Abstract
Male infertility is a multifactorial and heterogeneous pathological condition affecting 7% of the general male population. The genetic landscape of male infertility is highly complex as semen and testis histological phenotypes are extremely heterogeneous, and at least 2000 genes are predicted to be involved in spermatogenesis. Genetic factors have been described in each etiological category of male reproductive impairment: (1) hypothalamic–pituitary axis dysfunction; (2) quantitative and qualitative alterations of spermatogenesis; (3) ductal obstruction/dysfunction. In 25% of azoospermic and in 10% of oligozoospermic men, a genetic anomaly can be diagnosed with the current genetic testing. However, up to now, only a relatively low number of monogenic factors have a clear-cut cause–effect relationship with impaired reproductive function. Thanks to the widespread diffusion of Next-Generation Sequencing, a continuously increasing number of monogenic causes of male infertility are being discovered and their validation is currently ongoing. The identification of genetic factors is of outmost clinical importance since there is a risk of transmission of genetic defects through natural or assisted reproductive techniques. The benefit of the genetic diagnosis of infertility has an obvious clinical significance for the patient itself with implications not only for his reproductive health but in many instances also for his general health.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Male infertility
- Spermatogenesis
- Genetics
- Gene
- Hypogonadism
- Azoospermia
- Oligozoospermia
- Teratozoospermia
- NGS
- Exome
1 Introduction
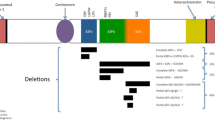
Infertility affects about 14% of the couples in the general population and globally male factor is contributing to it for about 50% of cases. In about 95% of cases, male factor implies quantitative or qualitative alterations of sperm parameters while in about 5% of cases it is related to semen deposition in vagina (aspermia, erectile dysfunction, retrograde ejaculation, etc.). Male infertility is a multifactorial complex pathological condition with highly heterogeneous phenotypic representations. Recently, male reproductive dysfunction has been classified into four etiologic categories: (1) hypothalamic–pituitary axis dysfunction; (2) quantitative alterations of spermatogenesis; (3) qualitative alterations of spermatogenesis; and (4) ductal obstruction/dysfunction (Tournaye et al. 2016). Genetic factors play an important role in each of these categories, with the highest prevalence in the severest form of quantitative alterations, i.e., azoospermia. In fact, karyotype (numerical and structural chromosomal anomalies) and Y chromosome microdeletions are found in about 15% of men affected by severe male factor infertility. In sharp contrast with the incidence of chromosomal anomalies, known monogenic alterations are relatively rare and their screening is restricted to congenital hypogonadotropic hypogonadism (CHH), absence of vas deferens, mild androgen insensitivity, and monomorphic terato/asthenozoospermia. In about 40% of quantitative disturbances of spermatogenesis, the etiology remains unknown and we refer to them as idiopathic infertility. Genetic factors are likely to play an important role in idiopathic testicular impairment/failure. A growing number of genes have been reported in idiopathic azoospermia/oligozoospermia but at the moment only a few of them have been validated by more than one study (Fig. 16.1). In this chapter, we are going to provide a detailed description of those genetic factors that are already included in the diagnostic workup of infertile men. In addition, a brief overview is given on genes with potential clinical interest.
Genotype–phenotype correlations, gene mutations versus semen phenotype. Genes in red with diagnostic value; genes in black with potential clinical value; cHH congenital Hypogonadotropic Hypogonadism, the complete list of genes is presented in Table 16.1. Asterisk mutations associated with unilateral congenital absence of vas deferens
2 Monogenic Causes with Diagnostic Value in the Four Etiologic Categories of Male Infertility
2.1 Genetic Causes of Hypothalamic–Pituitary Axis Dysfunction
A total of 35 candidate genes have been described in the literature to date (Boehm et al. 2015; Tournaye et al. 2016) with congenital hypogonadotropic hypogonadism (CHH) (Table 16.1). CHH is a rare, complex genetic disease (incidence of 1 in 8000 men) with variable expressivity, penetrance, and inheritance (Boehm et al. 2015). The classical phenotype of CHH is absent or delayed puberty, eunuchoid habitus, sparse or absent body hair, gynecomastia, cryptorchidism, micropenis, and very low testicular volume (<5 ml). However, in some cases, reduced spermatogenesis and mild hypoandrogenism are the only symptoms, resulting in a delayed CHH diagnosis after puberty. CHH can manifest itself with anosmia or hyposmia (Kallmann syndrome; KS) or as normosmic, isolated hypogonadotropic hypogonadism (Boehm et al. 2015). To note, KS can be associated with other developmental anomalies such as cleft lip or palate, dental agenesis, ear anomalies, congenital hearing impairment, renal agenesis, bimanual synkinesis, or skeletal anomalies (Boehm et al. 2015), whereas in normosmic CHH nonreproductive defects are absent. Some of the genes associated with CHH are also involved in different syndromic diseases (such as Gordon Holmes syndrome, CHARGE syndrome, and Waardenburg syndrome) (Boehm et al. 2015). Interestingly enough, “reversibility” of the gonadotropin deficiency after testosterone therapy has been described in about 10–15% of patients affected by KS or normosmic CHH (Quinton et al. 1999; Ribeiro et al. 2007; Raivio et al. 2007; Dwyer et al. 2016).
The 35 CHH genes are implicated either in the development/migration of the GnRH neurons or in the neuroendocrine regulation of GnRH secretion or action (Boehm et al. 2015). CHH presents a number of peculiar features from a genetic point of view: (1) in some cases the same gene (i.e., FGFR1, PROKR2) may cause both KS and normosmic CHH, implying that from a genetic point of view a clear distinction between the two clinical entities cannot be established; (2) it does not follow the rules of Mendelian inheritance since in about 20% of cases there is a digenic/oligogenic inheritance, i.e., two heterozygous mutations in two or more candidate genes.
2.1.1 Testing and Genetic Counseling
The indication for testing is restricted to patients with confirmed CHH after the exclusion of all secondary forms (pituitary tumors, empty sella, etc.). Currently, genetic testing is based on Next-Generation Sequencing (NGS) gene panel, which is able to provide the diagnosis in about 40% of cases (Boehm et al. 2015; Tournaye et al. 2016). Novel genes associated with CHH are expected to be discovered by whole-exome sequencing (WES) analysis in the near future.
Since in about 80% of CHH patients spermatogenesis can be induced by the administration of gonadotropins (Dwyer et al. 2015), gene mutations can be transmitted either spontaneously or by assisted reproductive techniques. Overall, the complexity of this disease (variable expressivity, penetrance, and inheritance pattern) makes predicting the exact health consequences for the offspring difficult. For this reason, the Preimplantation Genetic Diagnosis (PGD) or prenatal diagnosis should be offered to couples mainly for syndromic cases and for those cases where the gene mutation shows a clear-cut cause–effect relationship. A periodic suspension of the hormonal replacement therapy is recommended to all CHH patients in order to assess a potential recovery of the hypothalamic–pituitary axis. To note, genetic testing does not help in identifying patients with higher probability of “reversal,” since this condition has been described in association with mutations in different CHH candidates (Dwyer et al. 2016).
2.2 Genetic Causes of Quantitative Alterations of Spermatogenesis
2.2.1 AR (Androgen Receptor Gene)
AR (OMIM: 313700) is the only gene that is included in the diagnostic testing of specific cases of male infertility. Mutations in AR gene are associated with Androgen Insensitivity Syndrome (AIS) characterized by resistance to circulating testosterone. AIS is the most frequent cause of Disorders of Sexual Development (DSD) and based on the residual degree of functional capacity of the mutated AR the clinical phenotype can be divided into three categories: (1) Complete Androgen Insensitivity (CAIS; Morris syndrome) leading to a female phenotype in 46,XY individuals; (2) Partial Androgen Insensitivity (PAIS; Reifenstein syndrome) characterized by undervirilized male phenotype with ambiguous genitalia; (3) Mild Androgen Insensitivity (MAIS) associated with impaired sperm production in the presence of normal male genitalia (Krausz and Chianese 2014). Using conventional and Cre/Lox conditional Ar-null male mice recreated human disorders (Yeh et al. 2002). They showed female external sex development and testis atrophy with spermatocyte-stage arrest, resembling AIS human pathology. Sertoli cell-selective KO of the AR causes spermatocytic arrest, indicating an important role for intratesticular testosterone in meiosis (De Gendt et al. 2004).
The AR gene is situated on the X chromosome (Xq11–12) and contains eight exons that encode a protein of 920 amino acid residues. The protein functions as a steroid hormone-activated transcription factor and contains three major functional domains: the N-terminal domain (NTD, transcriptional activation region encoded by exon 1), the DNA-binding domain (DBD, encoded by exons 2 and 3), and the ligand-binding domain (LBD, encoded by exon 4–8). More than 1000 AR mutations have been described so far (Gottlieb et al. 2012) and the large majority of them are missense mutations located in the AR-DBD or AR-LBD leading to impairment in DNA or AR binding, respectively.
The AR gene also contains two polymorphic sites in the N-terminal transactivation domain (exon1) of the receptor: a polyglutamine tract -(CAG)n- and a polyglycine tract (GGC)n, which were the subject of many publications related to male infertility (Davis-Dao et al. 2007). The most extensively studied polymorphism concerns the trinucleotide CAG. Based on in vitro studies, it has been hypothesized that carriers of longer CAG repeat have a higher risk for infertility and cryptorchidism due to impaired androgen effect (Gao et al. 1996; Davis-Dao et al. 2007). This hypothesis has been challenged by novel functional and observational studies reporting that both a longer CAG tract and a shorter CAG tract might have a negative effect on the receptor function; hence, an optimal number of CAG repeats are necessary for the highest transcription (Nenonen et al. 2011; Davis-Dao et al. 2012). We can speculate that the optimum range may vary between the genomic and non-genomic actions, and also in different tissues, because the effect of polyQ repeat on transactivation is cell-specific, presumably due to distinct profiles of co-regulator proteins (Krausz 2012).
2.2.1.1 Testing and Genetic Counseling
Given the variable clinical phenotypes, indications are different for each type of AIS. CAIS is suspected in case of a 46,XY woman with primary amenorrhea, normal breast development and pubertal growth, reduced or absent sexual hair, and absent female internal genitalia. Clinical management is complex and involves a multidisciplinary approach including psychologists, endocrinologists, urologists, and gynecologists. Because of malignancy risk, gonads, usually located in the abdomen or inguinal canal, are commonly removed, requiring subsequent estrogen replacement to maintain feminization. In case of PAIS, the phenotype is highly dependent on the degree of residual AR function, ranging from male-appearing genitalia to severe undermasculinization resembling female genitalia (Mongan et al. 2015). Management of severe forms of PAIS, including gender assignment, is rather complex. The hormone profile of AIS is typically represented by high Androgen Sensitivity Index (ASI), calculated as the product of serum testosterone x serum luteinizing hormone, i.e., high LH with relatively high testosterone levels. In hypoandrogenized infertile men with high ASI, AR testing is indicted. However, a routine screening to all infertile men is not advised, since the frequency of AR mutations in unselected infertile men varies from 0–1.7% (Ferlin et al. 2006; Rajender et al. 2007). The frequency of AR mutations in PAIS is 41%, whereas no official estimate is given in the available mutation databases for the MAIS phenotype (Gottlieb et al. 2012). The role of CAG repeats in male infertility is probably more complex than it has been previously proposed; there are still important unanswered questions such as: what range of AR CAG repeat lengths predisposes to impaired sperm production and what risk of infertility is associated with each length (Davis-Dao et al. 2007). These open questions limit the clinical use of (CAG)n testing.
2.2.2 Y Chromosome Linked Male Infertility
The Y chromosome contains genes essential for testis development and function, such as the genes residing in the azoospermia factor (AZF) regions and the master gene for testis determination (SRY; OMIM:480000). Y chromosome microdeletions, removing the entire AZF regions (complete deletions), are one of the leading causes of spermatogenic failure and the screening for AZF deletions became part of the routine diagnostic workup of men with severe oligozoospermia/azoospermia (Krausz et al. 2014). A peculiar feature of the boundary of the AZF regions is the presence of repeated homologous sequences that are predisposed to deletion or duplication through a mechanism called nonallelic homologous recombination (NAHR). These deletions remove more than one gene in block; hence, they will not be further discussed in this chapter (for review see Krausz and Casamonti 2017). The only monogenic Y-chromosome-linked cause of male infertility concerns the SRY gene. SRY encodes the critical testis-determining transcription factor that activates a number of downstream transcription factors involved in testes formation. The gene is located below the pseudoautosomal region (PAR) of the short arm of the Y chromosome, and the erroneous translocation can occur during meiosis when the two sex chromosomes recombine between their PAR regions (Wu et al. 2014). This translocation leads to the 46,XX male syndrome (also known as de la Chapelle syndrome). This syndrome has a frequency of 1 in 20,000 children according to Genetics Home Reference (https://ghr.nlm.nih.gov/). Men with 46,XX male syndrome have smaller stature and a higher incidence of maldescended testes and gynecomastia, and are azoospermic with no exceptions (Vorona et al. 2007).
2.2.2.1 Testing and Genetic Counseling
Testicular sperm extraction (TESE) is not advised in XX male patients owing to the lack of Y chromosome-linked azoospermia factor (AZF) genes, meaning focal sperm production in the testis is not possible (Skaletsky et al. 2003). These patients can have hypoandrogenism, so a careful endocrine assessment (including analysis of FSH, LH, and testosterone levels) and follow-up monitoring of testosterone level are advised.
2.3 Genetic Causes of Qualitative Alterations of Spermatogenesis
2.3.1 DPY19L2 (Dpy-19 Like 2 Gene)
DPY19L2 (OMIM: 613893) is the only gene included in the routine genetic diagnostic workup of globozoospermia. Globozoospermia is very rare, affecting 0.1% of infertile men, and is characterized by the production of round-headed, acrosome-less spermatozoa that are unable to fertilize the oocyte, as no acrosome reaction can occur (Fig. 16.2a). In mouse models, globozoospermia has been observed as a consequence of >50 different gene mutations (Coutton et al. 2015), but in humans, only mutations in the DPY19L2 gene have been validated to be associated with this disorder. In fact, mutations in DPY19L2 have been found in 60–80% of globozoospermic patients. DPY19L2 is located on chromosome 12 and encodes a protein required during spermatogenesis for sperm head elongation and acrosome formation. The most frequent mutation is the complete deletion of the gene, caused by a similar mechanism to that observed for AZF deletions. DPY19L2 is located in a region flanked by two 28 kb segmental duplications, which predisposes it to NAHR. The complete deletion of DPY19L2 accounts for 80.4% of instances of DPY19L2-related globozoospermia, whereas the remaining instances are caused by intragenic deletions and point mutations (homozygous and compound heterozygous) (Ray et al. 2017).
2.3.1.1 Testing and Genetic Counseling
Mutations are mainly identified in patients with 100% globozoospermia; thus, genetic analysis should be restricted to this circumstance only. Given the high frequency of complete gene deletion, genetic testing can be easily performed using real-time quantitative PCR (Chianese et al. 2015) followed by breakpoint definition and mutation screening. Since DPY19L2 deletions are not exceptionally rare in the general population (heterozygous carriers 1:85), screening in the female partners of male carriers prior intracytoplasmic sperm injection (ICSI) should be performed. The lack of phospholipase C-ζ, an acrosome phospholipase, is responsible for the absence of oocyte activation. Consequently, artificial oocyte activation (AOA) has been proposed as an option for patients with complete globozoospermia undergoing ICSI. However, the safety of AOA has been questioned as continued increases in intracellular calcium concentration can affect downstream molecular events, and it should be restricted to selected cases of 100% globozoospermia in which finding spermatozoa with residual acrosome is impossible (Kuentz et al. 2013).
2.3.2 AURKC (Aurora Kinase C)
To date, AURKC (OMIM: 603495) gene mutations are the only validated genetic causes macrozoospermia. Macrozoospermia, also known as sperm macrocephaly, affects <1% of the male population and it was reported for the first time in 1977 by Nistal and colleagues (1977). This qualitative disturbance is characterized by a high percentage of spermatozoa with large, irregular heads and multiple flagella (Fig. 16.2b). AURKC gene is located in chromosome 19 and encodes for a component of the chromosomal passenger complex (CPC) in meiotic cells and is essential for correct meiotic chromosomal segregation and cytokinesis (Dieterich et al. 2007). The AURKC mutations are associated with alterations of meiotic divisions leading to tetraploid spermatozoa. The most common mutation is the deletion of a cytosine in the exon 3 (c.144delC), observed in more than 85% of patients affected by macrozoospermia (Ray et al. 2017). Interestingly enough, the mutation is relatively common in heterozygosis in the Maghrebian population (1/50 men) and it has been proposed that heterozygote carriers may have a selective advantage due to a more relaxed meiotic checkpoint (Ben Khelifa et al. 2012). In Europeans, a recurrent stop gain mutation in exon 6 (p.Y248∗) has been described (Ben Khelifa et al. 2012).
2.3.2.1 Testing and Genetic Counseling
All men with macrozoospermia should be tested for AURKC mutations before undergoing Assisted Reproductive Techniques (ART). After genetic testing, two different scenarios can occur: identification of homozygous or compound heterozygous mutations or an absence of mutations in this gene. In the first scenario, ICSI is not advised even after motile sperm organelle morphology examination, as all spermatozoa are polyploid (and are mostly tetraploid) and, therefore, normal embryonic development is not possible. By contrast, ART is not contraindicated in patients without mutations, but sperm FISH should be performed to evaluate the proportion of euploid sperm; hence, the likelihood of success. PGD can be proposed to those with intermediate rate of aneuploid spermatozoa.
2.3.3 DNAH1 (Dynein Axonemal Heavy Chain 1)
DNAH1 gene (OMIM: 603332) mutations seems to be the major cause of Multiple Morphological Abnormalities of the sperm flagella (MMAF) (Ben Khelifa et al. 2014; Amiri-Yekta et al. 2016; Wang et al. 2017; Sha et al. 2017a; Tang et al. 2017; Coutton et al. 2018). MMAF, previously reported as dysplasia of fibrous sheath (DFS), is a rare disease defined as an asthenoteratozoospermia resulting from a mosaic of morphological abnormalities concerning the sperm flagella, including absent, coiled, bent, angulated, irregular, or short flagella (Ben Khelifa et al. 2014). In addition, lack of central microtubules and/or dynein arms may also be observed by transmission electron microscopy (TEM) in the sperm flagella of the affected subjects (Ben Khelifa et al. 2014). The incidence of MMAF has not already been investigated precisely. DNAH1 is located on chromosome 3 and encodes an axonemal inner dynein arm heavy chain and when it is absent, the axoneme is grossly disorganized, often lacking the central pair (9 + 0 structure). Biallelic DNAH1 mutations seem to be responsible for 30% of MMAF patients (Coutton et al. 2018).
2.3.3.1 Testing and Genetic Counseling
The screening for DNAH1 mutations is recommended in patients affected by severe to complete asthenozoospermia due to sperm flagellar alterations. Flagellar abnormalities have been reported to be associated with an elevated frequency of aneuploidies and a poor ICSI outcome (Lewis-Jones et al. 2003; Baccetti et al. 2005; Collodel and Moretti 2006; Ghedir et al. 2014). However, patients with MMAF with mutated DNHA1 showed low aneuploidy rate and normal sperm DNA integrity, indicating that not all patients with MMAF are at risk of chromosomal anomalies (Wambergue et al. 2016).
In 2015, a homozygous mutation in DNAH1 was observed in two sisters affected by Primary ciliary dyskinesia (PCD) (Imtiaz et al. 2015), which is a disorder characterized by chronic respiratory tract infections, abnormally positioned internal organs, and infertility. This observation has prompted the novel hypothesis of a ‘phenotypic continuum’ ranging from infertile patients with PCD to patients with MMAF with no or mild PCD manifestations (Ray et al. 2017). Given the multitude of genes involved in ciliagenesis and function, MMAF could be a phenotypic variant of the classical form of PCD, and mutations affecting sperm flagella could be compensated for by other genes involved in other ciliated tissues. Therefore, it is still unclear the exact health consequences for the offspring and whether the female partner should be screened for DNAH1 mutations.
2.3.4 SUN5 (Sad1 and UNC84 Domain Containing 5 Gene)
The phenotype of the SUN5-mutated patients is characterized by acephalic spermatozoa with a variable but low proportion of abnormal head–tail junctions and tailless heads (Shang et al. 2017) (Fig. 16.2c). This sperm defect is due to the failure of centriole-tail attachment to the spermatid nucleus during the last phase of spermatogenesis. SUN5 (OMIM: 613942) encodes a testis-specific protein localized in the neck region of spermatids. The disease is extremely rare and it is transmitted through recessive inheritance. Homozygous and compound heterozygous mutations were reported by four different authors (Zhu et al. 2016; Elkhatib et al. 2017; Shang et al. 2017; Sha et al. 2018b).
2.3.4.1 Testing and Genetic Counseling
Patients affected by acephalic spermatozoa should be screened for SUN5 mutations. The only option for a biological paternity is ICSI through the selection of tailless sperm heads. The majority of papers report no pregnancy despite the presence of fertilized eggs. In five articles, nine couples obtained pregnancy after repeated ICSI attempts (Kamal et al. 1999; Porcu et al. 2003; Emery et al. 2004; Gambera et al. 2010; Shang et al. 2017).
2.4 Genetic Causes of Ductal Obstruction
2.4.1 CFTR (Cystic Fibrosis Transmembrane Conductance Regulator Gene)
Mutations in CFTR (OMIM: 602421) have been largely described in patients affected by Congenital Absence of Vas Deferens (CAVD). The CAVD may occur either as an isolated reproductive disorder or as an atypical symptom of Cystic Fibrosis, and accounts for up to 25% of patients with Obstructive Azoospermia (OA) (Oates and Amos 1994). It may affect one (CUAVD) or both vas deferens (CBAVD). The CUAVD is a rare condition associated with either oligo/or normozoospermia. In contrast, CBAVD associated with agenesis of seminal vesicles is characterized by typical semen alterations, such as low semen volume (<1 ml) with an acid pH (<7) and absence of spermatozoa. The CFTR gene is located on chromosome 7q31.2, contains 27 exons (Kerem et al. 1989; Riordan et al. 1989), and encodes a protein involved in chloride conduction across epithelial cell membranes. To date, more than 2000 variants have been identified in CFTR gene (http://www.genet.sickkids.on.ca/Home.html) and they are categorized in severe and mild mutations depending on their functional consequences. Although geographical and ethnic differences have been demonstrated in CFTR mutations, the most common mutations in CBAVD patients are F508del, 5T, and R117H (Yu et al. 2012). 5T is the shortest allele of IVS8-(T)n, which is a length variant of a polypyrimidine tract at the splice acceptor site of intron 8 of the CFTR gene. The length of the T tract (IVS8-5T, IVS8-7T, IVS8-9T) affects the splicing efficiency of exon 9 and thus the amount of normal CFTR mRNA. The phenotypic penetrance of 5 T allele depends on the length of adjacent TG repeats (12 or 13) and the M470 V missense mutation in exon 10, i.e., the 12TG-5T-V470 haplotype increases the risk of having CBAVD (de Meeus et al. 1998; Du et al. 2014).
2.4.1.1 Testing and Genetic Counseling
The screening for CFTR gene mutation is recommended in subjects with CAVD without renal agenesis (Jungwirth et al. 2012). In fact, subjects with CAVD and renal agenesis (in the majority of cases of unilateral agenesis of vas deferens) are considered to have different genetic basis, which may be attributed to defect of mesonephric duct development in the embryo (McCallum et al. 2001). This fact implies that all patients affected by CAVD should undergo an ultrasound scan of the pelvic region prior to genetic testing. Routine screening for CTFR variants is based on a panel of mutations (30–50 mutations) that are the most common for a given ethnic population (de Souza et al. 2017). In instances in which the two mutations are not identified using this panel (as CAVD is a recessive disease), the whole CFTR gene is subjected to sequencing in order to search for the second mutation. In those patients in whom pathogenic variants have been identified, genetic counseling is mandatory since patients with CBAVD are assumed to have normal testicular function, and they can undergo TESE–ICSI (as azoospermia is caused by obstruction) and generate their own biological children. Given that the carrier frequency of CFTR mutations in people of European descent is high (1 in 25), screening of the partner is mandatory in order to evaluate the risk of giving birth to a child affected by cystic fibrosis. If both parents are carriers, prenatal or PGD should be undertaken.
3 Additional Monogenic Causes of Male Infertility with Potential Clinical Interest
The diffusion of NGS platforms is allowing the identification of a number of genes involved in various semen phenotypes. However, only few of them have been validated by more than one independent study on a relatively high number of subjects (Table 16.2). In the following paragraphs we briefly describe those genes that were validated by more than one study and are potential candidates for diagnostic testing in the future.
3.1 Validated Candidate Genes Involved in Quantitative Alterations of Spermatogenesis
A total of six genes leading quantitative impairment of spermatogenesis have been validated by more than one independent study: NR5A1 (OMIM: 184757), TEX11 (OMIM: 300311), TEX14 (OMIM: 605792), TEX15 (OMIM: 605795), FANCM (OMIM: 609644), and XRCC2 (OMIM: 600375). With the exception of TEX11, which is X-linked, the remaining genes are mapping to autosomes. Apart from NR5A1, which follows the Autosomal Dominant inheritance pattern, only recessive mutations lead to quantitative alterations of spermatogenesis.
NR5A1 encodes steroidogenic factor 1 (SF1), crucial in male and female gonadal development and steroidogenesis. Heterozygosis mutations in NR5A1 have been associated with a variety of phenotypes ranging from primary adrenal insufficiency (AI) and complete 46,XY gonadal dysgenesis (Schimmer and White 2010; Ferraz-de-Souza et al. 2011) to 46,XY DSD including bilateral anorchia (Philibert et al. 2007; Brauner et al. 2011), hypospadias (Allali et al. 2011; Brandt et al. 2013), and hypogonadotropic hypogonadism (Hu et al. 2012).
Heterozygous mutations have also been reported in patients affected by severe oligozoospermia or azoospermia (Bashamboo et al. 2010; Ropke et al. 2013; Zare-Abdollahi et al. 2015; Ferlin et al. 2015; Tuttelmann et al. 2018) with a frequency ranging from 0.6% (Ropke et al. 2013) to 2.5% (Tuttelmann et al. 2018).
Concerning TEX11, TEX14, and TEX15, they belong to the family of Testis Expressed genes and, as their name indicates, they are over- or specifically expressed in the testis. While TEX15 is not associated with a clear-cut semen phenotype, since it has been found mutated both in patients with NOA and crypto/oligozoospermia (Okutman et al. 2015; Colombo et al. 2017; Wang et al. 2018), mutations in TEX11 and TEX14 are restricted to NOA (Yatsenko et al. 2015; Yang et al. 2015; Gershoni et al. 2017; Tuttelmann et al. 2018; Sha et al. 2018a; Fakhro et al. 2018). More precisely, TEX11 mutations seem to be more frequent in NOA men with testis histology of meiotic arrest (Yatsenko et al. 2015) and should be tested in cases of suspected meiotic arrest.
FANCA (OMIM:607139), FANCM, and XRCC2 are part of the Fanconi Anemia (FA) gene family. Proteins encoded by the FA gene family are involved in (DNA double strand breaks) DSB repair and are essential for mitosis and meiosis. By performing exome analysis, recessive FANCA mutations have been described in men affected by NOA (Sertoli Cell Only syndrome) and mild/borderline hematological alterations (Krausz et al. 2019). This finding underlies the importance of considering not only hormone dosage but also hematological parameters in the diagnostic workup of SCOS patients. Diagnosing occult FA is highly relevant since it is a medical condition that predisposes to specific FA-related cancers.
Mutations in FANCM are not associated with bone marrow failure and have been identified in both NOA and oligoasthenozoospermia (Kasak et al. 2018; Yin et al. 2019). Meiosis-specific mutations in the XRCC2 gene were reported in NOA patient with spermatocytic arrest (Yang et al. 2018; Zhang et al. 2019). Interestingly enough, mutations in this gene also cause Premature Ovarian Insufficiency (POI) in females (Zhang et al. 2019). Hence, the genetic counseling should involve not only male but also female relatives in case of XRCC2 mutations.
3.2 Validated Candidate Genes Involved in Qualitative Alterations of Spermatogenesis
Monomorhpic teratozoospermia defines a group of rare morphological anomalies, mainly globozoospermia, MMAF and sperm acephalia. The large majority of cases occur in consanguineous families since these are recessive diseases. In globozoospermia, besides the DPY19L2 gene (see paragragh on routine screening), mutations in SPATA16 (OMIM: 609856) have been reported in two independent studies (Elinati et al. 2016; Dam et al. 2007). This gene is located on chromosome 3 and encodes a testis-specific protein belonging to the tetratricopeptide repeat-like superfamily. The encoded protein localizes to the Golgi apparatus and is involved in the formation of sperm acrosome. Concerning the MMAF phenotype, besides DNAH1 mutations, homozygous, and compound heterozygous mutations in CFAP43 (OMIM: 617558), CFAP44 (OMIM: 617559), CFAP69 (OMIM: 617949), and WDR66 (OMIM: 618146) have been reported in more than one study (Sha et al. 2017b; Tang et al. 2017; Coutton et al. 2018; Dong et al. 2018; Auguste et al. 2018; Kherraf et al. 2018; He et al. 2019). These genes encode a family of proteins belonging to the cilia and flagella associated protein family (CFAP) and are necessary to produce functional flagella. Interestingly enough, CFAP43 and CFAP44, WDR66 encode WD-repeat proteins, which confirm the importance of these type of proteins in human diseases and especially in male infertility. Overall, mutations in these genes may explain up to 50% of cases of MMAF (Li et al. 2019). Finally, in relationship with acephalic spermatozoa two independent studies reported homozygous and compound heterozygous mutations in PMFBP1 (OMIM: 618085) (Zhu et al. 2018; Sha et al. 2019). PMFBP1 is localized at the head–tail coupling apparatus (HTCA) and cooperates with SUN5 and SPATA6 to connect sperm head to tail (Zhu et al. 2018). It has been postulated that mutations in SUN5 and PMFBP1 may explain 70% of cases of acephalic spermatozoa syndrome (Zhu et al. 2018); however, since mutations in PMFBP1 have been only found in patients from Asia, further validation in other ethnic populations is needed.
3.3 Validated Candidate Genes Involved in Ductal Obstruction
After a comprehensive analysis of the CFTR gene, in about 20% of cases the origin of CAVD remains unknown. Recently, the ADGRG2 gene (OMIM: 300572) has been identified as a new candidate gene in CBAVD in three independent studies (Patat et al. 2016; Yang et al. 2017; Khan et al. 2018). ADGRG2 is an X-linked gene encoding an adhesion-class G protein-coupled receptor and is highly expressed in the efferent ducts (Obermann et al. 2003). Adgrg2-mutant mice develop fluid accumulation in the testes ducts, leading to an obstructive infertility phenotype, which resembles that described in men with ADGRG2 mutation (Davies et al. 2004). Pathogenic ADGRG2 variants were reported, accounting for 11%–15% of the CBAVD patients who are CFTR-negative (Patat et al. 2016; Yang et al. 2017).
4 Conclusions
Although more than 2000 genes have been predicted to be involved in human spermatogenesis, there are relatively few monogenic mutations that have been conclusively demonstrated and validated to cause male infertility in humans. Consequently, the current diagnostic genetic testing is restricted to a relatively small set of genes. Genetic counseling of the couple is an absolute requirement prior to assisted reproduction and in some instances includes testing of the female partner and recommendation for PGD. The major breakthrough in the discovery of genetic factors involved in male infertility occurred more than 30 years ago with the identification of the Y-chromosome-linked AZF region deletions (Vogt et al. 1996), which implied the deletion in block of more than one gene. While candidate gene re-sequencing studies were relatively successful in uncovering the genetic basis of CHH, this approach did not lead to novel diagnostic tests in idiopathic oligo/azoospermia (Krausz and Riera-Escamilla 2018). Next-generation sequencing allowing the simultaneous analysis of several thousands of genes or the entire exome has contributed to major advances in the genetic diagnosis of monomorphic teratozoospermia, cHH, and in familial cases of idiopathic azoospermia. Exome studies are currently ongoing in quantitative impairment of spermatogenesis in large cohorts of sporadic patients. These studies have the potential to provide novel genetic diagnostic tests also in this category of patients. The clinical impact of discovering such factors became even more important in the era of in vitro fertilization, since these patients can now generate their own biological child through these techniques and the identification of transmissible genetic causes has relevance for their future children. Besides the consequences on reproductive health, an emerging issue is the possible genetic link between idiopathic impaired spermatogenesis and higher morbidity and mortality rates. This important topic is currently addressed by androgeneticists and will allow a more holistic clinical evaluation of our infertile patients.
Abbreviations
- AIS:
-
Androgen Insensitivity Syndrome
- AOA:
-
Artificial oocyte activation
- ART:
-
Assisted reproductive techniques
- AZF:
-
Azoospermia factor
- CAIS:
-
Complete Androgen Insensitivity
- CAVD:
-
Congentinal Absence of Vas Deferens
- CBAVD:
-
Congentinal Bilateral Absence of Vas Deferens
- CFAP:
-
Cilia and Flagella Associated Protein family
- CHARGE:
-
Coloboma, Heart defects, Atresia of choanae, Retardation of growth and/or development, Genital and/or urinary defects, Ear anomalies or deafness
- CHH:
-
Congenital hypogonadotropic hypogonadism
- CPC:
-
Chromosomal passenger complex
- CPHD:
-
Combined pituitary hormone deficiency
- CUAVD:
-
Congenital unilateral absence of vas deferens
- DBD:
-
DNA-binding domain
- DFS:
-
Dysplasia of fibrous sheath
- DNA-DSB:
-
DNA-Double Strand Breaks
- DSD:
-
Disorders of sexual development
- D-WS:
-
Dandy-Walker syndrome
- FA:
-
Fanconi anemia
- FISH:
-
Fluorescence in situ hybridization
- GHS:
-
Gordon Holmes syndrome
- HS:
-
Hartsfield syndrome
- HTCA:
-
Head-tail coupling apparatus
- ICSI:
-
Intracytoplasmic sperm injection
- KS:
-
Kallmann syndrome
- LBD:
-
Ligand-binding domain
- MAIS:
-
Mild androgen insensitivity
- MGS:
-
Morning glory syndrome
- MMAF:
-
Multiple Morphological Abnormalities of the sperm Flagella
- NAHR:
-
Nonallelic homologous recombination
- NGS:
-
Next-generation sequencing
- NOA:
-
Nonobstructive azoospermia
- NTD:
-
N-terminal domain
- OA:
-
Obstructive azoospermia
- PAIS:
-
Partial androgen insensitivity
- PAR:
-
Pseudoautosomal region
- PCD:
-
Primary ciliary dyskinesia
- PCR:
-
Polymerase chain reaction
- PEPNS:
-
Polyendocrine deficiencies and Polyneuropathies
- PGD:
-
Preimplantation genetic diagnosis
- POI:
-
Premature ovarian insufficiency
- SHFM:
-
Split-hand/foot malformation
- SOD:
-
Septo-optic dysplasia
- TEM:
-
Transmission electron microscopy
- TESE:
-
Testicular sperm extraction
- WES:
-
Whole-exome sequencing
- WS:
-
Waardenburg syndrome
References
Allali S, Muller J-B, Brauner R et al (2011) Mutation analysis of NR5A1 encoding steroidogenic factor 1 in 77 patients with 46, XY disorders of sex development (DSD) including hypospadias. PLoS One 6:e24117. https://doi.org/10.1371/journal.pone.0024117
Amiri-Yekta A, Coutton C, Kherraf Z-E et al (2016) Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum Reprod 31:2872–2880. https://doi.org/10.1093/humrep/dew262
Auguste Y, Delague V, Desvignes J-P et al (2018) Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am J Hum Genet 103:413–420. https://doi.org/10.1016/j.ajhg.2018.07.013
Baccetti B, Collodel G, Estenoz M et al (2005) Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum Reprod 20:2790–2794. https://doi.org/10.1093/humrep/dei126
Bashamboo A, Ferraz-de-Souza B, Lourenco D et al (2010) Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet 87:505–512. https://doi.org/10.1016/j.ajhg.2010.09.009
Ben Khelifa M, Coutton C, Blum MGB et al (2012) Identification of a new recurrent aurora kinase C mutation in both European and African men with macrozoospermia. Hum Reprod 27:3337–3346. https://doi.org/10.1093/humrep/des296
Ben Khelifa M, Coutton C, Zouari R et al (2014) Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 94:95–104. https://doi.org/10.1016/j.ajhg.2013.11.017
Boehm U, Bouloux P-M, Dattani MT et al (2015) Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism–pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 11:547–564. https://doi.org/10.1038/nrendo.2015.112
Brandt T, Blanchard L, Desai K et al (2013) 46,XY disorder of sex development and developmental delay associated with a novel 9q33.3 microdeletion encompassing NR5A1. Eur J Med Genet 56:619–623. https://doi.org/10.1016/j.ejmg.2013.09.006
Brauner R, Neve M, Allali S et al (2011) Clinical, biological and genetic analysis of anorchia in 26 boys. PLoS One 6:e23292. https://doi.org/10.1371/journal.pone.0023292
Chianese C, Fino MG, Riera Escamilla A et al (2015) Comprehensive investigation in patients affected by sperm macrocephaly and globozoospermia. Andrology 3:203–212. https://doi.org/10.1111/andr.12016
Collodel G, Moretti E (2006) Sperm morphology and aneuploidies: defects of supposed genetic origin. Andrologia 38:208–215. https://doi.org/10.1111/j.1439-0272.2006.00742.x
Colombo R, Pontoglio A, Bini M (2017) Two novel TEX15 mutations in a family with nonobstructive azoospermia. Gynecol Obstet Investig 82:283–286. https://doi.org/10.1159/000468934
Coutton C, Escoffier J, Martinez G et al (2015) Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update 21:455–485. https://doi.org/10.1093/humupd/dmv020
Coutton C, Vargas AS, Amiri-Yekta A et al (2018) Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun 9:686. https://doi.org/10.1038/s41467-017-02792-7
Dam AHDM, Koscinski I, Kremer JAM et al (2007) Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet 81:813–820. https://doi.org/10.1086/521314
Davies B, Baumann C, Kirchhoff C et al (2004) Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol Cell Biol 24:8642–8648. https://doi.org/10.1128/MCB.24.19.8642-8648.2004
Davis-Dao CA, Tuazon ED, Sokol RZ, Cortessis VK (2007) Male infertility and variation in CAG repeat length in the androgen receptor gene: a meta-analysis. J Clin Endocrinol Metab 92:4319–4326. https://doi.org/10.1210/jc.2007-1110
Davis-Dao C, Koh CJ, Hardy BE et al (2012) Shorter androgen receptor CAG repeat lengths associated with cryptorchidism risk among Hispanic white boys. J Clin Endocrinol Metab 97:E393–E399. https://doi.org/10.1210/jc.2011-2439
De Gendt K, Swinnen JV, Saunders PTK et al (2004) A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci U S A 101:1327–1332. https://doi.org/10.1073/pnas.0308114100
de Meeus A, Guittard C, Desgeorges M et al (1998) Linkage disequilibrium between the M470V variant and the IVS8 polyT alleles of the CFTR gene in CBAVD. J Med Genet 35:594–596
de Souza DAS, Faucz FR, Pereira-Ferrari L et al (2017) Congenital bilateral absence of the vas deferens as an atypical form of cystic fibrosis: reproductive implications and genetic counseling. Andrology. https://doi.org/10.1111/andr.12450
Dieterich K, Soto Rifo R, Faure AK et al (2007) Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet 39:661–665. https://doi.org/10.1038/ng2027
Dong FN, Amiri-Yekta A, Martinez G et al (2018) Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am J Hum Genet 102:636–648. https://doi.org/10.1016/j.ajhg.2018.03.007
Du Q, Li Z, Pan Y et al (2014) The CFTR M470V, intron 8 poly-T, and 8 TG-repeats detection in Chinese males with congenital bilateral absence of the vas deferens. Biomed Res Int 2014:689185. https://doi.org/10.1155/2014/689185
Dwyer AA, Raivio T, Pitteloud N (2015) Gonadotrophin replacement for induction of fertility in hypogonadal men. Best Pract Res Clin Endocrinol Metab 29:91–103. https://doi.org/10.1016/j.beem.2014.10.005
Dwyer AA, Raivio T, Pitteloud N (2016) Management of endocrine disease: reversible hypogonadotropic hypogonadism. Eur J Endocrinol 174:R267–R274. https://doi.org/10.1530/EJE-15-1033
ElInati E, Fossard C, Okutman O et al (2016) A new mutation identified in SPATA16 in two globozoospermic patients. J Assist Reprod Genet 33:815–820. https://doi.org/10.1007/s10815-016-0715-3
Elkhatib RA, Paci M, Longepied G et al (2017) Homozygous deletion of SUN5 in three men with decapitated spermatozoa. Hum Mol Genet 26:3167–3171. https://doi.org/10.1093/hmg/ddx200
Emery BR, Thorp C, Malo JW, Carrell DT (2004) Pregnancy from intracytoplasmic sperm injection of a sperm head and detached tail. Fertil Steril 81:686–688. https://doi.org/10.1016/j.fertnstert.2003.07.025
Fakhro KA, Elbardisi H, Arafa M et al (2018) Point-of-care whole-exome sequencing of idiopathic male infertility. Genet Med 20:1365–1373. https://doi.org/10.1038/gim.2018.10
Ferlin A, Vinanzi C, Garolla A et al (2006) Male infertility and androgen receptor gene mutations: clinical features and identification of seven novel mutations. Clin Endocrinol 65:606–610. https://doi.org/10.1111/j.1365-2265.2006.02635.x
Ferlin A, Rocca MS, Vinanzi C et al (2015) Mutational screening of NR5A1 gene encoding steroidogenic factor 1 in cryptorchidism and male factor infertility and functional analysis of seven undescribed mutations. Fertil Steril 104:163–9.e1. https://doi.org/10.1016/j.fertnstert.2015.04.017
Ferraz-de-Souza B, Lin L, Achermann JC (2011) Steroidogenic factor-1 (SF-1, NR5A1) and human disease. Mol Cell Endocrinol 336:198–205. https://doi.org/10.1016/j.mce.2010.11.006
Gambera L, Falcone P, Mencaglia L et al (2010) Intracytoplasmic sperm injection and pregnancy with decapitated sperm. Fertil Steril 93:1347.e7–1347.12. https://doi.org/10.1016/j.fertnstert.2008.12.087
Gao T, Marcelli M, McPhaul MJ (1996) Transcriptional activation and transient expression of the human androgen receptor. J Steroid Biochem Mol Biol 59:9–20
Gershoni M, Hauser R, Yogev L et al (2017) A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genet Med. https://doi.org/10.1038/gim.2016.225
Ghedir H, Mehri A, Mehdi M et al (2014) Meiotic segregation and sperm DNA fragmentation in Tunisian men with dysplasia of the fibrous sheath (DFS) associated with head abnormalities. J Assist Reprod Genet 31:1167–1174. https://doi.org/10.1007/s10815-014-0290-4
Gottlieb B, Beitel LK, Nadarajah A et al (2012) The androgen receptor gene mutations database: 2012 update. Hum Mutat 33:887–894. https://doi.org/10.1002/humu.22046
He X, Li W, Wu H et al (2019) Novel homozygous CFAP69 mutations in humans and mice cause severe asthenoteratospermia with multiple morphological abnormalities of the sperm flagella. J Med Genet 56:96–103. https://doi.org/10.1136/jmedgenet-2018-105486
Hu SC, Ye J, Fathi AK et al (2012) Mutations in NR5A1 and PIN1 associated with idiopathic hypogonadotropic hypogonadism. Genet Mol Res 11:4575–4584. https://doi.org/10.4238/2012.October.9.6
Imtiaz F, Allam R, Ramzan K, Al-Sayed M (2015) Variation in DNAH1 may contribute to primary ciliary dyskinesia. BMC Med Genet 16:14. https://doi.org/10.1186/s12881-015-0162-5
Jungwirth A, Giwercman A, Tournaye H et al (2012) European Association of Urology guidelines on Male Infertility: the 2012 update. Eur Urol 62:324–332. https://doi.org/10.1016/j.eururo.2012.04.048
Kamal A, Mansour R, Fahmy I et al (1999) Easily decapitated spermatozoa defect: a possible cause of unexplained infertility. Hum Reprod 14:2791–2795
Kasak L, Punab M, Nagirnaja L et al (2018) Bi-allelic recessive loss-of-function variants in FANCM cause non-obstructive azoospermia. Am J Hum Genet 103:200–212. https://doi.org/10.1016/j.ajhg.2018.07.005
Kerem B, Rommens JM, Buchanan JA et al (1989) Identification of the cystic fibrosis gene: genetic analysis. Science 245:1073–1080
Khan MJ, Pollock N, Jiang H et al (2018) X-linked ADGRG2 mutation and obstructive azoospermia in a large Pakistani family. Sci Rep 8:16280. https://doi.org/10.1038/s41598-018-34262-5
Kherraf Z-E, Amiri-Yekta A, Dacheux D et al (2018) A homozygous ancestral SVA-insertion-mediated deletion in WDR66 induces multiple morphological abnormalities of the sperm flagellum and male infertility. Am J Hum Genet 103:400–412. https://doi.org/10.1016/j.ajhg.2018.07.014
Krausz C (2012) An encore for the repeats: new insights into an old genetic variant. J Clin Endocrinol Metab 97:764–767
Krausz C, Casamonti E (2017) Spermatogenic failure and the Y chromosome. Hum Genet 136(5):637–655. https://doi.org/10.1007/s00439-017-1793-8
Krausz C, Chianese C (2014) Genetic testing and counselling for male infertility. Curr Opin Endocrinol Diabetes Obes 21:244–250. https://doi.org/10.1097/MED.0000000000000058
Krausz C, Riera-Escamilla A (2018) Genetics of male infertility. Nat Rev Urol 15(6):369–384. https://doi.org/10.1038/s41585-018-0003-3
Krausz C, Hoefsloot L, Simoni M, Tuttelmann F (2014) EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology 2:5–19. https://doi.org/10.1111/j.2047-2927.2013.00173.x
Krausz C, Riera-Escamilla A, Chianese C et al (2019) From exome analysis in idiopathic azoospermia to the identification of a high-risk subgroup for occult Fanconi anemia. Genet Med Genet Med 21(1):189–194. https://doi.org/10.1038/s41436-018-0037-1
Kuentz P, Van den Meerschaut F, Elinati E et al (2013) Assisted oocyte activation overcomes fertilization failure in globozoospermic patients regardless of the DPY19L2 status. Hum Reprod 28:1054–1061. https://doi.org/10.1093/humrep/det005
Lewis-Jones I, Aziz N, Seshadri S et al (2003) Sperm chromosomal abnormalities are linked to sperm morphologic deformities. Fertil Steril 79:212–215
Li W, He X, Yang S et al (2019) Biallelic mutations of CFAP251 cause sperm flagellar defects and human male infertility. J Hum Genet 64:49–54. https://doi.org/10.1038/s10038-018-0520-1
McCallum T, Milunsky J, Munarriz R et al (2001) Unilateral renal agenesis associated with congenital bilateral absence of the vas deferens: phenotypic findings and genetic considerations. Hum Reprod 16:282–288
Mongan NP, Tadokoro-Cuccaro R, Bunch T, Hughes IA (2015) Androgen insensitivity syndrome. Best Pract Res Clin Endocrinol Metab 29:569–580. https://doi.org/10.1016/j.beem.2015.04.005
Nenonen HA, Giwercman A, Hallengren E, Giwercman YL (2011) Non-linear association between androgen receptor CAG repeat length and risk of male subfertility–a meta-analysis. Int J Androl 34:327–332. https://doi.org/10.1111/j.1365-2605.2010.01084.x
Nistal M, Paniagua R, Herruzo A (1977) Multi-tailed spermatozoa in a case with asthenospermia and teratospermia. Virchows Arch B Cell Pathol 26:111–118
Oates RD, Amos JA (1994) The genetic basis of congenital bilateral absence of the vas deferens and cystic fibrosis. J Androl 15:1–8
Obermann H, Samalecos A, Osterhoff C et al (2003) HE6, a two-subunit heptahelical receptor associated with apical membranes of efferent and epididymal duct epithelia. Mol Reprod Dev 64:13–26. https://doi.org/10.1002/mrd.10220
Okutman O, Muller J, Baert Y et al (2015) Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet 24:5581–5588. https://doi.org/10.1093/hmg/ddv290
Patat O, Pagin A, Siegfried A et al (2016) Truncating mutations in the adhesion G protein-coupled receptor G2 gene ADGRG2 cause an X-linked congenital bilateral absence of vas deferens. Am J Hum Genet 99:437–442. https://doi.org/10.1016/j.ajhg.2016.06.012
Philibert P, Zenaty D, Lin L et al (2007) Mutational analysis of steroidogenic factor 1 (NR5a1) in 24 boys with bilateral anorchia: a French collaborative study. Hum Reprod 22:3255–3261. https://doi.org/10.1093/humrep/dem278
Porcu G, Mercier G, Boyer P et al (2003) Pregnancies after ICSI using sperm with abnormal head-tail junction from two brothers: case report. Hum Reprod 18:562–567
Quinton R, Cheow HK, Tymms DJ et al (1999) Kallmann’s syndrome: is it always for life? Clin Endocrinol 50:481–485
Raivio T, Falardeau J, Dwyer A et al (2007) Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med 357:863–873. https://doi.org/10.1056/NEJMoa066494
Rajender S, Singh L, Thangaraj K (2007) Phenotypic heterogeneity of mutations in androgen receptor gene. Asian J Androl 9:147–179. https://doi.org/10.1111/j.1745-7262.2007.00250.x
Ray PF, Toure A, Metzler-Guillemain C et al (2017) Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin Genet 91:217–232. https://doi.org/10.1111/cge.12905
Ribeiro RS, Vieira TC, Abucham J (2007) Reversible Kallmann syndrome: report of the first case with a KAL1 mutation and literature review. Eur J Endocrinol 156:285–290. https://doi.org/10.1530/eje.1.02342
Riordan JR, Rommens JM, Kerem B et al (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073
Ropke A, Tewes A-C, Gromoll J et al (2013) Comprehensive sequence analysis of the NR5A1 gene encoding steroidogenic factor 1 in a large group of infertile males. Eur J Hum Genet 21:1012–1015. https://doi.org/10.1038/ejhg.2012.290
Schimmer BP, White PC (2010) Minireview: steroidogenic factor 1: its roles in differentiation, development, and disease. Mol Endocrinol 24:1322–1337. https://doi.org/10.1210/me.2009-0519
Sha Y, Yang X, Mei L et al (2017a) DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil Steril 107:1312–1318.e2. https://doi.org/10.1016/j.fertnstert.2017.04.007
Sha Y-W, Wang X, Xu X et al (2017b) Novel mutations in CFAP44 and CFAP43 cause multiple morphological abnormalities of the sperm flagella (MMAF). Reprod Sci. https://doi.org/10.1177/1933719117749756
Sha Y, Zheng L, Ji Z et al (2018a) A novel TEX11 mutation induces azoospermia: a case report of infertile brothers and literature review. BMC Med Genet 19:63. https://doi.org/10.1186/s12881-018-0570-4
Sha Y-W, Xu X, Ji Z-Y et al (2018b) Genetic contribution of SUN5 mutations to acephalic spermatozoa in Fujian China. Gene 647:221–225. https://doi.org/10.1016/j.gene.2018.01.035
Sha Y-W, Wang X, Xu X et al (2019) Biallelic mutations in PMFBP1 cause acephalic spermatozoa. Clin Genet 95:277–286. https://doi.org/10.1111/cge.13461
Shang Y, Zhu F, Wang L et al (2017) Essential role for SUN5 in anchoring sperm head to the tail. Elife 6. https://doi.org/10.7554/eLife.28199
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837. https://doi.org/10.1038/nature01722
Tang S, Wang X, Li W et al (2017) Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 100:854–864. https://doi.org/10.1016/j.ajhg.2017.04.012
Tournaye H, Krausz C, Oates RD (2016) Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol 5(7):544–553. https://doi.org/10.1016/S2213-8587(16)30040-7
Tuttelmann F, Ruckert C, Ropke A (2018) Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Genet 30:12–20. https://doi.org/10.1007/s11825-018-0181-7
Vogt PH, Edelmann A, Kirsch S et al (1996) Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 5:933–943
Vorona E, Zitzmann M, Gromoll J et al (2007) Clinical, endocrinological, and epigenetic features of the 46,XX male syndrome, compared with 47,XXY Klinefelter patients. J Clin Endocrinol Metab 92:3458–3465. https://doi.org/10.1210/jc.2007-0447
Wambergue C, Zouari R, Fourati Ben Mustapha S et al (2016) Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod 31:1164–1172. https://doi.org/10.1093/humrep/dew083
Wang X, Jin H, Han F et al (2017) Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin Genet 91:313–321. https://doi.org/10.1111/cge.12857
Wang X, Jin H-R, Cui Y-Q et al (2018) Case study of a patient with cryptozoospermia associated with a recessive TEX15 nonsense mutation. Asian J Androl 20:101–102
Wu Q-Y, Li N, Li W-W et al (2014) Clinical, molecular and cytogenetic analysis of 46, XX testicular disorder of sex development with SRY-positive. BMC Urol 14:70. https://doi.org/10.1186/1471-2490-14-70
Yang F, Silber S, Leu NA et al (2015) TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol Med 7:1198–1210. https://doi.org/10.15252/emmm.201404967
Yang B, Wang J, Zhang W et al (2017) Pathogenic role of ADGRG2 in CBAVD patients replicated in Chinese population. Andrology 5:954–957. https://doi.org/10.1111/andr.12407
Yang Y, Guo J, Dai L et al (2018) XRCC2 mutation causes meiotic arrest, azoospermia and infertility. J Med Genet 55:628–636. https://doi.org/10.1136/jmedgenet-2017-105145
Yatsenko AN, Georgiadis AP, Ropke A et al (2015) X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 372:2097–2107. https://doi.org/10.1056/NEJMoa1406192
Yeh S, Tsai M-Y, Xu Q et al (2002) Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci U S A 99:13498–13503. https://doi.org/10.1073/pnas.212474399
Yin H, Ma H, Hussain S et al (2019) A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med 21:62–70. https://doi.org/10.1038/s41436-018-0015-7
Yu J, Chen Z, Ni Y, Li Z (2012) CFTR mutations in men with congenital bilateral absence of the vas deferens (CBAVD): a systemic review and meta-analysis. Hum Reprod 27:25–35. https://doi.org/10.1093/humrep/der377
Zare-Abdollahi D, Safari S, Mirfakhraie R et al (2015) Mutational screening of the NR5A1 in azoospermia. Andrologia 47:395–401. https://doi.org/10.1111/and.12274
Zhang Y-X, Li H-Y, He W-B et al (2019) XRCC2 mutation causes premature ovarian insufficiency as well as non-obstructive azoospermia in humans. Clin Genet 95(3):442–443
Zhu F, Wang F, Yang X et al (2016) Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am J Hum Genet 99:942–949. https://doi.org/10.1016/j.ajhg.2016.08.004
Zhu F, Liu C, Wang F et al (2018) Mutations in PMFBP1 cause acephalic spermatozoa syndrome. Am J Hum Genet 103:188–199. https://doi.org/10.1016/j.ajhg.2018.06.010
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Krausz, C., Riera-Escamilla, A. (2019). Monogenic Forms of Male Infertility. In: Igaz, P., Patócs, A. (eds) Genetics of Endocrine Diseases and Syndromes. Experientia Supplementum, vol 111. Springer, Cham. https://doi.org/10.1007/978-3-030-25905-1_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-25905-1_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-25904-4
Online ISBN: 978-3-030-25905-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)