Abstract
Efficient and accurate segmentation of prostate gland facilitates the prediction of the pathologic stage and treatment response. Recently, deep learning methods have been proposed to tackle this issue. However, the effectiveness of these methods is often limited by inadequate semantic discrimination and spatial context modeling. To address these issues, we propose the Hybrid Discriminative Network (HD-Net), which consists of a 3D segmentation decoder using channel attention block to generate semantically consistent volumetric features and an auxiliary 2D boundary decoder guiding the segmentation network to focus on the semantically discriminative intra-slice features. Meanwhile, we further design the pyramid convolution block and residual refinement block for HD-Net to fully exploit multi-scale spatial contextual information of the prostate gland. In addition, to reduce the information loss in propagation and fully fuse the multi-scale feature maps, we introduce inter-scale dense shortcuts for both decoders. We evaluated our model on the Prostate MR Image Segmentation 2012 (PROMISE12) challenge dataset and achieved a synthetic score of 90.34, setting a new state of the art.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Accurate segmentation of the prostate gland using magnetic resonance (MR) imaging is critical to the diagnosis and treatment of prostate diseases. Due to the anatomical variation across subjects and the interference of adjacent structures with similar appearances, approaches based on the traditional machine learning algorithms [3, 9] still suffer from low accuracy and poor generalization.
Recent works based on the 3D fully convolutional encoder-decoder [5] have shown convincing performance on this task. The design focus has been on effective ways of exploiting spatial context, which is an important cue for prostate segmentation. Milletari et al. [6] incorporated the Dice loss and unique data augmentation into a volumetric and fully convolutional network called V-Net to improve the segmentation result. Yu et al. [11] introduced both long and short residual connections to a 3D fully convolutional encoder-decoder to exploit the spatial contextual information, and achieved the top performance in the PROMISE 12 challenge [1] at that time. Zhu et al. [13] adopted dense connections and long connections to alleviate the vanishing gradient and overfitting issues encountered in the training process. Nie et al. [7] proposed an adversarial model ASDNet, which adopted region-attention based semi-supervised loss to address the insufficient data problem for training the complex networks. However, due to the large variability in the appearance of the gland capsule and its low contrast and high similarity to adjacent structures, these methods still reveal inadequate intra-class consistency and inter-class discrimination, which typically reflects in poor segmentation performance on the regions with the same semantic label but different appearances (usually inside the prostate gland) or the regions with similar appearances but different semantic labels (usually on the boundary of the prostate gland). Furthermore, prostate MR images normally have complex spatial contextual information due to the widely existed anisotropic voxel resolution. 3D isotropic convolutions [5, 6] tend to be less-capable of segmenting the volumetric prostate gland accurately.
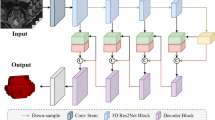
In this paper, we propose a Hybrid Discriminative Network (HD-Net) for automated prostate gland segmentation in MR images, as illustrated in Fig. 1. This HD-Net model has a 2D + 3D encoder-decoder structure, including a ResNet-101-based 3D encoder, a 3D segmentation decoder, and a 2D boundary decoder. In the training phase, the encoder and two decoders are trained in an end-to-end manner; whereas in the inference phase, the trained HD-Net without the boundary decoder is employed to segment prostate MR images. We evaluated our model against several state-of-the-art segmentation algorithms on the PROMISE 12 challenge dataset [1], and our HD-Net outperformed all the other methods. The further ablation experiment results demonstrate the effectiveness of each component of HD-Net.
The contributions of this work are three-fold: (1) we design a 3D ResNet encoder to characterize prostate MR images, a 3D segmentation decoder with channel attention to extract semantically consistent features, and an auxiliary 2D boundary decoder to guide the segmentation decoder with the intra-slice discriminative features on both sides of the prostate boundary; (2) we use a modified design of the pyramid convolution block and residual refinement block, and incorporate them into both decoders to fully exploit the multi-scale spatial contextual information of the prostate gland; and (3) we introduce inter-scale dense shortcuts to both decoders, aiming to reduce the information loss and to fuse multi-scale feature maps effectively.
2 Method
2.1 3D ResNet Encoder
In the proposed HD-Net, we use the ImageNet pre-trained ResNet-101 [4] as the backbone encoder model (Fig. 1). Here, considering the limited image size and uncontrollable computing complexity, we only utilize the first three res-blocks of ResNet-101, which means our encoder has four stages of feature map scale. Since the original ResNet-101 was constructed for 2D natural images, we need to extend it to 3D medical image segmentation. Specifically, we use \(7\times 7\times 3\) 3D convolution to replace \(7\times 7\) 2D convolution in the first convolutional layer of ResNet-101. For the other convolutional layers, we directly transform \(x\times x\) 2D convolutions to \(x\times x\times 1\) 3D convolutions. Such extension makes it feasible to use the pre-trained ResNet-101 to initialize our encoder to transfer the knowledge learned on the large-scale ImageNet images to characterize prostate MR images. In addition, similar to [10], we also add a global average pooling layer [12] on top of the segmentation network to get the strongest semantic consistency.
2.2 3D Segmentation Decoder
Channel Attention Block. For segmentation architecture with encoder-decoder style, it was observed that the features produced at the early stages of the network reveal fine spatial information but have weak semantic guidance, whereas the later-stage features have strong semantic consistency and discrimination but give coarse spatial prediction. Some studies [2, 10] introduced channel-wise semantic attention to generate features with more discriminative capability. Based on these considerations, in the decoder of the segmentation network, we propose a channel attention block to provide up-bottom semantic discriminative guidance. As illustrated in Fig. 2(a), the channel attention block has two input feature maps from two adjacent stages. We first concatenate these two feature maps together and use global average pooling to generate the channel-wise attention. Then we multiply the obtained channel-wise attention vector with the early-stage feature map to enhance its semantic discrimination. Last, we combine the enhanced feature map with the later-stage feature map as the output feature map. With this design, the segmentation network not only integrate multi-scale context information, but also can pay more attention to the feature maps that are associated with more semantic information.
Detailed structures of the channel attention block, pyramid convolution block, residual refinement block and 2D residual refinement block of boundary network. C, \(\times \) and + represent concatenation, matrix multiplication and matrix summation, respectively. At each scale stage, the number of large convolutions and the corresponding kernel size are (\(31\times 31\times 11\), \(23\times 23\times 9\), \(19\times 19\times 7\), \(15\times 15\times 5\)), (\(19\times 19\times 7\), \(15\times 15\times 5\), \(9\times 9\times 3\)), (\(9\times 9\times 3\), \(7\times 7\times 3\)) and \(3\times 3\times 1\), respectively.
Pyramid Convolution Block. According to [8], in semantic segmentation task, large kernel convolution can promote voxel classification ability of deep network besides its inherent spatial localization capability. However, considering the variability in size and shape of the gland capsule, here we design the pyramid convolutional block to generate more discriminative features. Specifically, inside the pyramid convolution block, several 3D convolutions with different kernel sizes are constructed in parallel and the numbers of 3D convolutions are adaptive to the scale of feature map, as illustrated in Fig. 2(b). Pyramid kernels can fuse local and global image contents at multiple scales and reduce information loss. In addition, to reduce computational complexity, we further apply convolution decomposition to reduce the parameters of the large kernel 3D convolution.
Residual Refinement Block. To address the anisotropic spatial context, within each stage of the decoder, we introduce the residual refinement block instead of the traditional isotropic block with \(3\times 3\times 3\) convolutions. As illustrated in Fig. 2(c), the input to residual refinement block is passed to a \(3\times 3\times 1\) and then a \(1\times 1\times 3\) 3D convolutions in turn. Inspired by the residual connection [4], the outputs of both 3D convolutions are then summed with the input together as the output. With this design, the \(3\times 3\times 1\) convolution helps to capture the 2D features inside the x-y planes, and the \(1\times 1\times 3\) convolution can focus on between-slices features.
2.3 2D Boundary Decoder
Besides the intra-class consistency, the inter-class discrimination of adjacent region is also crucial for accurate semantic segmentation of the prostate. We suggest that a feasible solution to address this issue is to add the guidance of semantic boundary. To do this, we construct an auxiliary boundary network in a bottom-up fashion, so that the boundary network can use early-stage features to extract accurate prostate boundary and subsequently represent the extracted boundary with later-stage semantic information, as shown in Fig. 1. In addition, due to anisotropic spatial voxel resolution of prostate MR images, in each stage, we use 2D pyramid convolution blocks (of the same architecture as the 3D one but without the convolution kernel in z dimension) and 2D residual refinement block (shown in Fig. 2(d)) rather than 3D ones to obtain the prostate boundary on each x-y plane of the 3D feature maps. Since both decoders share one 3D encoder, the boundary decoder can guide the segmentation decoder to generate features on both sides of the prostate boundary with more semantic discrimination.
Note that we choose to model the 2D boundary information instead of 3D because compared to the whole image volume, the proportion of the boundary is low and the boundary of adjacent slices may have large variety in both shape and location, which all make it hard to model the 3D boundary information with deep network.
2.4 Further Refinements
To further reduce the potential information loss in the propagation and make full use of the inter-scale features, we introduce the inter-scale dense shortcuts in both decoders (Fig. 1). Since we construct segmentation decoder and boundary decoder in a up-bottom and bottom-up fashion, respectively. We apply different dense shortcuts strategies. Specifically, for segmentation network, the high stage input of the channel attention block is a combination of upsampled feature maps from all higher stages. In each stage of the boundary network, the input of the second refinement block further is the summation of the upsampled feature maps from all lower stages. With these specific inter-scale dense shortcuts, the HD-Net can fully fuse the features from different scales and directly propagates the forward and backward information from one scale stage to anther scale stage.
Throughout the whole HD-Net, we apply \(1\times 1\times 1\) convolutions to adjust the channel number of feature maps. In addition, for explicit refinement and effective model optimization, instead of only using a final prediction supervision, we further add multi-scale side output supervisions to the segmentation decoder, which is accomplished by upsampling the multi-scale output feature maps of the residual refinement blocks of both decoders to the size of ground truths as segmentation results (Fig. 1). To reduce the influence of class imbalance in a dynamic way, we use mini-batch class weighted cross entropy loss for both sub-models:
where x is the network prediction and the class weight \(r_b\) is the ratio of the numbers of voxels in the prostate and non-prostate regions in mini-batch b. The final loss of the HD-Net is \(L=L_s+\lambda L_b\), a combination of the segmentation network loss \(L_s\) and boundary network loss \(L_b\) with the balance parameter \(\lambda \).
3 Experiments and Results
Datasets: We used the PROMISE12 challenge database [1] to evaluate the proposed HD-Net. The dataset contains 50 training and 30 test T2-weighted MR images. The corresponding ground truths of the whole prostate annotated by the experts are available in the training set and that of the test set is withheld for online independent evaluation. We trained the proposed method on the training set and submitted the segmentation of the test set to the ongoing challenge for an evaluation score.
Implementation Details: We implemented the proposed method based on the Pytorch framework with two Nvidia Geforce GTX 1080Ti 11GB GPUs. We performed some simple image preprocessing, including bias field correction, voxel spacing unification to a fixed size of \(0.625 \times 0.625 \times 1.5\) mm and intensity normalization into zero mean and unit variance. We employed several online data augmentations to reduce the potential overfitting caused by limited training images, including random flipping (up-down or left-right in x-y planes), random rotation (±25, 90, 180, or 270\(^{\circ }\) in x-y planes), random Gaussian noise addition (\(\sigma \) from 0.3 to 0.7) and random 3D scaling (±0.2). We trained the network using the Adam optimizer with a batch size of 16 and betas of (0.9, 0.999). The initial learning rate was \(1e^{-3}\) and decayed by multiplied with \((1-\frac{iteration}{max_iteration})^{0.9}\). The loss combination weight \(\lambda \) was set to 0.5 after some comparisons. In the inference phase, for each MR image, we extracted the sub-volumes with a fixed stride of \(24\times 24\times 8\) and averaged the predictions from the overlapping volumes to get the final segmentation.
Comparison with State-of-the-Art Methods: In Table 1, we compared our HD-Net with nine top-ranking methods listed on the PROMISE12 challenge leaderboard. Note that the dice similarity coefficient (DSC), average boundary distance (ABD), 95% Hausdorff distance (95HD) and absolute relative volume difference (aRVD) of the whole prostate gland, apex part (first 1/3 of the prostate volume), and base part (last 1/3 of the prostate volume) and a synthetic score were generated by the online validation system. At the writing of this paper, our proposed approach is ranked the first out of 291 entries. Due to the limitation of space, we can only list the results of all metrics on the whole prostate gland. From the results in Table 1, we can see that although some methods achieved better performance on a single metric, our HD-Net gained the overall best performance. Our HD-Net also has the best result on 95HD and second best result on aRVD, which indicates gland volumes segmented by our HD-Net have few outliers and globally match the ground truths, respectively.
Ablation Study: We also conducted an ablation study on the training set with a 5-fold cross validation to evaluate the contributions of the pyramid convolution block, channel attention block, inter-scale dense shortcuts, multi-scale deep supervision and boundary decoder to the overall performance of the proposed HD-Net. The results are given in Table 2. It shows that all components in the HD-Net are beneficial to the overall segmentation performance. In particular, it is clear to see that using pyramid convolution block and further incorporating channel attention block significantly increased DSC by 0.68% and 0.75%. Such results demonstrate that our HD-Net is highly effective in the fully exploitation of spatial image contextual information and of semantic discrimination between the prostate and surrounding tissues. Meanwhile, the results also show that adding inter-scale dense shortcuts can increase the DSC by more than 0.4%, which indicates its effectiveness in the reduction the information loss and the further fusion of the inter-scale features. In addition, we can observe the boundary decoder and multi-scale deep supervision can contribute to a further performance improvement by 0.15% and 0.13%, respectively.
4 Conclusions
In this paper, we propose the HD-Net, a novel fully convolutional encoder-decoder with a 3D segmentation decoder and an auxiliary 2D boundary decoder, for the segmentation of prostate gland in volumetric MR images. Specifically, we introduce the channel attention block to enhance semantic discrimination, use both pyramid convolution block and residual refinement block to fully exploit the spatial contextual information, and adopt the multi-scale deep supervision to further improve the performance. Moreover, we incorporate inter-scale dense shortcuts into both decoders to reduce the information loss and fuse the multi-scale features. Our experimental results suggest that the proposed HD-Net outperforms nine recent methods and sets the new state of the art on the PROMISE12 challenge dataset.
References
MICCAI grand challenge: Prostate MR image segmentation 2012 (2012). https://promise12.grand-challenge.org/Home/
Chen, L., et al.: SCA-CNN: spatial and channel-wise attention in convolutional networks for image captioning. In: CVPR, pp. 5659–5667 (2017)
Guo, Y., Gao, Y., Shen, D.: Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE TMI 35(4), 1077–1089 (2016)
He, K., Zhang, X., Ren, S., et al.: Deep residual learning for image recognition. In: CVPR, pp. 770–778 (2016)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: CVPR, pp. 3431–3440 (2015)
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 3DV, pp. 565–571. IEEE (2016)
Nie, D., Gao, Y., Wang, L., Shen, D.: ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 370–378. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_43
Peng, C., Zhang, X., Yu, G., Luo, G., Sun, J.: Large kernel matters-improve semantic segmentation by global convolutional network. In: CVPR, pp. 4353–4361 (2017)
Toth, R., Madabhushi, A.: Multifeature landmark-free active appearance models: application to prostate MRI segmentation. IEEE TMI 31(8), 1638–1650 (2012)
Yu, C., Wang, J., Peng, C., Gao, C., Yu, G., Sang, N.: Learning a discriminative feature network for semantic segmentation. In: CVPR, pp. 1857–1866 (2018)
Yu, L., Yang, X., Chen, H., Qin, J., Heng, P.A.: Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images. In: AAAI, pp. 66–72 (2017)
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: CVPR, pp. 2881–2890 (2017)
Zhu, Q., Du, B., Wu, J., Yan, P.: A deep learning health data analysis approach: automatic 3D prostate MR segmentation with densely-connected volumetric ConvNets. In: IJCNN, pp. 1–6. IEEE (2018)
Acknowledgement
This work was supported in part by the National Natural Science Foundation of China under Grants 61771397, in part by the Science and Technology Innovation Committee of Shenzhen Municipality, China, under Grants JCYJ20180306171334997, in part by Synergy Innovation Foundation of the University and Enterprise for Graduate Students in Northwestern Polytechnical University under Grants XQ201911, in part by the Project for Graduate Innovation team of Northwestern Polytechnical University, and in part by the US NIH R01 AG049371 and the US NSF IIS 1836938, DBI 1836866, IIS 1845666, IIS 1852606, IIS 1838627, IIS 1837956.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Jia, H., Song, Y., Huang, H., Cai, W., Xia, Y. (2019). HD-Net: Hybrid Discriminative Network for Prostate Segmentation in MR Images. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11765. Springer, Cham. https://doi.org/10.1007/978-3-030-32245-8_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-32245-8_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32244-1
Online ISBN: 978-3-030-32245-8
eBook Packages: Computer ScienceComputer Science (R0)