Abstract
This paper presents a novel method for detection of irregular tissues in mammography images. Previous works solved such irregularity detection tasks with binary classifiers. Here, we propose to detect irregularities by only observing the healthy samples and describe anything largely different from them as irregularity (i.e., unhealthy or cancerous tissues in terms of demographic breast images). This is particularly of great interest as it is very complicated to acquire datasets with all types of cancer cell shapes and tissues for building binary classifiers. Our modeling allows for learning an irregularity detector without any supervising signal from the irregular class. To this end, we propose an architecture with two deep convolutional networks (\(\mathbf {R}\) and \(\mathbf {M}\)) that are trained adversarially. \(\mathbf {R}\) learns to Reconstruct regular mammography images by only observing healthy tissues and \(\mathbf {M}\) (a Matching network) to detect if its input is healthy or not. The experimental results confirm the reliability and superior performance of our methods for detecting cancer tissues in mammography images in comparison with state-of-the-art irregularity detection methods. The code is available at https://github.com/milad-ahmadi/GAID.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Invasive ductal carcinoma (also known as breast cancer) is the most common type of cancer threatening the lives of women worldwide [20]. Its early detection is very critical and one of the most important aspects of its treatment planning [8]. Mammography is the most common method for screening and diagnosing breast cancer before biopsy or utilizing any other types of imaging [25]. In standard breast examination procedures, X-Ray images form two different views are taken and one or two expert radiologists investigate these data. Exploring mammography images by experts is an expensive, challenging, and time-consuming task. Furthermore, due to the low contrast of such images, the interpretations may be troublesome from time to time.
Previous work for detecting cancer in mammography images modeled the problem in a binary classification setting. Earlier methods developed approaches based on traditional machine learning techniques [6, 14] and more recently deep learning methods have led to great improvements for detecting irregularities and delineating cancerous regions in mammography images [2, 9, 10, 22]. However, cancer regions in mammography images are by nature irregularities materialized in between normal and healthy tissues. Modeling this as a two-class classification problem is ill-posed. Specifically for this problem, cancer regions may appear with different shapes or tissues and may have widely different levels of progression. Modeling this as a binary classification problem may lead to good results when used on datasets that contain only specific views and types of images with similar irregularity characteristics. We argue that the appropriate way is to learn what the healthy and normal tissues should look like and single out the abnormalities. On the other hand, manually labeling and annotating medical images is an expensive and time-consuming task. In this paper, we propose a method based on Generative Adversarial Networks (GAN) [5] with an end-to-end trainable architecture to identify irregularities in mammography images under unsupervised settings. Our proposed method is comprised of two networks, \(\mathbf {R}\) and \(\mathbf {M}\), which are trained adversarially against each other. \(\mathbf {R}\) is trained to reconstruct images while it fools \(\mathbf {M}\) that its output does not contain any irregularity. In the meantime, \(\mathbf {M}\) is trained to identify if its input is normal (healthy) tissue or is irregularity (i.e., cancer). After these two networks are trained, we define a scoring function, \(\mathcal {S}(\cdot )\), based on the outputs of both to quantify the likeliness of being irregularity for each input image. With this definition, our method is related to the recent advance in anomaly detection in computer vision applications, such as Adversarial Visual Irregularity Detection (AVID) [18], GANomaly [1], AnoGAN [19], and Adversarial Learned One-Class Classifier (ALOCC) [17]. Although these methods are designed for anomaly detection, they needed major modifications to accommodate the needs of our application. Specifically, ALOCC and GANomaly methods assumed that there exists a major difference between the concept of regular samples and irregular ones. This assumption is often reasonable for outlier detection and anomaly detection in RGB images. Medical images, however, are different and the difference between regular and irregular concepts is very minor. For instance, the noise element used in the previous works, such as ALOCC disrupts medical images. Also, as later shown by our experiments, GANomaly cannot properly reconstruct mammographic images and AnoGAN is too slow and has a hard time operating on limited training samples (since it exploits a residual loss function). In summary, the main contributions of this work are: (1) We propose an end-to-end trainable deep network able to detect irregularities (unhealthy cancerous tissues) in mammography images. To the best of our knowledge, this is the first approach for unsupervised irregularity detection in mammography images, i.e., the method detects irregular tissues in images without any supervising signal from the irregularity class. (2) Our method is tailored for detecting and localizing cancerous regions in mammography images. It obtains considerable performance which are comparable to fully-supervised methods and better than state-of-the-art irregularity detection methods, i.e., GANomaly [1], AnoGAN [19], and ALOCC [17].
2 Proposed Method
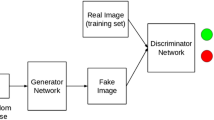
Detecting irregular tissues in mammography images can be defined as discovering regions that do not comply with normal (healthy) tissues present in the training set. To this end, we propose a method based on adversarial training composed of two important modules. The first module, denoted by \(\mathbf {R}\), discovers the distribution of the healthy tissues by learning to reconstruct them. The second module, \(\mathbf {M}\), learns to detect if its input is healthy or irregular (real or fake). In a nutshell, given an input image, \(\mathbf {R}\) learns how to reconstruct it in a way that \(\mathbf {M}\) does not identify it as irregular. Training \(\mathbf {R}\)+\(\mathbf {M}\) in a joint adversarial manner, we propose a procedure to interpret mammography images and identify irregular cancerous regions. With respect to \(\mathbf {R}\) and \( \mathbf {M}\), we define an irregularity score function \(\mathcal {S}(\cdot )\). The details of our Generative Adversarial Irregularity Detection (GAID) method are outlined in Fig. 1 and explained in the following subsections.
Outline of proposed method: (a) \(\mathbf {R}\)+\(\mathbf {M}\) are jointly learned to identify irregularity in mammography images. \(\mathbf {R}\) learns to reconstruct healthy tissues. Whereas, \(\mathbf {M}\) is trained to distinguish between regular (healthy) and irregular tissues (real and fake in GAN setting). During inference for a testing sample X, we use the final feature layer learned in \(\mathbf {M}\), denoted by \(\mathbf {M}_\mathcal {F}(X)\), to define the scoring function. (b) The scoring function \(\mathcal {S}(X)\) is defined based on \(||X\,-\,\mathbf {R}(X)||^2\) and \(||\mathbf {M}_\mathcal {F}(X)\,-\,\mathbf {M}_\mathcal {F}(\mathbf {R}(X))||^2\).
R: Reconstruction Network: Encoder-decoder networks are widely used for tasks such as segmentation [3], inpainting [26], and anomaly detection [15,16,17]. Recently, similar structures were utilized for visual irregularity detection [18]. It was demonstrated that encoder-decoder networks can reconstruct normal samples present in the training set, when trained in an adversarial manner together with a discriminator. Similarly, we use an encoder-decoder network for \(\mathbf {R}\), whose parameters, \(\theta _\mathbf {R}\), are optimized to reconstruct healthy tissues. After training, \(\mathbf {R}_{\theta _{\mathbf {R}}}(X)\) (or \(\mathbf {R}(X)\) for short) should be similar or close to X. High deviations of \(\mathbf {R}(X)\) with respect to X may conclude that X is not similar to the pool of training samples. Based on the previous work [1, 18], \(\mathbf {R}\) will be robust against some levels of noise in the input [23] while larger amounts of deviations will be considered as irregularity. We construct the training data such that it only involves images with healthy (normal) tissues. Therefore, \(\mathbf {R}\) only learns to reconstruct healthy images, and will be incapable of reconstructing images with cancerous or irregular tissues. This incapability which is relative to irregularity likelihood can be measured by \(||X-\mathbf {R}(X)||\). Figure 2(a) shows the details of \(\mathbf {R}\), which is composed of two sub-networks (1) encoder and (2) decoder. The encoder maps X to a latent space, which is then mapped back to the original image space using the decoder. For better stability, max-pooling layers are not used in this network, and instead kernel size of \(5\times 5\) with stride 2 are exploited in all convolutional layers. Except the last layer, Leaky ReLU activation function is applied on the output of convolutional layers of the encoder, and ReLU activation function on up-convolutional layers of the decoder. The last layer uses a \(\tanh (\cdot )\) activation function.
M: Matching Network: As mentioned before, \(\mathbf {R}\) is optimized to reconstruct regular tissues with a minimum error. Besides, \(\mathbf {R}\) tries to fool \(\mathbf {M}\) and trick it to accept its generated image as original and not fake. On the other hand, \(\mathbf {M}\) focuses on discriminating between the reconstructed (fake) and original (real) images in the dataset. In some previous work [1, 17], it is demonstrated that after the training of such discriminator in a GAN setting, it can detect irregular samples. As a result, similarly we train the the parameters of \(\mathbf {M}\), \(\theta _{\mathbf {M}}\), which can distinguish between regular and irregular tissues at the inference time. In summary, if X contains irregular tissues, \(\mathbf {M}_{\theta _{\mathbf {M}}}(X)\) (or \(\mathbf {M}(X)\) for short) provides a good guidance to detect its irregularity. See Fig. 2(b) for the structure of \(\mathbf {M}\), which involves convolution and fully connected layers.
Irregularity Detector: Both \(\mathbf {R}\) and \(\mathbf {M}\) are trained to manipulate and analyze mammography images from different aspects. \(\mathbf {R}\) learns to reconstruct its input and finally generate an output that not only has small reconstruction error, but it also can fool \(\mathbf {M}\). As a result, for input samples even with moderate to low amounts of noise, \(\mathbf {R}\) can reconstruct them. But when there is an irregularity or high amounts of noise in the image (which were not present in the training images), \(\mathbf {R}\) fails to reconstruct them and as a side effect decimates the input. Consequently, the reconstruction error is very high and hence it is a good discriminative measure for detecting irregular tissues in mammography images. On the other hand, \(\mathbf {M}\) is learned to detect the irregular (fake) images. As shown by the previous work [17], after training the two networks in a similar setting as ours, \(\mathbf {M}(X)\) defines a good detector for irregularities. However, we define a scoring function to reliably single out irregularities using both \(\mathbf {R}\) and \(\mathbf {M}\) trained networks.
Network structures. Batch normalization is included after some layers (denoted by the green bar). Every layer is defined using five elements (kernel-size-width \(\times \) kernel-size-height \(\times \) stride \(\times \) input-feature-map-size \(\times \) output-feature-map-size). (Color figure online)
To define this scoring function, suppose X and \(X'\) are normal and irregular samples, respectively. Based the on above intuitions, it is anticipated that \(||X\,-\,\mathbf {R}(X)||^2\le ||X'-\mathbf {R}(X')||^2\). This means that the difference of an irregular sample compared to its reconstruction is more than the same difference metric for a regular sample. Let \(\mathbf {M}_\mathcal {F}\) define the last fully connected layer of \(\mathbf {M}\) (before the decision layer). Therefore, it is likely that \(||\mathbf {M}_\mathcal {F}(X)\,-\,\mathbf {M}_\mathcal {F}(\mathbf {R}(X))||^2\le ||\mathbf {M}_\mathcal {F}(X')\,-\,\mathbf {M}_\mathcal {F}(\mathbf {R}(X'))||^2\). Based on these two useful directives from the \(\mathbf {R}\) and \(\mathbf {M}\) networks, a function for irregularity score can be defined as the following, such that a larger score will imply a larger likelihood for irregularity:
where \(\lambda \in [0, 1]\) is a regularization hyperparameter. With this scoring function, Eq. (2) summarizes the irregularity detector (ID) used for labeling of samples:
where \(\tau \) is a threshed value. subsectionTraining \(\mathbf {R}\)+\(\mathbf {M}\) \(\mathbf {R}\)+\(\mathbf {M}\) are adversarially trained on a training set containing only healthy mammography images. They are learned similar to GANs [5]. These two networks compete to each other, i.e., \(\mathbf {R}\) tries to provide a lossless reconstruction from healthy images, and confuse \(\mathbf {M}\) in distinguish between reconstructed and original images. Therefore, the training of \(\mathbf {R}\)+\(\mathbf {M}\) can be formulated as:
where \(p_{data}\) is the distribution of the training images (with healthy tissues). Parameters of \(\theta _\mathbf {R}\) are optimized to efficiently reconstruct the images (with the loss term \(\mathcal {L}_{rec}\)) while being able to fool \(\mathbf {M}\) that \(\mathbf {R}(X)\) is a real image and not a reconstructed one (\(\mathcal {L}_{real}\)). Consequently, the loss function of \(\mathbf {R}\) can defined as the combination of these two terms:
where \(f_\sigma (s,t)\) is sigmoid cross-entropy function, i.e., the cross-entropy over s with the desired label of t. \(\beta _{rec}\) and \(\beta _{real}\) are two hyperparameters to adjust the importance of each term (we set them equal to 1 and 4, respectively). On the other hand, \(\mathbf {M}\) trains to identify \(\mathbf {R}(X)\) as fake and X as real. Therefore, the objective function of \(\mathbf {M}\), \(\mathcal {L}_\mathbf {M}\), is defined as:
3 Experiments
In this section, we evaluate the proposed Generative Adversarial Irregularity Detection (GAID) method for detecting mammography image regions that contain irregular tissues. Comparison with the state-of-the-art in the scope of irregularity detection in mammography images is challenging due to two important difficulties: (1) Most of the previous methods solve this problem as a binary classification problem. This setting is different than ours while ours is more realistic, since obtaining enough irregular or cancerous samples may be very hard. Our method is the first that detects irregularities in mammography images by only observing and hence modeling healthy tissues; (2) The previous methods are often evaluated on private data, which make the comparison very complicated. To this end, for a fair set of comparisons, we re-implemented some binary classifiers on this dataset (AlexNet [11] and VGG-Net [24]) and compare with the fully-supervised settings, similar to the ones in [7]. Furthermore, we compare the performance of GAID with state-of-the-art anomaly detection methods (AnoGan [19], GANomaly [1], and ALOCC [17]) on the same mammography dataset. For a fair comparison among different methods, we execute them all with similar setting. We used the open-source shared source codes of these methods.
Experiment Setting: We implemented our method using of the Tensor-Flow framework on a 1080 Ti GPU. \(\lambda \) in the score function Eq. (1) is set to 0.4. All networks are learned with a batch size of 64. For the fully-supervised tests, since the number of irregular samples are very limited, available irregular patches are repeated to make equal sets of samples across the two classes. We Use the Contrast limited adaptive histogram equalization on mammography images for improving their contrast. We report the Area Under ROC Curve (AUC) and F1 score as evaluation metrics widely used by the previous works of irregularity detection. AUC is calculated by varying the \(\tau \) threshold as introduced in (2). For our method and the other anomaly detection methods, the training set is composed of all normal samples (image patches), and there is no assumption about the context of irregular tissues during of training. For the binary classifiers, the irregular samples in the dataset are split equally between training and testing.
Datasets: Mammographic Image Analysis Society (MIAS) Dataset: This dataset [21] contains 322 mammography images in mediolateral-oblique (MLO) view with a 1024 \(\times \) 1024 resolution. We consider all the benign and malignant cases in this dataset as irregular versus the normal class present in the dataset. The ground-truth of irregular (i.e., benign or malignant tumor) regions are annotated in the dataset using the center and the diameter of those regions. We partition all images into patches with three of size for different experiments (1) 217,866 patches of size 64 \(\times \) 64, (2) 52,519 patches of size 128 \(\times \) 128, and (3) 4,346 patches of size 256 \(\times \) 256. These patches only contain healthy tissues and are used for training. For testing, in all three experiments 119 healthy and 119 irregular patches are considered (not included in the training set). Note that for building the supervised methods, 50% of irregularity patches are used for training.
INbreast Database: This dataset [13] contains 410 mammography images in mediolateral-oblique (MLO) and cranial-caudal (CC) views with a 3000 \(\times \) 4000 resolution. We consider all the mass cases in this dataset as irregular versus the normal class present in the dataset.
We partition all images into patches with three of size for different experiments (1) 2,267,643 patches of size 64 \(\times \) 64, (2) 541,341 patches of size 128 \(\times \) 128, and (3) 135,155 patches of size 256 \(\times \) 256. For testing, for different experiments (1) 177 healthy and 177 irregular (2) 1,596 healthy and 1,596 irregular (3) 10,259 healthy and 10,259 irregular.
Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM) Dataset: This dataset [12] contains 2,620 scanned film mammography studies from both cranial-caudal (CC) and mediolateral-oblique (MLO) views. The labels in this dataset also include benign, malignant, and normal with verified pathology information. We use this dataset only in a testing scenario and qualitatively evaluate the pretrained model on MIAS on this data. This test can demonstrate the generalizability of the proposed method, specially when it is tested on an entirely new dataset.
Testing results of the proposed irregularity detector on the CBIS-DDSM dataset [12], trained on MIAS [21] and INBreast [13] datasets. Brighter areas of heat-map indicate higher likelihood of irregularity; The heat-map1 and heat-map2 are for training the GAID on MIAS and INBreast datasets, respectively.
Results: To evaluate the generalizability of GAID, we train it on MIAS [21] and INBreast, and test it on the CBIS-DDSM dataset [12]. Some of these results are visualized in Fig. 3. For each input image (X), two heat-maps are generated based on the irregularity score \(\mathcal {S}(\cdot )\), defined in Eq. (1). Comparing the heat-maps with the ground-truth confirms that our method can delineate regions containing irregular tissues, i.e., cancerous regions. Note that the MIAS dataset contains images with only the CC view. This can be a reason why our method perform a bit worse on the images with the MLO view. Domain adaptation techniques can resolve such issues, which can define directions for future works.
Comparison with the State-of-the-Art Irregularity Detection Methods: Table 1 lists the comparison of our method with baseline and state-of-the-art methods on MIAS and INBreast datasets. In this table, the performance of the methods for detection of irregularities are provided in terms of AUC, F1-score, as well as their computational complexity (run-time at testing phase). As can be seen, our method GAID outperforms all baseline and state-of-the-art methods, in some cases by a large margin. In terms of running time, our proposed method is better or as fast as other methods, except for GANomaly [1]. Considering both of aspects of run-time and detection accuracy, our method yields a good compromise. Note, patch size and number of training epochs are important for these results. Here, all models are trained with similar settings. Compared to the fully-supervised, although our method does not see any irregular samples during training, it obtains acceptable results. Note that accuracy scores of all unsupervised methods are significantly better than chance (i.e., p-value <0.01 in Fisher’s exact test [4]). In this experiment, the supervised methods work well, as the diversity among the irregular samples in this dataset is rather low. On the other hand, if we test the trained models on entirely new datasets the supervised models perform worse in identifying the abnormalities. In real-world applications, imaging protocols and tumors can form in different shapes and with different progressing patterns. To show the generalizability of our method, we use the pretrained GAID and the supervised baseline, AlexNet, on the INBreast dataset, and test both on MIAS dataset [12]. Table 2 shows their performance.
4 Conclusion
In this paper, we proposed a simple yet effective method for detecting irregular tissues in mammography images. Our methods, GAID, was inspired by the adversarial learning techniques in the widely used GANs. We proposed two networks that were adversarially trained for interpreting mammography images. After training, based on these two networks, we defined an irregularity scoring function that was able to detect irregular tissues. Our method outperformed fully-supervised and the state-of-the-art methods for irregularity detection. Note, all previous methods for detecting irregular tissues in mammography were learned in a fully-supervised manner at the presence of both regular and irregular samples during training. Our method is the first that is trained without any supervision signal from the irregular class. In this work, our method operated on patches (i.e., it is a patch-based method). As a direction for the future work, one can extend the model to process the whole image at one-step, similar to AVID [18].
References
Akcay, S., Atapour-Abarghouei, A., Breckon, T.P.: GANomaly: semi-supervised anomaly detection via adversarial training. arXiv preprint arXiv:1805.06725 (2018)
Arevalo, J., González, F.A., Ramos-Pollán, R., Oliveira, J.L., Lopez, M.A.G.: Representation learning for mammography mass lesion classification with convolutional neural networks. Comput. Methods Programs Biomed. 127, 248–257 (2016)
Fayyaz, M., Saffar, M.H., Sabokrou, M., Fathy, M., Huang, F., Klette, R.: STFCN: spatio-temporal fully convolutional neural network for semantic segmentation of street scenes. In: Chen, C.-S., Lu, J., Ma, K.-K. (eds.) ACCV 2016. LNCS, vol. 10116, pp. 493–509. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-54407-6_33
Fisher, R.A.: The logic of inductive inference. J. Roy. Stat. Soc. 98(1), 39–82 (1935)
Goodfellow, I., et al.: Generative adversarial nets. In: Advances in Neural Information Processing Systems, pp. 2672–2680 (2014)
Gubern-Mérida, A., et al.: Automated localization of breast cancer in DCE-MRI. Med. Image Anal. 20(1), 265–274 (2015)
Huynh, B.Q., Li, H., Giger, M.L.: Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 3(3), 034501 (2016)
Jafari, S.H., et al.: Breast cancer diagnosis: Imaging techniques and biochemical markers. J. Cell. Physiol. 233(7), 5200–5213 (2018)
Jamieson, A.R., Drukker, K., Giger, M.L.: Breast image feature learning with adaptive deconvolutional networks. In: Medical Imaging 2012: Computer-Aided Diagnosis, vol. 8315, p. 831506. International Society for Optics and Photonics (2012)
Jiao, Z., Gao, X., Wang, Y., Li, J.: A deep feature based framework for breast masses classification. Neurocomputing 197, 221–231 (2016)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Lee, R.S., Gimenez, F., Hoogi, A., Miyake, K.K., Gorovoy, M., Rubin, D.L.: A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 4, 170177 (2017)
Moreira, I.C., Amaral, I., Domingues, I., Cardoso, A., Cardoso, M.J., Cardoso, J.S.: INbreast: toward a full-field digital mammographic database. Acad. Radiol. 19(2), 236–248 (2012)
Petrick, N., Chan, H.P., Sahiner, B., Wei, D.: An adaptive density-weighted contrast enhancement filter for mammographic breast mass detection. IEEE-TMI 15(1), 59–67 (1996)
Sabokrou, M., Fathy, M., Hoseini, M.: Video anomaly detection and localisation based on the sparsity and reconstruction error of auto-encoder. Electron. Lett. 52(13), 1122–1124 (2016)
Sabokrou, M., Fayyaz, M., Fathy, M., Klette, R.: Deep-cascade: cascading 3D deep neural networks for fast anomaly detection and localization in crowded scenes. IEEE Trans. Image Process. 26(4), 1992–2004 (2017)
Sabokrou, M., Khalooei, M., Fathy, M., Adeli, E.: Adversarially learned one-class classifier for novelty detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3379–3388 (2018)
Sabokrou, M., et al.: AVID: adversarial visual irregularity detection. arXiv preprint arXiv:1805.09521 (2018)
Schlegl, T., Seeböck, P., Waldstein, S.M., Schmidt-Erfurth, U., Langs, G.: Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In: Niethammer, M., et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 146–157. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59050-9_12
Stewart, B., Wild, C.P., et al.: World cancer report 2014. Self (2018)
Suckling, J., et al.: Mammographic image analysis society (MIAS) database v1.21 [dataset] (2015). https://www.repository.cam.ac.uk/handle/1810/250394
Sun, W., Tseng, T.L.B., Zhang, J., Qian, W.: Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput. Med. Imaging Graph. 57, 4–9 (2017)
Vincent, P., Larochelle, H., Bengio, Y., Manzagol, P.A.: Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th International Conference on Machine Learning, pp. 1096–1103. ACM (2008)
Wang, L., Guo, S., Huang, W., Qiao, Y.: Places205-VGGNet models for scene recognition. arXiv preprint arXiv:1508.01667 (2015)
Weissleder, R.: Molecular imaging in cancer. Science 312(5777), 1168–1171 (2006)
Xie, J., Xu, L., Chen, E.: Image denoising and inpainting with deep neural networks. In: Advances in Neural Information Processing Systems, pp. 341–349 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ahmadi, M., Sabokrou, M., Fathy, M., Berangi, R., Adeli, E. (2019). Generative Adversarial Irregularity Detection in Mammography Images. In: Rekik, I., Adeli, E., Park, S. (eds) Predictive Intelligence in Medicine. PRIME 2019. Lecture Notes in Computer Science(), vol 11843. Springer, Cham. https://doi.org/10.1007/978-3-030-32281-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-32281-6_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32280-9
Online ISBN: 978-3-030-32281-6
eBook Packages: Computer ScienceComputer Science (R0)