Abstract
Pituitary adenomas are rare intracranial tumors that are often found incidentally in MR images. On the other hand, radiomics is a new field whose aim is converting images in mineable data; particularly, texture analysis is a postprocessing technique extracting quantitative parameters from the heterogeneity of pixel grey level. In this scenario, machine learning can be applied in order to classify these adenomas into functional and non-functional starting from features extracted through texture analysis on MRI images acquired through a protocol including a coronal T2-weighted Turbo Spin Echo sequence. The boosting of J48, a multinomial logistic regression and K nearest neighbour are implemented employing Knime analytics platform. Excluding J48 whose accuracy was 83.0%, multinomial logistic regression and K nearest neighbour achieved accuracies beyond 92.0% and the Area Under the Curve Receiving Characteristic Operator till 98.4%. Diagnosing correctly this delicate disease is crucial in order to achieve the best management as well as the most appropriate cure for patients. The novelty of this paper lies in proving the ability of the combination of radiomics and machine learning to pre-operatively predict tumoral behavior. Prior to this analysis it was believed that only blood tests or histopathological analysis could provide this information.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Pituitary adenomas are rare intracranial tumors, presenting with a prevalence of 1/1500 in the general population. In most cases, they are benign lesions, whose clinical manifestations are related to mass effect signs - depending on tumor size and/or to hormone hypersecretion syndromes. On the other hand, low dimension intrasellar adenomas may be clinically silent and their diagnosis often comes as an incidental finding on MR scans [1,2,3,4,5].
Radiomics, consisting in the conversion of images into mineable data and subsequent analysis for decision support, is an emerging field allowing tumor classification [6]. In particular, texture analysis is a postprocessing technique for quantitative parameter extraction from pixel grey level heterogeneity. It consists of statistical analysis based on both simple intensity value distribution histograms and more complex gray level distribution matrix analyses which also retain information on spatial distribution of voxel intensities [7].
In this setting, machine learning can be applied in order to predict the outcome of patients and help clinicians in decision-making [8,9,10,11]. There is a wide range of applications of machine learning in different areas of medicine, from cardiology to radiology [12, 13]. In particular, studies applying machine learning on texture analysis according to the “radiomic process” were described by Kumar et al. [14]: Zacharaki et al. classified brain tumor type and grade using MRI texture and shape through Linear Discriminant Analysis with Fisher’s discriminant rule, k-nearest neighbour (KNN), nonlinear Support Vector Machine (SVM) and employing leave one out cross-validation [15]; Juntu et al. differentiated benign from malignant soft‐tissue tumours in T1‐MRI images testing three classifiers (neural networks, decision tree and SVM) [16]; Romeo et al. characterized adrenal lesions on unenhanced MRI images [17]; finally, Stanzione and colleagues have recently demonstrated the potential of this approach in prostate cancer local staging [18]. Moreover, recent studies investigated the relevance of first and second order histogram features obtained by diffusion-weighted imaging magnetic resonance in differentiating functional from non-functional pituitary macro-adenoma through a classic statistical analysis [19].
Therefore, the aim of this study is to apply machine learning algorithms on parameters obtained by texture analysis on MRI images in order to distinguish functional from non-functional pituitary macroadenomas.
2 Materials and Methods
2.1 Subjects
We retrospectively reviewed data of 50 patients, who received so-called standard endoscopic endonasal approach for the removal of a pituitary adenoma, between January 2013 and December 2017, at the Division of Neurosurgery of the University of Naples ‘Federico II’ in Italy. All of them underwent preoperative MRI at our Institution prior to the surgical procedure. Demographic data, preoperative assessment - i.e. endocrinological and visual status and presenting signs - tumor features, prior treatments, surgical results and complications, were retrieved from our electronic database (Filemaker Pro 11 - File Maker Inc, Santa Clara, CA, USA).
2.2 MRI Acquisition and Texture Analysis
All exams were acquired on a 1.5-Tesla scanner (Gyroscan Intera, Philips, Eindhoven, The Netherlands). The imaging protocol always included a coronal T2-weighted Turbo Spin Echo sequence (TR/TE: 2600/89 ms, FOV: 180 × 180 mm; matrix: 288 × 288; thk: 3 mm) used for the following radiomic feature extraction.
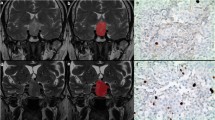
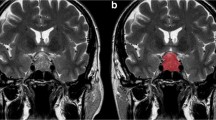
First of all, lesions were detected by an expert neuroradiologist who then performed their manual contouring by means of a bidimensional polygonal ROI after selection of the slice where it showed maximum extension (Fig. 1). Further editing with a brush tool was performed, when needed. This process was carried on using a freely available segmentation software (ITKSnap v3.6.0) [20].
Image pre-processing and feature extraction were performed on an open-source Python radiomics software (Pyradiomics v2.1.2) [21]. The first step consisted of image gray level normalization with a scale of 100). This step was mandatory since T2-weighted images are not quantitative and intensity values are not absolute in contrast to T2 maps. The latter were not available as only routine clinical scans were selected for the analysis, also in order to guarantee reproducibility of the results in the clinical setting. Subsequently, all volumes and corresponding lesion masks were resampled to a 2 × 2 × 2 mm voxel resolution. The next pre-processing step consisted of intensity value discretization. For this task, a fixed bin width approach was chosen, obtaining an ideal bin count between 16 and 128, as suggested in previous studies [22].
The use of wavelet decomposition, yielding all possible combinations of High and Low pass filtering in the x, y and z dimensions, and edge enhancement Laplacian of Gaussian (LoG) filters, emphasizing gray level change at different texture coarseness, allowed additional feature extraction from the derived images.
Finally, in relation to texture parameter extraction, together with bidimensional shape and first order statistics, we also obtained higher order class parameters. In detail, these were derived from the symmetrical Gray Level Co-occurrence Matrix (GLCM), Gray Level Size Zone Matrix (GLSZM), Gray Level Run Length Matrix (GLRLM, Neighboring Gray Tone Difference Matrix (NGTDM) and Gray Level Dependence Matrix (GLDM).
2.3 Tool
Knime analytics platform (v. 3.7.1) was chosen to conduct this machine learning study, as it is a well-known open source platform implementing a wide range of machine learning algorithms and integrated with Weka, Python and other software; moreover, it was already employed in literature for other studies [23, 24]. The algorithms used in this paper are briefly presented in the next section.
2.4 Algorithms and Evaluation Metrics
J48 is the Java implementation of a C4.5 decision tree [25], which consists of the evolution of the ID3 algorithm. It is an easy structure made up of leaves, representing classes, and nodes, representing test phases over an attribute. Multinomial Logistic Regression (MLR) with ridge estimator is applied through the “Logistic” node of Weka that follows the implementation of Le Cessie, and van Houwelingen [26, 27]. K Nearest Neighbour (KNN) is an easy instance-based classifier that assigns a label basing its choice on the dominance of a class in the nearest neighbours [28]. For all these algorithms, “smote” (Synthetic Minority Over-sampling Technique) was applied [29]. Smote generates artificial data by extrapolating between a real object of a given class and one of its nearest neighbours (of the same class). Boosting was implemented for J48, it converts weak learners into strong learners that predict with higher accuracy; it selects only the parameters that can improve the predictive ability of algorithms during the training phase, making the complexity in terms of dimension decrease and improving execution time [30]. The evaluation metrics employed in this study are:
-
Accuracy: correct classifications over the total;
-
Error: misclassifications over the total;
-
Recall: the ratio of positives correctly classified;
-
Precision: the ratio of positives correctly predicted in the positive class;
-
Sensitivity: capacity to detect true positives;
-
Specificity: capacity to detect true negatives.
Moreover, Area Under the Curve Receiving Characteristic Operator (AUCROC) was computed for each algorithm and for both bagging and boosting groups.
3 Results
Of the included lesions, 25 were functioning adenomas (5 Adreno Cortico Tropic Hormone, 8 Growth Hormone, 5 Growth Hormone/Prolactin, 6 Prolactin and 1 Thyroid-Stimulating Hormone secreting) and 25 non-functioning. A total of 1128 features was extracted from each patient.
Due to the small number of patients, smote technique was applied to make the number of records rise from 50 to 100. Then, a procedure of feature selection was applied to reduce the number of features extracted by the images: the matrix of correlation was computed among all variables and a threshold of correlation of 0.4 was chosen: all the variables with a correlation higher than the threshold were excluded because they did not add information to the classifiers. It allowed us to reduce the number of features from more than one thousand to 28. As the number of patients was not so high, leave one out was applied for all the implemented algorithms. J48, MLR and KNN were implemented together with the boosting node of Knime. Results are summarized in Table 1 while Table 2 shows the features used to build the models.
MLR obtained the highest accuracy, recall, precision, sensitivity, specificity and AUCROC among the three implemented algorithms. Despite getting the lowest accuracy (83.0%), J48 reached an AUCROC comparable to the KNN’s one.
4 Discussion
First, the MRI acquisition of 50 patients was performed at the department of Advanced Biomedical Sciences of the University Hospital “Federico II” of Naples. Furthermore, a texture analysis was conducted to extract more than one thousand quantitative features from the MRI images. The machine learning analysis was finally performed in order to carry out some evaluation metrics as regards the algorithms.
Mentioning other studies that employed radiomics and machine learning, Romeo et al. [17] characterized adrenal lesions with a diagnostic accuracy of 80%, while Juntu et al. [16] distinguished benign from malignant tumors with an accuracy of 93%; Zacharaki et al. [15] obtained 85% of accuracy classifying type and grade of brain tumours. Although a direct comparison with other studies would not be completely fair (due to the use of different datasets), this study shows greater capacity to correctly make classifications (functional and non-functional pituitary macroadenomas), exploiting features extracted through texture analysis. A comparison may be done with the study of Sanei et al. [19] who distinguished functional from non-functional pituitary macroadenomas with lower scores than those obtained through a machine learning analysis.
The functional status of pituitary lesions has a significant influence on the clinical manifestations of disease: the correct diagnosis and management is crucial for the selection of the correct therapeutic strategy and therefore cure this multifaceted disease. Although a previous study has shown the promise of Apparent Diffusion Coefficient values of pituitary lesions in this assessment [20], Diffusion Weighted Imaging is not routinely performed in the imaging of the sellar region. It is known that this area is potentially more prone to artefacts on echo-planar imaging and this technique is time consuming. For these reasons, an approach that obtains similar results while employing routine MRI sequences has more potential for its application in the current clinical setting.
Of course, this study is affected by some limitations: the dataset was augmented with artificial data in order to improve its size allowing us to perform the analysis. Major dataset could be studied, and machine learning analysis could be performed to reach 100% accuracy in this classification. Nevertheless, machine learning proved to be the best way to distinguish functional from non-functional pituitary macroadenomas using texture analysis on MRI images.
This paper proved that the combination of radiomics and machine learning can be used to predict tumoral behaviour pre-operatively while only blood tests or histopathological analysis were known as providers of this information.
References
Gittleman, H., Ostrom, Q.T., Farah, P.D., et al.: Descriptive epidemiology of pituitary tumors in the United States, 2004–2009. J. Neurosurg. 121(3), 527–535 (2014)
Ostrom, Q.T., Gittleman, H., Farah, P., et al.: CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15(Suppl 2), ii1-56 (2013)
Thapar, K., Laws, E.R.J.: Pituitary tumors. In: Kaye, A.W., Jr. LER, (eds.) Brain Tumors 2001, pp. 804–854. Churchill Livingstone, London (2001)
Wilson, P.J., Omay, S.B., Kacker, A., et al.: Endonasal endoscopic pituitary surgery in the elderly. J. Neurosurg. 128, 1–8 (2017)
Solari, D., Cavallo, L.M., Cappabianca, P.: Surgical approach to pituitary tumors. Handb. Clin. Neurol. 124, 291–301 (2014)
Gillies, R.J., Kinahan, P.E., Hricak, H.: Radiomics: images are more than pictures, they are data. Radiology 278(2), 563–577 (2015). https://doi.org/10.1148/radiol.2015151169
Lubner, M.G., Smith, A.D., Sandrasegaran, K., et al.: CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 37(5), 1483–1503 (2017). https://doi.org/10.1148/rg.2017170056
Malik, M.M., Abdallah, S., Ala’raj, M.: Data mining and predictive analytics applications for the delivery of healthcare services: a systematic literature review. Ann. Oper. Res. 270(1–2), 287–312 (2018)
Santini, S., Pescapè, A., Valente, A., et al.: Using fuzzy logic for improving clinical daily-care of β-thalassemia patients. In: 2017 IEEE International Conference on Fuzzy Systems (FUZZ-IEEE), pp. 1–6. IEEE (2017).. https://doi.org/10.1109/fuzz-ieee.2017.8015545
Improta, G., Russo, M.A., Triassi, M., et al.: Use of the AHP methodology in system dynamics: modelling and simulation for health technology assessments to determine the correct prosthesis choice for hernia diseases. Math. Biosci. 299, 19–27 (2018). https://doi.org/10.1016/j.mbs.2018.03.004
Romano, M., D’Addio, G., Clemente, F., et al.: Symbolic dynamic and frequency analysis in foetal monitoring. In: 2014 IEEE International Symposium on Medical Measurements and Applications (MeMeA), pp. 1–5. IEEE (2014). https://doi.org/10.1109/memea.2014.6860122
Johnson, K.W., Soto, J.T., Glicksberg, B.S., et al.: Artificial intelligence in cardiology. J. Am. Coll. Cardiol. 71(23), 2668–2679 (2018)
Choy, G., Khalilzadeh, O., Michalski, M., et al.: Cur-rent applications and future impact of machine learning in radiology. Radiology 288(2), 318–328 (2018)
Kumar, V., Gu, Y., Basu, S., et al.: Radiomics: the process and the challenges. Magn. Reson. Imaging 30(9), 1234–1248 (2012). https://doi.org/10.1016/j.mri.2012.06.010
Zacharaki, E.I., Wang, S., Chawla, S., et al.: Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 62(6), 1609–1618 (2009)
Juntu, J., Sijbers, J., De Backer, S., et al.: Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J. Magn. Reson. Imaging 31(3), 680–689 (2010)
Romeo, V., Maurea, S., Cuocolo, R., et al.: Characterization of adrenal lesions on unenhanced MRI using texture analysis: a machine-learning approach. J. Magn. Reson. Imaging 48(1), 198–204 (2018)
Stanzione, A., Cuocolo, R., Cocozza, S., et al.: Detection of extraprostatic extension of cancer on biparametric MRI combining texture analysis and machine learning: preliminary results. Acad. Radiol. (2019). https://doi.org/10.1016/j.acra.2018.12.025
Sanei, M.T., Kimia, F., Mehrnahad, M., et al.: Accuracy of diffusion-weighted imaging-magnetic resonance in differentiating functional from non-functional pituitary macro-adenoma and classification of tumor consistency. Neuroradiol. J. 32, 74–85 (2018). https://doi.org/10.1177/1971400918809825
Yushkevich, P.A., Piven, J., Hazlett, H.C., et al.: User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3), 1116–1128 (2006). https://doi.org/10.1016/j.neuroimage.2006.01.015
Van Griethuysen, J.J., Fedorov, A., Parmar, C., et al.: Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77(21), e104–e107 (2017). https://doi.org/10.1158/0008-5472.CAN-17-0339
Leijenaar, R.T., Nalbantov, G., Carvalho, S., et al.: The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci. Rep. 5, 11075 (2015). https://doi.org/10.1038/srep11075
Mannarino, T., Assante, R., Ricciardi, C., et al.: Head-to-head comparison of diagnostic accuracy of stress-only myocardial perfusion imaging with conventional and cadmium-zinc telluride single-photon emission computed tomography in women with suspected coronary artery disease. J. Nucl. Cardiol. (2019). https://doi.org/10.1007/s12350-019-01789-7
Romeo, V., Ricciardi, C., Cuocolo, R., et al.: Machine learning analysis of MRI-derived texture features to predict placenta accreta spectrum in patients with placenta previa. Magnetic Resonance Imaging (2019). https://doi.org/10.1016/j.mri.2019.05.017. ISSN 0730-725X
Quinlan, J.R.: Bagging, boosting, and C4. 5. In: AAAI/IAAI, vol. 1, pp. 725–730, August 1996
le Cessie, S., van Houwelingen, J.C.: Ridge estimators in logistic regression. Appl. Stat. 41(1), 191–201 (1991)
Aha, D.W., Kibler, D., Albert, M.K.: Instance-based learning algorithms. Mach. Learn. 6(1), 37–66 (1992)
Keller, J.M., Gray, M.R., Givens, J.A.: A fuzzy k-nearest neighbor algorithm. IEEE Trans. Syst. Man, Cybern. 4, 580–585 (1985)
Chawla, N., Bowyer, K., Hall, L., et al.: SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. (JAIR) 16, 321–357 (2002). https://doi.org/10.1613/jair.953
Freund, Y., Schapire, R.E.: A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55(1), 119–139 (1997)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
The authors have no conflict of interests.
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Carlo, R. et al. (2020). Distinguishing Functional from Non-functional Pituitary Macroadenomas with a Machine Learning Analysis. In: Henriques, J., Neves, N., de Carvalho, P. (eds) XV Mediterranean Conference on Medical and Biological Engineering and Computing – MEDICON 2019. MEDICON 2019. IFMBE Proceedings, vol 76. Springer, Cham. https://doi.org/10.1007/978-3-030-31635-8_221
Download citation
DOI: https://doi.org/10.1007/978-3-030-31635-8_221
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31634-1
Online ISBN: 978-3-030-31635-8
eBook Packages: EngineeringEngineering (R0)