Abstract
Accurate lesion detection in computer tomography (CT) slices benefits pathologic organ analysis in the medical diagnosis process. More recently, it has been tackled as an object detection problem using the Convolutional Neural Networks (CNNs). Despite the achievements from off-the-shelf CNN models, the current detection accuracy is limited by the inability of CNNs on lesions at vastly different scales. In this paper, we propose a Multi-Scale Booster (MSB) with channel and spatial attention integrated into the backbone Feature Pyramid Network (FPN). In each pyramid level, the proposed MSB captures fine-grained scale variations by using Hierarchically Dilated Convolutions (HDC). Meanwhile, the proposed channel and spatial attention modules increase the network’s capability of selecting relevant features response for lesion detection. Extensive experiments on the DeepLesion benchmark dataset demonstrate that the proposed method performs superiorly against state-of-the-art approaches.
Q. Shao and L. Gong contribute equally and share the first authorship. This work was done when Q. Shao was an intern in Tencent Youtu Lab. The source code and results are available at https://github.com/shaoqb/multi_scale_booster.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Automatically detecting lesions in CT slices is important to computer-aided detections/diagnosis (CADe/CADx). The identification and analysis of lesions in the clinic practice benefit the diagnosis of diseases at the early stage. The recent progress of the CADx mainly focuses on the visual recognition. By using the Convolutional Neural Networks (CNNs), the automatic detection of lesions has reduced the workload of the manual examinations. These lesion detection approaches arise from the object detection frameworks such as Faster R-CNN [8] and Feature Pyramid Network (FPN) [7], which typically employ a two-stage process. First, they draw a set of bounding box samples indicating the potential region-of-interest (ROI) on the feature maps of CT slices. Then, each sample is classified as either lesion or background by a binary classifier. The two-stage CNN based detection frameworks have been trained in an end-to-end fashion and achieved the state-of-the-art performance.
To further improve the detection accuracy of CT data where blur and artifacts rarely exist [9, 11, 12], several methods [2, 3, 6, 13] have been proposed to leverage the 3D spatial information. Ding et al. [2] proposed a 3D-CNN classifier to refine the detection results of the pulmonary cancer from the 2D-CNN framework. Furthermore, Dou et al. [3] explored a 3D-CNN for false positive reduction in pulmonary nodules detections. On the other side, Liao et al. [6] extended the region proposal network (RPN) [8] to 3D-RPN to generate 3D proposals. Although spatial representations extracted from 3D space improve the network performance on certain tasks, these methods suffer from tremendous memory and computational consumption. To tackle the computation efficiency problem, Yan et al. [13] proposed a 3D context enhanced region-based CNN (3DCE) to produce 3D context from feature maps of 2D input images. It achieved similar performance to 3D-CNN while consuming the same speed of the traditional 2D-CNN, which deserves further improvement with more advanced networks.
In real-world scenarios, body lesions usually have arbitrary size. For instance, in the DeepLesion [14] dataset, the lesion size ranges from 0.21 mm to 342.5 mm. Since most of the established CNNs are not robust to handle such spatial scale variations, they have unpredictable behavior in the varying cases. As shown in Fig. 1, both Faster R-CNN and FPN fail to detect tiny lesions in the first row, while they produce small false positive lesions around the actual large lesion locations in the second and third rows.
In this paper, we propose a fine-grained lesion detection approach with a novel multi-scale attention mechanism. We use 2D FPN as the backbone to construct the feature pyramid in a relatively coarse scale. Within each level of the feature pyramid, we propose to use a Multi-Scale Booster (MSB) to facilitate lesion detection across fine-grained scales. Given the feature maps from one pyramid level, MSB first performs Hierarchically Dilated Convolution (HDC) that consists of several dilated convolution operations with different dilation rates [15]. The feature responses from HDC contain fine-grained information that is complementary to the original feature pyramid, which is achieved by extensive feature extraction in 2D space. The over-sampled feature responses are then concatenated and further exploited by channel-wise and spatial-wise attention. The channel attention module in MSB explores different lesion responses from the subchannels of the concatenated feature maps. The spatial attention module in MSB locates lesion response within each attentive channel. The channel-wise and spatial-wise attention modules enable the network to focus on particular lesion responses offered by the fine-grained features, while annealing the irrelevant and interference information. Thorough experiments demonstrate that MSB improves the deep pyramid prediction results and performs favorably against state-of-the-art approaches on the DeepLesion benchmarks.
2 Proposed Method
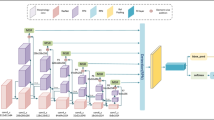
Figure 2 shows an overview of the pipeline. Our method uses a pre-trained FPN network to extract features from the input image at different pyramid levels. The extracted features are further processed by channel and spatial attention modules to capture fine-grained information to handle large spatial scale variations. The output of the MSB modules is used to make the final prediction at each pyramid feature map respectively.
2.1 Revisiting FPN
The FPN [7] consists of three components for object detection: the bottom-up pathway, top-down pathway and skip connections in between. The bottom-up pathway computes feature maps at several different scales with a down-sampling factor of 2. We use \(C_i^D\) to denote the feature maps at the i-th down-sampled pyramid. The \(C_i^D\) has strides of \(2^{i+1}\) pixels with respect to the input image. In the top-down pathway, the feature maps from the coarse levels are upsampled gradually to the finer resolutions with an up-sampling factor of 2. We denote the upsampled feature maps at the i-th upsampled pyramid as \(C_i^U\). The skip connections merge the downsampled and upsampled feature maps together at each pyramid level and the fused feature maps can be written as:
where \(\oplus \) is the element-wise addition operation. After generating the feature maps \(P_i\), the potential objects are then detected at each feature pyramid level.
2.2 Hierarchically Dilated Convolution
The dilated convolution is commonly used to expand the reception fields without loss of the original resolution. In ASPP [1], dilated convolution provided precise scale estimations for pixel-level semantic segmentation. Given the input feature map \(C_i^D\), the dilated convolution can be written as:
where x is the location of the current pixel under processing; k is the supporting pixels in the convolution process; W is the filter weight; and r is the dilation rate. The dilation rate corresponds to the stride that we use to sample input feature map \(C_i^D\). We denote the dilated convolution in a general form as \(\mathcal {D}_r(C_i^D)\) where \(\mathcal {D}_r\) is the dilated convolution operator with dilation rate r.
The HDC performs multiple dilated convolutions with different dilation rates. In our method, we use three dilated convolutions (i.e., \(d_1\), \(d_2\), and \(d_3\)) and keep the filter weight W fixed. The HDC output of the input feature map \(C_i^D\) can be formulated in the following:
where \(\mathcal {H}\) is the concatenation of the dilated convolution results \(\mathcal {D}(C_i^D)\) and dimension mapping results \(\mathcal {M}(C_i^D)\). We denote the concatenated results as \(H_i^D\). The dimension mapping operation \(\mathcal {M}\) is a \(1\times 1\) convolution on the input feature maps to ensure the channel consistency with respect to the dilated convolution results, while maintaining the original feature information from the FPN. We use different dilation rates to capture the lesion responses from each pyramid feature map respectively. These fine-grained feature responses of HDC contain multiple scales of reception fields within each feature pyramid level \(C_i^D\). In order to only capture the scale variation responses on the pyramid feature maps, we share weights among HDC to overcome other interferences such as rotation and deformation.
2.3 Channel Attention
We refine the HDC result using a squeeze-and-excitation network as shown in Fig. 2 following [5]. The HDC result \(H_i^D\) captures the feature responses of the potential lesions from the multi-scale perspective. For a particular lesion with a certain dimension, high feature response may reside in one of the dilated convolution scales. Therefore it is intuitive to attend the network to the subchannels of \(H_i^D\). We propose a channel attention module as shown in Fig. 2. It first squeezes \(H_i^D\) by a global pooling operation and then activates the reduced feature maps by a \(1\times 1\) convolution layer. The channel attention can be written as:
where \(\mathcal {P}_{avg}\) and \(W_{1\times 1}\) represent the global pooling and the convolution operation, respectively. The channel attention output \(\mathcal {F}_{ch}(H_i^D)\) is a one dimensional vector re-weighting \(C_i^D\). The network is learned to pay more attention to the subchannels of \(H_i^D\) where the precise scale response of the lesion region resides. The reweighted feature maps from channel attention can be written as:
where \(\otimes \) is the element-wise multiplication operation.
2.4 Spatial Attention
The channel attention ensures the network to focus on \(H_i^{D ch}\), where the response of the scale estimation from HDC resides. To increase the network’s attention to the lesion response within \(H_i^{D ch}\), we propose a spatial attention module that reduces the distraction outside of the ROIs. The proposed spatial attention module first squeezes \(H_i^{D ch}\) by using a max pooling operation along channel axis to generate the spatial feature map \(\mathcal {F}_{sp}(H_i^{D ch})\), which encodes where to emphasize. The spatial attention activation process can be written as:
where \(\mathcal {P}_{max}\) is the max pooling. The spatial attention \(\mathcal {F}_{sp}(H_i^{D ch})\) is a one-channel feature map with size \(H \times W\) used to filer out the irrelevant information of \(H_i^{D ch}\). As a result, the network will attentively focus around the lesion region. The refined output feature map can be formulated as:
where \(\otimes \) is the same as that in Eq. 1. The output feature map \(\hat{P}_i\) is then used for lesion detection.
3 Experiments
We evaluate the proposed method on the large-scale benchmark dataset DeepLesion [14]. It includes 32,735 lesions from 32,120 CT slices, which are captured from 4,427 patients. The lesion areas cover liver, lung nodules, bone, kidney, and other organs. We follow the dataset configuration to split into the training, validation and test sets. In the training process, we use ResNet50 [4] as the feature extraction backbone. The initial weights from conv1 to conv5 are from the ImageNet pretrained model [10] and the remaining weights are randomly initialized. We resize the CT slices to \(512\times 512\) pixels and concatenate three consecutive CT slides as the input to predict lesions of the central slice. The five anchor scales and three anchor ratios are set as (8, 16, 32, 64, 128), \(\{1:2, 1:1, 2:1\}\) respectively at each level while training RPN. The learning rate is set as 0.01 and the learning process is around 10 epochs.
3.1 Ablation Study
The proposed network consists of four major components. They are FPN, HDC, CH (channel attention), and SP (spatial attention). To evaluate the effectiveness of each module and weights sharing, we ablatively study on the DeepLesion dataset. The evaluation metric is the average sensitivity values at different false positives rates of the whole test set. The evaluation configuration is shown in Table 1. The comparisons among different configurations demonstrate that the proposed MSB achieves highest sensitivity under different false positives rates.
3.2 Comparisons with State-of-the-Art
We compare the proposed method with state-of-the-art approaches including 3DCE [13], Faster R-CNN [8] and FPN [7]. Yan et al. [13] sent multiple slices into the 2D detection network (i.e., Faster R-CNN [8]) to generate feature maps separately, and then aggregated them to incorporate 3D context information for final prediction. We note that the results of 3DCE [13] are the only available results reported on this dataset. We perform the evaluation from two perspectives. The first one is to compute the sensitivity values at different false positives rates as illustrated in Sect. 3.1. It reflects the averaged performance of each method for test set. The other one is to compute the sensitivity values generated based on different sizes of lesions. It reflects how effective each method is to detect lesions at different scales.
Table 2 shows the evaluation results. It demonstrates that the proposed method performs superiorly against existing methods. We note that there are different numbers of CT slices used as input for 3DCE to produce different sensitivity values. The result shows that sensitivity value increases when more CT slices are taken as input. As these CT slices are captured on the same organ of the patient, using more slices will provide sufficient information to the network to detect. Nevertheless, we show that the proposed method achieves higher sensitivity values when using only three slices as input.
To evaluate how the proposed method performs when detecting different size of lesions, we divide the test set into five categories. Each category consists of lesions in a fixed range of size and the range does not overlap with each other. Table 3 shows the evaluation results. The proposed method shows better performance to detect lesions in different sizes. Meanwhile, the sensitivity values of the proposed method exceed those of existing methods more when the size of the testing lesions becomes extremely large or small (i.e., the diameters of the lesions are above 100 mm or below 10 mm). It indicates that the proposed method is more effective to detect extreme scales of the input lesions.
4 Conclusion
We proposed a multi-scale booster (MSB) to detect lesion in large scale variations. We use FPN to decompose the feature map response into several coarse-grained pyramid levels. Within each level, we increase the network awareness of the scale variations by using HDC. The HDC offers fine-grained scale estimations to effectively capture the scale responses. To effectively select meaningful responses, we proposed a cascaded attention module consists of channel and spatial attentions. Evaluations on the DeepLesion benchmark indicated the effectiveness of the proposed method to detect lesions at vastly different scales.
References
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40, 834–848 (2017)
Ding, J., Li, A., Hu, Z., Wang, L.: Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 559–567. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_64
Dou, Q., Chen, H., Yu, L., Qin, J., Heng, P.A.: Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans. Biomed. Eng. 64, 1558–1567 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (2016)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: IEEE Conference on Computer Vision and Pattern Recognition (2018)
Liao, F., Liang, M., Li, Z., Hu, X., Song, S.: Evaluate the malignancy of pulmonary nodules using the 3D deep leaky noisy-or network. IEEE Trans. Neural Netw. Learn. Syst. PP (2017). https://doi.org/10.1109/TNNLS.2019.2892409
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: International Conference on Computer Vision (2017)
Ren, S., He, K., Girshick, R., Sun, J.: Faster R-CNN: towards real-time object detection with region proposal networks. In: Neural Information Processing Systems (2015)
Roy, A.G., Navab, N., Wachinger, C.: Concurrent spatial and channel ‘squeeze & excitation’ in fully convolutional networks. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 421–429. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00928-1_48
Russakovsky, O., et al.: ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015)
Song, Y., Zhang, J., Bao, L., Yang, Q.: Fast preprocessing for robust face sketch synthesis. In: International Joint Conference on Artificial Intelligence (2017)
Song, Y., et al.: Joint face hallucination and deblurring via structure generation and detail enhancement. Int. J. Comput. Vis. 127, 785–800 (2018)
Yan, K., Bagheri, M., Summers, R.M.: 3D context enhanced region-based convolutional neural network for end-to-end lesion detection. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 511–519. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00928-1_58
Yan, K., Wang, X., Lu, L., Summers, R.M.: DeepLesion: automated deep mining, categorization and detection of significant radiology image findings using large-scale clinical lesion annotations. arXiv:1710.01766 (2017)
Yu, F., Koltun, V.: Multi-scale context aggregation by dilated convolutions. arXiv:1511.07122 (2015)
Acknowledgments
This work was founded by the Key Area Research and Development Program of Guangdong Province, China (No. 2018B010111001).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Shao, Q., Gong, L., Ma, K., Liu, H., Zheng, Y. (2019). Attentive CT Lesion Detection Using Deep Pyramid Inference with Multi-scale Booster. In: Shen, D., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science(), vol 11769. Springer, Cham. https://doi.org/10.1007/978-3-030-32226-7_34
Download citation
DOI: https://doi.org/10.1007/978-3-030-32226-7_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32225-0
Online ISBN: 978-3-030-32226-7
eBook Packages: Computer ScienceComputer Science (R0)