDACs have been observed in a number of cancers and play a driver role in cancer initiation and growth [173]. We will next discuss the role of these enzymes in the onset and metastatic progression of prostate cancer.
Acetylation Writers: HATs in Prostate Cancer
CBP/p300: The CBP/p300 family of nuclear phosphoproteins contain the ubiquitously expressed and homologous (~61% homology) CBP (KAT3A) and p300 (KAT3B) proteins that participate in a number of physiological processes such as cell proliferation, differentiation, and apoptosis [174, 175]. Both CBP and p300 contain three CH and two ZZ zinc finger domains, a bromodomain that recognizes acetylated residues, and a catalytic acetyltransferase domain that catalyzes histone acetylation [175]. CBP/p300 is one of the well-studied histone acetyltransferases that acetylates H3 lysines 14 and 18, H4 lysines 5 and 8, H2A lysine 5, and H2B lysine 12 and 15 [21]. CBP/p300 HAT also acetylates other proteins; a recent study estimated that 411 proteins were part of the CBP/p300 interactome, and that 615 genes were part of the p300/CPB cistrome [175]. A majority of these proteins include components of the transcription complex and major signal transduction pathways, thus making CBP/p300 a central component of the cellular machinery that coordinates signal flow to regulate gene transcription [174].
CBP/p300 proteins are broadly linked to cancer as well as several human pathologies. The role of CBP/p300 in cancer as an oncogene or tumor suppressor is debatable and may be context dependent. In prostate cancer, CBP/p300 displays oncogenic properties by acting as a co-activator of the AR, which is the main oncogenic driver in prostate cancer. The formation of this AR–coactivator complex at AR-binding sites promotes chromatin opening and recruitment of the transcriptional machinery to target genes [176]. Expression of histone acetyltransferase p300 and AR also correlates positively in human prostate cancer specimens, especially those marked with PTEN loss. Mechanistically, PTEN loss induces AR phosphorylation at serine 81 (Ser81) to promote p300 binding and acetylation of AR, thereby precluding its polyubiquitination and degradation. Thus, p300 acetyltransferase regulates AR degradation and PTEN-deficient prostate tumorigenesis [177,178,179,180].
Acetylation Erasers: HDACs in Prostate Cancer
Histone Deacetylases (HDACs) belong to a family of enzymes that catalyze the removal of the acetyl group from ε-N-acetyl lysine amino acid. The 18 known HDACs are grouped into four major families: class I (reduced potassium dependency 3 (RPD3)-like proteins HDAC1, HDAC2, HDAC3, and HDAC8), class II (Histone deacetylase 1 (HDA1)-like proteins HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10), class III (silent information regulator 2 (Sir2)-like proteins SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, SIRT7), and class IV (protein HDAC11) [181].
The catalytic activity in class I, II and IV HDACs is driven by zinc (Zn2+)-dependent deacetylation reaction, while that in class III HDACs is driven by a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylation mechanism. HDAC catalyzed deacetylation of histone and non-histone proteins affect key signaling pathways including cell cycle, apoptosis, DNA damage response, metastasis, angiogenesis, and autophagy. Dysregulated HDAC functions affect one or more of the mentioned pathways that could serve as a driver of cancer development. Though genetic alterations in HDACs are rare, most hematological malignancies and solid tumors display aberrant expression of various HDACs (see reference [182] for the full list). In most cases, higher levels of HDAC expression are associated with advanced disease and poor prognosis. Studies on prostate cancer samples, cell lines, and mouse models have shown that HDAC1, 2, and 3 are overexpressed in prostate cancer and that increased HDAC2 expression correlates with shorter PSA relapse after radical prostatectomy [183].
The ubiquitous role of HDACs in cell function makes them attractive targets for therapeutic interventions in prostate cancer patients. So far, many compounds known as HDAC inhibitors (HDACi) have been developed to inhibit the activity of these HDAC complexes. Trichostatin A (TSA) was one of the first natural compounds found to be a potent HDACi. Some of the early studies tested the efficacy of TSA and sodium butyrate as HDAC inhibitors in the prostate cancer line PC3 and found them to induce differentiation and apoptosis [184]. Another compound, suberoylanilide hydroxamic acid (SAHA ; Vorinostat) was the first HDACi approved by the US FDA for the treatment of cancer and was found to inhibit proliferation of the prostate cancer cell lines LNCaP, DU-145 and PC3 as well as prostate tumors in animal models of prostate cancer [185]. More recently, a compound called PAC-320 was shown to induce G2/M arrest and apoptosis in human prostate cancer cells 125178668 in vitro. After the initial success with these agents in other cancer types, there have been a number of Phase I and Phase II clinical trials on HDACi conducted in individuals with advanced prostate cancer where they have been tested as single agent or in combination with other anti-cancer agents [186,187,188]. These clinical trial results have been mixed with only marginal response and slightly improved outcomes. Therefore, further investigation is necessary to clarify the benefits and drawbacks of these medications.
Sirtuins : The Silent Information Regulator 2 (SIR2) proteins, Sirtuins (SIRT1–7), belong to the Class III family of Nicotinamide Adenine dinucleotide (NAD+)-dependent histone deacetylases. Sirtuins deacetylate both histone (SIRT1–3, 6, 7) and nonhistone substrates (SIRT1–3, 5, 7). They differ greatly in their functions and localizations. SIRT1 is the best-characterized member of mammalian sirtuins and is involved in several cellular processes such as metabolism, DNA repair, recombination, aging, apoptosis and cellular senescence. Aberrant expression of sirtuin proteins has been reported in many diseases, including Bowen’s disease, type I diabetic nephropathy, Alzheimer disease and amyotrophic lateral sclerosis, nonalcoholic fatty liver disease and cancer. SIRT1 and SIRT2 levels are upregulated in many cancers including prostate cancer, thus potentially functioning as oncogenes. One study [189] compared the expressions of SIRT1 and SIRT2 in a variety of CRPC cell lines (LNCaP, 22Rv1, PC-3 and DU145), with normal prostate epithelial PrEC cells, and normal prostate stromal PrSC cells. Their data demonstrated that SIRT1 and SIRT2 are significantly upregulated in all CRPC cell lines compared to normal prostate cells. Moreover, immunohistochemical analysis of human tissues showed that SIRT2 was significantly upregulated in prostate cancer compared to normal prostate. They also observed that chemical inhibition and/or genetic knockdown of Sirt1 caused a FoxO1-mediated inhibition in the growth and viability of human PCa cells [189]. Another study showed that SIRT1 levels are significantly elevated in mouse and human prostate cancer [190]. Overexpression of SIRT1 induces epithelial-mesenchymal transition (EMT) by inducing EMT transcription factors like ZEB1 to promote prostate cancer cell migration and metastasis [191]. Furthermore, SIRT1 associates with and deacetylates matrix metalloproteinase-2, and regulates its expression by controlling protein stability through the proteasomal pathway, and enhances tumor cell invasion in prostate cancer cells [192].
One study showed that SIRT1, by physically interacting and cooperating with MPP8, represses E-cadherin expression and promotes EMT in prostate cancer cells [193]. Nicotinamide N-methyltransferase (NNMT) is an important activator and stabilizer of SIRT1, and overexpression of NNMT in PC-3 prostate cancer cells upregulates SIRT1 expression, leading to enhanced cell invasion and migration [194].
Despite mounting evidences on the involvement of sirtuins in cancer development, their role as oncogenes or tumor suppressor genes is not well established. Evidence supporting the latter was provided in a study where mesenchymal stem cells overexpressing SIRT1 significantly suppressed prostate cancer growth by promoting the recruitment of Natural Killer cells and macrophages as anti-tumor effectors [195]. Another study showed that SIRT1 repressed androgen responsive gene expression and induced autophagy in the prostate, and that disruption of this SIRT1-dependent autophagy checkpoint in the prostate resulted in prostatic intraepithelial neoplasia (PIN) lesion formation [196]. Thus, these reports further highlight the role of SIRT1 as a tumor suppressor in prostate cancer, which is contradictory to previous reports suggesting its role in oncogenesis.
Acetylation Readers
BET bromodomain proteins: The bromodomain and extra-terminal (BET) family of proteins are an important class of epigenetic readers of acetylated histones regulating a vast network of protein expression across many different cancers. The BET subfamily is made up of the four members BRD2, BRD3, and BRD4, and BRDT all of which contain two bromodomains (BD1 and BD2) at the N terminal and an extra-terminal domain and a C-terminal domain [171, 197]. The bromodomain motifs are histone acetylation readers due to their ability to recognize and bind to acetylated lysines on histone tails (primarily on H4) and form a scaffold for the assembly of multi-protein complexes. They also recognize acetylated non-histone proteins. Well known examples of BET non-histone activity include its role in regulating the transcriptional activity of NF-kappaB, and of ERG in acute myeloid leukemia. BRD4 also binds to FLI1, MYB, SPI1, CEBPA, and p53 in a bromodomain independent manner [197].
BET proteins are usually part of large nuclear complexes and play decisive roles in cellular processes such as transcription, replication, chromatin remodeling, DNA damage and cell growth. Embryonic lethality upon knockdown of these proteins highlights their indispensable role in normal physiological processes. For example, BRD4 is associated with a coactivator complex of transcription factors [198] and promotes transcriptional elongation by increasing the processivity of RNA polymerase II, leading to expression of growth-promoting genes [199, 200]. The critical requirement of BET proteins in these basic cellular processes explains their dysregulation in terms of overexpression or recurrent translocations in many human cancers such as B-cell lymphoma and NUT midline carcinoma. BET proteins are essential components of the AR transcription machinery that drives AR signaling in both PCa and mCRPC [201] and are significantly over-expressed in mCRPC [202]. Overexpression of BET family proteins increase DNA accessibility, which could be used to identify advanced mCRPC from primary prostate tumors.
Therapeutic targeting of BET proteins with BET inhibitors (BETi) is an attractive area of clinical development [197]. BET inhibitors target bromodomains on BET proteins and abrogate oncogenic signaling commonly mediated by distal regulatory regions such as enhancers and superenhancers. The centrality of BET proteins in signaling mediated by AR, ETS fusions, and MYC, makes it an attractive candidate for BETi therapy. Treatment with the bromodomain inhibitor JQ1 suppressed c-Myc function and suppressed ligand-independent prostate cancer cell survival [203]. Our group recently demonstrated that JQ1 and I-BET762, two selective small-molecule inhibitors that target the dual N-terminal bromodomains of BRD4, exhibit anti-proliferative effects in prostate cancer cells as well as xenograft mouse models [201]. We further showed that BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists such as enzalutamide in the treatment of mCRPC as observed by enhanced prostate tumor growth inhibition when enzalutamide and JQ1 were combined together [204]. The next generation BET inhibitors, such as biBET, MT1, and AZD5153, aim to target both the BET bromodomains in a bivalent mode that could lead to stronger inhibitor activity. These compounds have shown promising result in vitro, and their clinical translation is underway. None-the-less, resistance to BETi therapy eventually develops, and mechanisms of BETi resistance have been documented [197]. For instance, BET resistance in prostate cancer patients carrying SPOP mutations has been shown to be due to increased or decreased degradation of BET proteins. In a recent study, we demonstrated that efficacy of BETi in prostate cancer might be limited due to acquired resistance mechanisms such as reactivation of AR signaling by CDK9 and PRC2 mediated silencing of DDR genes [205].
Targeted induced degradation is another effective approach to inhibit protein activity and function. Degradation of BET proteins using proteolysis targeting chimeras (PROTACs) is emerging as an effective strategy to inhibit their function [64, 206, 207]. The first BET PROTACs, including dBET1, MZ1 and ARV-825 [208,209,210], used JQ1 for the BET inhibitor and Thalidomide, VHL-ligand, and Thalidomide, respectively, as the small molecule targeting the E3-ubiquitin ligase. Of these, ARV-825 showed higher potency causing BET protein degradation in cell line models of Burkitt’s lymphoma and led to MYC suppression and apoptosis induction [210]. Similarly, dBET1 also triggered apoptosis in primary human AML cells and tumor inhibition in xenograft studies [208]. Modifications to ARV-825, wherein the small molecule component was changed to a VHL-ligand, led to the development of ARV-771 with superior PK/PD and efficacy in 22Rv1 and VCaP xenograft models of castration-resistant prostate cancer [211].
Epigenetic Therapies Targeting Histone Acetylation
Histone acetylases and deacteylases have been a major target of epigenetics-based therapies. In Table 4 we provide a list of drugs presently in pre-clinical development or clinical trials for use as therapeutic agents in mCRPC and other cancers. The inherent dependency of AR activity on CBP/p300 coactivators makes them attractive targets for therapeutic interventions to treat prostate cancer. Inhibitors such as MS2126, MS7972, and Ischemin were developed to target the CBP/p300 bromodomain, which in turn restores levels of the tumor suppressor protein p53. Second generation compounds such as SGC-CBP30, PF-CBP1 and I-CBP112 to target CBP/p300 bromodomains and restore p53 activity showed better selectivity at nanomolar concentrations. I-CBP112 confirmed potential involvement of CBP/p300 in self-renewal of leukemia cells, and more recently it has been reported to stimulate the catalytic activity of CBP/p300 proteins with loss of function mutations in tumors with inherently low acetylation levels. A more advanced analogue, GNE-049 which showed low nanomolar potency and over 4000-fold selectivity for CBP/p300, demonstrated improved in vitro and in vivo activity in preclinical models of mCRPC [212]. Similarly, A-485, a potent, selective and drug-like catalytic inhibitor of p300 and CBP, selectively inhibited proliferation in lineage-specific tumor types, including AR-positive prostate cancer and several hematological malignancies. A-485 inhibited the AR transcriptional program in both androgen-sensitive and castration-resistant prostate cancer and inhibited tumor growth in a castration-resistant xenograft model [213]. Taken together, these data strongly support CBP/p300 inhibition as a therapeutic strategy in mCRPC. To validate these findings in a clinical setting, CellCentric has initiated and is currently recruiting for a Phase I/IIa clinical trial (NCT03568656) for their lead CBP/p300 bromodomain inhibitor CCS1477 as a mono or combination therapy with Enzalutamide and/or Abiraterone in metastatic prostate cancer and other solid tumors.
Multiple early phase clinical trials of BET inhibitors are currently ongoing in hematological malignancies as well as solid tumors. These include a study of the novel BET inhibitor FT-1101 (ClinicalTrials.gov Identifier: NCT02543879) in patients with relapsed or refractory hematologic malignancy and a study of RO6870810 (ClinicalTrials.gov Identifier: NCT03068351) as single agent and combination therapy in advanced multiple myeloma. Another Phase I study of CPI0610 in patients with previously treated multiple myeloma and lymphoma, demonstrated changes in the expression of MYC and other genes in malignant tumor cells; changes in cellular proliferation and in the extent of apoptosis (ClinicalTrials.gov Identifier: NCT02157636). Many novel BET inhibitors are also being tested for safety, tolerability and efficacy for metastatic prostate cancer. For instance, a novel and highly potent small molecule BET inhibitor from Zenith Epigenetics called ZEN003694 is currently in Phase 1b/2a alone (ClinicalTrials.gov Identifier: NCT02705469) as well as in combination with enzalutamide in patients with abiraterone refractory but enzalutamide naïve mCRPC (ClinicalTrials.gov Identifier: NCT02711956).
Chromatin Remodeling Complexes
DNA accessibility to transcription factors can be modulated either by the deposition/removal of histone PTMs, as discussed earlier, or by the activity of chromatin remodeling enzymes/remodeler complexes. ATP-dependent chromatin remodeling enzymes include multi-subunit complexes of the Snf2 family, which are evolutionarily conserved from yeast to human. They are highly abundant in the cell with roughly one enzyme per ten nucleosomes [214]. Chromatin remodeling complexes utilize energy from ATP hydrolysis to disrupt DNA-histone contacts and slide, eject, or alter the position of histone octamers that allows for transcription of previously inaccessible DNA regions. They are recruited to specific sites through their interactions with cell specific transcriptional regulators. Remodeler complexes are broadly grouped into four subfamilies, namely: ISWI (imitation switch), CHD (chromodomain, helicase, DNA binding), INO80 (inositol requiring 80) and SWI/SNF (switching defective/sucrose nonfermenting). These proteins contain a conserved ATPase domain that facilitates nucleic acid binding and ATP hydrolysis, but differ in their flanking domains. For example, proteins of the SWI/SNF family contain a bromodomain that recognizes acetylated histone lysines, the CHD family contains two chromodomains that recognize methylated histone lysines, and the ISWI family contains HAND, SANT, and SLIDE domains that all recognizes nucleosomes and internucleosomal DNA. For more details on the mechanism of remodeler complexes, see references [214, 215]. Remodeler complexes have been implicated in a variety of cellular processes such as gene expression, DNA replication, repair, chromosomal recombination and mitosis, and their dysregulation, particularly SWI/SNF, has been observed in ~20% of human cancers. Even though mutations in SWI/SNF genes are not common in prostate cancer, accumulating evidence suggests its influence on AR signaling, ERG mediated transcription, cell cycle and DNA methylation. Understanding the role of these remodeler complexes and their interactions with other epigenetic marks and their enzymes would facilitate our understanding of how chromatin is regulated by non-genetic factors and their role in various pathologies including cancer.
Conclusions and Outlook
In this chapter, we have presented a brief overview of how histone methylation and acetylation based epigenetic modifications govern cell function through their ability to modulate DNA accessibility and recruit other effector proteins to regulate gene transcription. We have discussed the role of the major histone marks and their regulatory enzymes in prostate cancer development and metastasis. It is becoming increasingly clear that the epigenetic code, contained in the reversible, heritable, covalent modifications of DNA and histones, is as significant as the genetic code in diversity, complexity, and functionality. The advent of high throughput sequencing methods, particularly ChIP-seq, has greatly helped us gain a genome-wide view of the mechanisms and dynamics of histone post translational modifications. Interestingly, the role of epigenetic modifications has been reported in nearly all cellular functions including cell proliferation, differentiation, motility, cytoskeletal reorganization, apoptosis, chemoresistance and embryonic development.
Aberrant expression of PTM-regulating enzymes, specifically HMTs, HATs, HDACs and demethylases, has been reported in many types of blood and solid tumors. It is believed that the initial stages of tumor development are entirely regulated by epigenetic mechanisms until a stabilizing genomic mutation or structural variation appears and defines a tumor phenotype. Most histone PTMs play dual roles, both as tumor suppressors and oncogenes, and their interaction with nuclear receptors drive the progression of hormone-dependent cancers. In prostate cancer, AR interactions with acetylation and methylation enzymes regulate ligand-dependent and independent transcription of AR target genes. Epigenetic mechanisms play a central role in rewiring of the AR transcriptional network that drives a primary prostate tumor to mCRPC and also endows them with resistance to androgen deprivation therapies [216]. Given their importance in cancer, epigenetic regulatory enzymes have become major targets for therapeutic intervention. There is a heavy focus on the development of epigenetic drugs, and their number in clinical trials is steadily increasing. Presently there are nearly 250 monotherapy or combination therapies in phase I/II trials with epigenetic drugs (see ClinicalTrials.gov).
References
S.A. Grigoryev, C.L. Woodcock, Chromatin organization - the 30 nm fiber. Exp. Cell Res. 318(12), 1448–1455 (2012)
D.V. Fyodorov et al., Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19(3), 192–206 (2018)
J. Ellinger et al., Global levels of histone modifications predict prostate cancer recurrence. Prostate 70(1), 61–69 (2010)
S. Venkatesh, J.L. Workman, Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16(3), 178–189 (2015)
J.M. Belton et al., Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58(3), 268–276 (2012)
J. Dekker et al., The 4D nucleome project. Nature 549(7671), 219–226 (2017)
G. Andrey, S. Mundlos, The three-dimensional genome: regulating gene expression during pluripotency and development. Development 144(20), 3646–3658 (2017)
F. Spitz, E.E. Furlong, Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13(9), 613–626 (2012)
J.O. Carlsten, X. Zhu, C.M. Gustafsson, The multitalented Mediator complex. Trends Biochem. Sci. 38(11), 531–537 (2013)
B. Bonev, G. Cavalli, Organization and function of the 3D genome. Nat. Rev. Genet. 17(11), 661–678 (2016)
A. Barski et al., High-resolution profiling of histone methylations in the human genome. Cell 129(4), 823–837 (2007)
S.-H. Song, T.-Y. Kim, CTCF, cohesin, and chromatin in human cancer. Genomics Inform. 15(4), 114–122 (2017)
M. Nowacka-Zawisza, E. Wiśnik, DNA methylation and histone modifications as epigenetic regulation in prostate cancer (Review). Oncol. Rep. 38(5), 2587–2596 (2017)
C.E. Massie, I.G. Mills, A.G. Lynch, The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol. 166, 1–15 (2017)
Y. Wu, M. Sarkissyan, J.V. Vadgama, Epigenetics in breast and prostate cancer. Methods Mol. Biol. 1238, 425–466 (2015)
R. Zelic et al., Global DNA hypomethylation in prostate cancer development and progression: a systematic review. Prostate Cancer Prostatic Dis. 18(1), 1–12 (2015)
M. Ngollo et al., Epigenetic modifications in prostate cancer. Epigenomics 6(4), 415–426 (2014)
S.B. Baylin, P.A. Jones, A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 11(10), 726–734 (2011)
X. Wu, Y. Zhang, TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18(9), 517–534 (2017)
K. Ruggero et al., Epigenetic regulation in prostate cancer progression. Curr. Mol. Biol. Rep. 4(2), 101–115 (2018)
T. Kouzarides, Chromatin modifications and their function. Cell 128(4), 693–705 (2007)
P. Chi, C.D. Allis, G.G. Wang, Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10(7), 457–469 (2010)
S.R. Bhaumik, E. Smith, A. Shilatifard, Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14(11), 1008–1016 (2007)
H. Huang et al., SnapShot: histone modifications. Cell 159(2), 458–458.e1 (2014)
B.D. Strahl, C.D. Allis, The language of covalent histone modifications. Nature 403(6765), 41–45 (2000)
B.A. Benayoun et al., H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158(3), 673–688 (2014)
D.B. Seligson et al., Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435(7046), 1262–1266 (2005)
F. Valdés-Mora et al., Acetylated histone variant H2A.Z is involved in the activation of neo-enhancers in prostate cancer. Nat. Commun. 8(1), 1346 (2017)
E.L. Greer, Y. Shi, Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13(5), 343–357 (2012)
R.F. Luco et al., Regulation of alternative splicing by histone modifications. Science 327(5968), 996–1000 (2010)
K. Hyun et al., Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49(4), e324 (2017)
V.K. Rao, A. Pal, R. Taneja, A drive in SUVs: from development to disease. Epigenetics 12(3), 177–186 (2017)
I. Hoffmann et al., The role of histone demethylases in cancer therapy. Mol. Oncol. 6(6), 683–703 (2012)
X. Zhang, H. Wen, X. Shi, Lysine methylation: beyond histones. Acta Biochim. Biophys. Sin. Shanghai 44(1), 14–27 (2012)
A. D’Oto et al., Histone demethylases and their roles in cancer epigenetics. J. Med. Oncol. Therap. 1(2), 34–40 (2016)
A. Janardhan et al., Prominent role of histone lysine demethylases in cancer epigenetics and therapy. Oncotarget 9(76), 34429–34448 (2018)
R.M. Labbé, A. Holowatyj, Z.-Q. Yang, Histone lysine demethylase (kdm) subfamily 4: structures, functions and therapeutic potential. Am. J. Transl. Res. 6(1), 1–15 (2014)
N.D. Heintzman et al., Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39(3), 311–318 (2007)
N.D. Heintzman et al., Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459(7243), 108–112 (2009)
L.M. Soares, Determinants of histone H3K4 methylation patterns Rpb4-Set1 fusion. Mol. Cell 68, 773–785 (2017)
G. Liang et al., Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. U. S. A. 101(19), 7357–7362 (2004)
B. Jin, Y. Li, K.D. Robertson, DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2(6), 607–617 (2011)
D. Faucher, R.J. Wellinger, Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 6(8), e1001082 (2010)
D.B. Seligson et al., Global levels of histone modifications predict prognosis in different cancers. Am. J. Pathol. 174(5), 1619–1628 (2009)
T. Bianco-Miotto et al., Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol. Biomark. Prev. 19(10), 2611–2622 (2010)
Q. Wang et al., Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138(2), 245–256 (2009)
X.S. Ke et al., Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One 4(3), e4687 (2009)
H.H. He et al., Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 42(4), 343–347 (2010)
M.S. Geybels et al., PTEN loss is associated with prostate cancer recurrence and alterations in tumor DNA methylation profiles. Oncotarget 8(48), 84338–84348 (2017)
J.M. Spangle et al., PI3K/AKT signaling regulates H3K4 methylation in breast cancer. Cell Rep. 15(12), 2692–2704 (2016)
C.S. Grasso et al., The mutational landscape of lethal castration-resistant prostate cancer. Nature 487(7406), 239–243 (2012)
R. Malik et al., Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 21(4), 344–352 (2015)
T. Kouzarides, SnapShot: histone-modifying enzymes. Cell 131(4), 822 (2007)
A. Shilatifard, The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81(1), 65–95 (2012)
T. Miller et al., COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 98(23), 12902–12907 (2001)
J.J. Meeks, S. Ali, Multiple roles for the MLL/COMPASS family in the epigenetic regulation of gene expression and in cancer. Annu. Rev. Cancer Biol. 1(1), 425–446 (2017)
C. Kandoth et al., Mutational landscape and significance across 12 major cancer types. Nature 502(7471), 333–339 (2013)
R.C. Rao, Y. Dou, Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15(6), 334–346 (2015)
M.G. Guenther et al., Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 102(24), 8603–8608 (2005)
M. Wu et al., Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28(24), 7337–7344 (2008)
L. Wu et al., ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol. Cell 49(6), 1108–1120 (2013)
S.S. Dhar et al., Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 26(24), 2749–2762 (2012)
R. Mo, S.M. Rao, Y.-J. Zhu, Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J. Biol. Chem. 281(23), 15714–15720 (2006)
S. Lv et al., Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene 37(10), 1354–1368 (2018)
C. Deng et al., USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet. 9(6), e1003524 (2013)
R.A. Varier, H.T. Timmers, Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta 1815(1), 75–89 (2011)
P.A. Boriack-Sjodin, K.K. Swinger, Protein methyltransferases: a distinct, diverse, and dynamic family of enzymes. Biochemistry 55(11), 1557–1569 (2016)
K. Leinhart, M. Brown, SET/MYND lysine methyltransferases regulate gene transcription and protein activity. Genes (Basel) 2(1), 210–218 (2011)
G.S. Van Aller et al., Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics 7(4), 340–343 (2012)
F.Q. Vieira et al., SMYD3 contributes to a more aggressive phenotype of prostate cancer and targets Cyclin D2 through H4K20me3. Oncotarget 6(15), 13644–13657 (2015)
S.J. Du, X. Tan, J. Zhang, SMYD proteins: key regulators in skeletal and cardiac muscle development and function. Anat. Rec. (Hoboken) 297(9), 1650–1662 (2014)
R. Hamamoto et al., SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6(8), 731–740 (2004)
C. Liu et al., SMYD3 as an oncogenic driver in prostate cancer by stimulation of androgen receptor transcription. J. Natl. Cancer Inst. 105(22), 1719–1728 (2013)
A.M. Cock-Rada et al., SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 72(3), 810–820 (2012)
T. Hara et al., Androgen receptor and invasion in prostate cancer. Cancer Res. 68(4), 1128–1135 (2008)
G. Rajajeyabalachandran et al., Therapeutical potential of deregulated lysine methyltransferase SMYD3 as a safe target for novel anticancer agents. Expert Opin. Ther. Targets 21(2), 145–157 (2017)
A. Peserico et al., A SMYD3 small-molecule inhibitor impairing cancer cell growth. J. Cell. Physiol. 230(10), 2447–2460 (2015)
L.H. Mitchell et al., Novel oxindole sulfonamides and sulfamides: EPZ031686, the first orally bioavailable small molecule SMYD3 inhibitor. ACS Med. Chem. Lett. 7(2), 134–138 (2016)
G.S. Van Aller et al., Structure-based design of a novel SMYD3 inhibitor that bridges the SAM-and MEKK2-binding pockets. Structure 24(5), 774–781 (2016)
J. McGrath, P. Trojer, Targeting histone lysine methylation in cancer. Pharmacol. Ther. 150, 1–22 (2015)
S. Hino, K. Kohrogi, M. Nakao, Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci. 107(9), 1187–1192 (2016)
L. Ellis, M. Loda, LSD1: a single target to combat lineage plasticity in lethal prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 115(18), 4530–4531 (2018)
A. Sehrawat et al., LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc. Natl. Acad. Sci. U. S. A. 115(18), E4179–E4188 (2018)
S. Regufe da Mota et al., LSD1 inhibition attenuates androgen receptor V7 splice variant activation in castration resistant prostate cancer models. Cancer Cell Int. 18, 71 (2018)
A. Ketscher et al., LSD1 controls metastasis of androgen-independent prostate cancer cells through PXN and LPAR6. Oncogene 3, e120 (2014)
P.A. Cloos et al., Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22(9), 1115–1140 (2008)
J. Plch, J. Hrabeta, T. Eckschlager, KDM5 demethylases and their role in cancer cell chemoresistance. Int. J. Cancer 144(2), 221–231 (2019)
F. Crea et al., The emerging role of histone lysine demethylases in prostate cancer. Mol. Cancer 11, 52 (2012)
Y. Xiang et al., JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 104(49), 19226–19231 (2007)
J.R. Horton et al., Characterization of a linked Jumonji domain of the KDM5/JARID1 family of histone H3 lysine 4 demethylases. J. Biol. Chem. 291(6), 2631–2646 (2016)
R. Liefke et al., Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 24(6), 590–601 (2010)
F.L. Carvalho et al., Notch signaling in prostate cancer: a moving target. Prostate 74(9), 933–945 (2014)
Q. Su, L. Xin, Notch signaling in prostate cancer: refining a therapeutic opportunity. Histol. Histopathol. 31(2), 149–157 (2016)
L. Marignol et al., Hypoxia, notch signalling, and prostate cancer. Nat. Rev. Urol. 10(7), 405–413 (2013)
K.T. Kuo et al., Histone demethylase JARID1B/KDM5B promotes aggressiveness of non-small cell lung cancer and serves as a good prognostic predictor. Clin. Epigenetics 10(1), 107 (2018)
M.I. Khan et al., AKT inhibition modulates H3K4 demethylase levels in PTEN-null prostate cancer. Mol. Cancer Ther. 18(2), 356–363 (2019)
J. Taylor-Papadimitriou, J. Burchell, JARID1/KDM5 demethylases as cancer targets? Expert Opin. Ther. Targets 21(1), 5–7 (2017)
J. Stein et al., KDM5C is overexpressed in prostate cancer and is a prognostic marker for prostate-specific antigen-relapse following radical prostatectomy. Am. J. Pathol. 184(9), 2430–2437 (2014)
Z. Hong et al., KDM5C is transcriptionally regulated by BRD4 and promotes castration-resistance prostate cancer cell proliferation by repressing PTEN. Biomed. Pharmacother. 114, 108793 (2019)
B. Rondinelli et al., Histone demethylase JARID1C inactivation triggers genomic instability in sporadic renal cancer. J. Clin. Invest. 125(12), 4625–4637 (2015)
N. Li et al., JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis. Cancer Res. 76(4), 831–843 (2016)
G. Perinchery et al., Deletion of Y-chromosome specific genes in human prostate cancer. J. Urol. 163(4), 1339–1342 (2000)
K.M. Sinha et al., Oncogenic and osteolytic functions of histone demethylase NO66 in castration-resistant prostate cancer. Oncogene 38(25), 5038–5049 (2019)
M. Vedadi et al., Targeting human SET1/MLL family of proteins. Protein Sci. 26(4), 662–676 (2017)
F. Cao et al., Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 53(2), 247–261 (2014)
H. Karatas et al., High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein–protein interaction. J. Am. Chem. Soc. 135(2), 669–682 (2013)
G. Senisterra et al., Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 449(1), 151–159 (2013)
J. Grembecka et al., Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 8(3), 277–284 (2012)
A. Jambhekar, J.N. Anastas, Y. Shi, Histone lysine demethylase inhibitors. Cold Spring Harb. Perspect. Med. 7(1), a026484 (2017)
C. Mozzetta et al., Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 16(8), 499–513 (2015)
H. Wu et al., Structural biology of human H3K9 methyltransferases. PLoS One 5(1), e8570 (2010)
D. Wang et al., Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc. Natl. Acad. Sci. U. S. A. 110(14), 5516–5521 (2013)
J.C. Rice et al., Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12(6), 1591–1598 (2003)
A.H. Peters et al., Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12(6), 1577–1589 (2003)
M. Tachibana et al., Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19(7), 815–826 (2005)
R.A. Rao et al., KMT1 family methyltransferases regulate heterochromatin-nuclear periphery tethering via histone and non-histone protein methylation. EMBO Rep. 20(5), e43260 (2019)
S. Nakanishi et al., A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15(8), 881–888 (2008)
C.M. Milner, R.D. Campbell, The G9a gene in the human major histocompatibility complex encodes a novel protein containing ankyrin-like repeats. Biochem. J. 290(Pt 3), 811–818 (1993)
C. Chaturvedi et al., Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc. Natl. Acad. Sci. U. S. A. 109(46), 18845–18850 (2012)
A. Dutta, Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science 352(6293), 1576–1580 (2016)
D.J. Purcell et al., Recruitment of coregulator G9a by Runx2 for selective enhancement or suppression of transcription. J. Cell. Biochem. 113(7), 2406–2414 (2012)
Y. Kondo et al., Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One 3(4), e2037 (2008)
E. Metzger et al., LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437(7057), 436–439 (2005)
N.R. Rose et al., Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases. J. Med. Chem. 55(14), 6639–6643 (2012)
J. Yang et al., The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 70(16), 6456–6466 (2010)
K. Coffey et al., The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 41(8), 4433–4446 (2013)
M. Wissmann et al., Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9(3), 347–353 (2007)
T. Chiba et al., Histone lysine methyltransferase SUV39H1 is a potent target for epigenetic therapy of hepatocellular carcinoma. Int. J. Cancer 136(2), 289–298 (2015)
Z. Lu et al., Histone-lysine methyltransferase EHMT2 is involved in proliferation, apoptosis, cell invasion, and DNA methylation of human neuroblastoma cells. Anti-Cancer Drugs 24(5), 484–493 (2013)
M. Vedadi et al., A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 7(8), 566–574 (2011)
Y. Yuan et al., A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem. Biol. 7(7), 1152–1157 (2012)
E.T. Wiles, E.U. Selker, H3K27 methylation: a promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43, 31–37 (2017)
S. Aranda, G. Mas, L. Di Croce, Regulation of gene transcription by Polycomb proteins. Sci. Adv. 1(11), e1500737 (2015)
E. Conway, E. Healy, A.P. Bracken, PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 37, 42–48 (2015)
R. Margueron, D. Reinberg, The Polycomb complex PRC2 and its mark in life. Nature 469(7330), 343–349 (2011)
S. Varambally et al., The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419(6907), 624–629 (2002)
S. Varambally et al., Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322(5908), 1695–1699 (2008)
K. Xu et al., EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science (New York, N.Y.) 338(6113), 1465–1469 (2012)
D. Wang et al., LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 6(38), 41045–41055 (2015)
M.C. Donaldson-Collier et al., EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat. Genet. 51(3), 517–528 (2019)
H. Kaniskan, M.L. Martini, J. Jin, Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 118(3), 989–1068 (2018)
W.A. Schulz et al., The histone demethylase UTX/KDM6A in cancer: progress and puzzles. Int. J. Cancer 145(3), 614–620 (2019)
S.H. Jung et al., Genetic progression of high grade prostatic intraepithelial neoplasia to prostate cancer. Eur. Urol. 69(5), 823–830 (2016)
V.M. Morozov et al., Inhibitor of H3K27 demethylase JMJD3/UTX GSK-J4 is a potential therapeutic option for castration resistant prostate cancer. Oncotarget 8(37), 62131–62142 (2017)
W. Yu et al., Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 3, 1288 (2012)
J. Tan et al., Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 21(9), 1050–1063 (2007)
M.T. McCabe et al., EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492(7427), 108–112 (2012)
S. Schmähling et al., Regulation and function of H3K36 di-methylation by the trithorax-group protein complex AMC. Development 145(7), dev163808 (2018)
D.K. Pokholok et al., Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122(4), 517–527 (2005)
T. Vacík, D. Lađinović, I. Raška, KDM2A/B lysine demethylases and their alternative isoforms in development and disease. Nucleus 9(1), 431–441 (2018)
I.A. Asangani et al., Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol. Cell 49(1), 80–93 (2013)
N. Li et al., AKT-mediated stabilization of histone methyltransferase WHSC1 promotes prostate cancer metastasis. J. Clin. Invest. 127(4), 1284–1302 (2017)
T. Ezponda et al., The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial–mesenchymal transition and invasive properties of prostate cancer. Oncogene 32(23), 2882–2890 (2013)
M. Yan et al., The critical role of histone lysine demethylase KDM2B in cancer. Am. J. Transl. Res. 10(8), 2222–2233 (2018)
N. Zacharopoulou et al., The epigenetic factor KDM2B regulates cell adhesion, small rho GTPases, actin cytoskeleton and migration in prostate cancer cells. Biochim. Biophys. Acta, Mol. Cell Res. 1865(4), 587–597 (2018)
A.T. Nguyen, Y. Zhang, The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25(13), 1345–1358 (2011)
M.I. Valencia-Sanchez et al., Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. Mol. Cell 74(5), 1010–1019.e6 (2019)
E.J. Worden et al., Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176(6), 1490–1501.e12 (2019)
M.S. Singer et al., Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150(2), 613–632 (1998)
M. Annala et al., DOT1L-HES6 fusion drives androgen independent growth in prostate cancer. EMBO Mol. Med. 6(9), 1121–1123 (2014)
W. Kim et al., Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J. Biol. Chem. 287(8), 5588–5599 (2012)
L. Yang et al., LncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500(7464), 598–602 (2013)
S.R. Daigle et al., Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20(1), 53–65 (2011)
C.R. Klaus et al., DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. J. Pharmacol. Exp. Ther. 350(3), 646–656 (2014)
Y. Zhao et al., Prodrug strategy for PSMA-targeted delivery of TGX-221 to prostate cancer cells. Mol. Pharm. 9(6), 1705–1716 (2012)
S.R. Daigle et al., Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122(6), 1017–1025 (2013)
J.L. Anglin et al., Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J. Med. Chem. 55(18), 8066–8074 (2012)
C.A. Musselman et al., Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19(12), 1218–1227 (2012)
T.G. Kutateladze, SnapShot: histone readers. Cell 146(5), 842–842.e1 (2011)
Z. Chen et al., Histone modifications and chromatin organization in prostate cancer. Epigenomics 2(4), 551–560 (2010)
M. Pérez-Salvia, M. Esteller, Bromodomain inhibitors and cancer therapy: from structures to applications. Epigenetics 12(5), 323–339 (2017)
Z. Wang et al., Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40(7), 897–903 (2008)
M. Han et al., Epigenetic enzyme mutations: role in tumorigenesis and molecular inhibitors. Front. Oncol. 9, 194 (2019)
H.M. Chan, N.B. La Thangue, p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114(Pt 13), 2363–2373 (2001)
B.M. Dancy, P.A. Cole, Protein lysine acetylation by p300/CBP. Chem. Rev. 115(6), 2419–2452 (2015)
M. Fu et al., Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol. Cell. Biol. 23(23), 8563–8575 (2003)
M. Fu et al., p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 275(27), 20853–20860 (2000)
R.M. Attar, C.H. Takimoto, M.M. Gottardis, Castration-resistant prostate cancer: locking up the molecular escape routes. Clin. Cancer Res. 15(10), 3251–3255 (2009)
B. Comuzzi et al., The androgen receptor co-activator CBP is up-regulated following androgen withdrawal and is highly expressed in advanced prostate cancer. J. Pathol. 204(2), 159–166 (2004)
J. Zhong et al., P300 acetyltransferase regulates androgen receptor degradation and pten-deficient prostate tumorigenesis. Cancer Res. 74(6), 1870–1880 (2014)
E. Seto, M. Yoshida, Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6(4), a018713 (2014)
Y. Li, E. Seto, HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 6(10), a026831 (2016)
W. Weichert et al., Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br. J. Cancer 98(3), 604–610 (2008)
H. Huang et al., Carboxypeptidase A3 (CPA3): a novel gene highly induced by histone deacetylase inhibitors during differentiation of prostate epithelial cancer cells. Cancer Res. 59(12), 2981–2988 (1999)
L.K. Gediya et al., Improved synthesis of histone deacetylase inhibitors (HDIs) (MS-275 and CI-994) and inhibitory effects of HDIs alone or in combination with RAMBAs or retinoids on growth of human LNCaP prostate cancer cells and tumor xenografts. Bioorg. Med. Chem. 16(6), 3352–3360 (2008)
P.N. Munster et al., Phase I trial of vorinostat and doxorubicin in solid tumours: histone deacetylase 2 expression as a predictive marker. Br. J. Cancer 101(7), 1044–1050 (2009)
D. Rathkopf et al., A phase I study of oral panobinostat alone and in combination with docetaxel in patients with castration-resistant prostate cancer. Cancer Chemother. Pharmacol. 66(1), 181–189 (2010)
L.R. Molife et al., Phase II, two-stage, single-arm trial of the histone deacetylase inhibitor (HDACi) romidepsin in metastatic castration-resistant prostate cancer (CRPC). Ann. Oncol. 21(1), 109–113 (2009)
B. Jung-Hynes, N. Ahmad, Role of p53 in the anti-proliferative effects of Sirt1 inhibition in prostate cancer cells. Cell Cycle 8(10), 1478–1483 (2009)
D.M. Huffman et al., SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 67(14), 6612–6618 (2007)
V. Byles et al., SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene 31(43), 4619–4629 (2012)
J.D. Lovaas et al., SIRT1 enhances matrix metalloproteinase-2 expression and tumor cell invasion in prostate cancer cells. Prostate 73(5), 522–530 (2013)
L. Sun et al., MPP8 and SIRT1 crosstalk in E-cadherin gene silencing and epithelial-mesenchymal transition. EMBO Rep. 16(6), 689–699 (2015)
Z. You, Y. Liu, X. Liu, Nicotinamide N-methyltransferase enhances the progression of prostate cancer by stabilizing sirtuin 1. Oncol. Lett. 15(6), 9195–9201 (2018)
Y. Yu et al., Mesenchymal stem cells overexpressing Sirt1 inhibit prostate cancer growth by recruiting natural killer cells and macrophages. Oncotarget 7(44), 71112–71122 (2016)
M.J. Powell et al., Disruption of a Sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer Res. 71(3), 964–975 (2011)
A. Stathis, F. Bertoni, BET proteins as targets for anticancer treatment. Cancer Discov. 8(1), 24–36 (2018)
S. Malik, R.G. Roeder, The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11(11), 761–772 (2010)
K.J. Moon et al., The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19(4), 523–534 (2005)
Z. Yang et al., Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19(4), 535–545 (2005)
I.A. Asangani et al., Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510(7504), 278–282 (2014)
A. Urbanucci et al., Androgen receptor deregulation drives bromodomain-mediated chromatin alterations in prostate cancer. Cell Rep. 19(10), 2045–2059 (2017)
L. Gao et al., Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS One 8(5), e63563 (2013)
I.A. Asangani et al., BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol. Cancer Res. 14(4), 324–331 (2016)
A. Pawar et al., Resistance to BET inhibitor leads to alternative therapeutic vulnerabilities in castration-resistant prostate cancer. Cell Rep. 22(9), 2236–2245 (2018)
D.P. Bondeson, C.M. Crews, Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 (2016)
P.M. Cromm, C.M. Crews, Targeted protein degradation: from chemical biology to drug discovery. Cell Chem. Biol. 24(9), 1181–1190 (2017)
G.E. Winter et al., Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348(6241), 1376–1381 (2015)
M. Zengerle, K.H. Chan, A. Ciulli, Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10(8), 1770–1777 (2015)
J. Lu et al., Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22(6), 755–763 (2015)
K. Raina et al., PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 113(26), 7124–7129 (2016)
L. Jin, J. Garcia, E. Chan, Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration-resistant prostate cancer. Cancer Res. 77(20), 5564–5575 (2017)
L.M. Lasko et al., Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550(7674), 128–132 (2017)
G. Längst, L. Manelyte, Chromatin remodelers: from function to dysfunction. Genes (Basel) 6(2), 299–324 (2015)
S.V. Saladi, I.L. de la Serna, ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 6(1), 62–73 (2010)
D.P. Labbe, M. Brown, Transcriptional regulation in prostate cancer. Cold Spring Harb. Perspect. Med. 8(11), a030437 (2018)
Acknowledgements
We thank Reyaz Ur Rasool and Qu Deng for stimulating discussions. Research in the Asangani Laboratory is supported by the Department of Defense Idea Development Award (W81XWH17-1-0404 to I.A.A).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Natesan, R., Aras, S., Effron, S.S., Asangani, I.A. (2019). Epigenetic Regulation of Chromatin in Prostate Cancer. In: Dehm, S., Tindall, D. (eds) Prostate Cancer. Advances in Experimental Medicine and Biology, vol 1210. Springer, Cham. https://doi.org/10.1007/978-3-030-32656-2_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-32656-2_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32655-5
Online ISBN: 978-3-030-32656-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)