Abstract
The lignocellulose-rich plant biomass is a readily available renewable resource having immense potential to be utilized as a sustainable alternative to ever-limiting fossil fuels. It, however, resists abiotic and biotic degradation due to its complex, recalcitrant, and intricate structure comprising primarily of lignocellulose. Interestingly, few specific microbial groups have evolved unparalleled capabilities to degrade and utilize these recalcitrant biopolymers through highly coordinated and genetically evolved enzymatic processes. Of these, wood-degrading fungi and anaerobic rumen fungi are endowed with exceptional enzymatic capabilities. White rot fungi, brown rot fungi, anaerobic rumen fungi, termite gut wood-decaying fungi, and other related fungi are considered as the major natural biomass utilization systems displaying immense contributions for degradation and mineralization of recalcitrant plant biomass in an array of terrestrial habitats. Several fungal species from phyla Basidiomycota, Neocallimastigomycota, and Ascomycota mainly Phanerochaete chrysosporium, Postia placenta, Neocallimastix spp., Orpinomyces, Gloeophyllum trabeum, Trametes versicolor, Agaricus bisporus, Pleurotus ostreatus, Serpula lacrimans, and many others are capable of degrading plant cell wall constituents through secretion of hydrolytic and oxidative enzymes, collectively called carbohydrate-active enzymes (CAZymes). These enzymes are broadly classified into glycoside hydrolases (GHs), carbohydrate esterases (CEs), glycosyltransferases (GTs), polysaccharide lyases (PLs), auxiliary activities (AAs), and lytic polysaccharide monooxygenases (LPMOs). The most crucial enzymes in lignocellulose degradation are β-glucosidases, glucanases, cellobiohydrolases, xylanases, endomannanases, feruloyl esterases, laccases, lignin peroxidases, manganese peroxidases, versatile peroxidases, etc. Comparative secretome studies elucidated considerable variations in lignocellulolytic enzyme repertoire of white rot fungi, brown rot fungi, and rumen fungi. In this chapter, we discuss the fungal secretomes associated with degradation of plant matter by wood-decaying fungi and anaerobic rumen fungi. A greater insight on their remarkable enzymatic capabilities is poised to open new avenues for their future biotechnological applications in the areas of animal nutrition, biofuel, biorefinery, and bioremediation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Anaerobic rumen fungi
- Carbohydrate-active enzymes
- Cellulose
- Glycoside hydrolases
- Phanerochaete chrysosporium
- Secretome
- White rot fungi
1.1 Introduction
Lignocellulose is a widely available recalcitrant plant biopolymer composed of polymeric polysaccharides cellulose, hemicellulose, and heteropolymeric lignin (Lewis and Yamamoto 1990; Eastwood et al. 2011; Bugg et al. 2011; Janusz et al. 2017; dos Santos et al. 2018; Bissaro et al. 2018; Brink et al. 2019; Ralph et al. 2019). According to an estimate, 550 billion tons of carbon are present in vegetation in terrestrial ecosystems including forest ecosystems where dead wood is the major form of the plant biomass (Siegenthaler and Sarmiento 1993; Krah et al. 2018). This recalcitrant plant biomass is recognized as the most abundant carbon source in terrestrial ecosystem. In woody plants (angiosperms and gymnosperms), cellulose generally constitutes 40–50% of the dry weight, whereas the amount of hemicelluloses and lignin ranges from 15% to 30% (Krah et al. 2018; Adesogan et al. 2019). Cellulose is a macropolymer of numerous glucose units attached linearly by β-1,4-glycosidic linkages. It is responsible for rigidity and crystalline form of plant cell walls (Baldrian and Valaskova 2008; McFarlane et al. 2014; Bissaro et al. 2018). Hemicelluloses, on the other hand, are complex and heterogeneous plant polysaccharides consisting of xylose, mannose, arabinose, glucose, galactose, and sugar acids. Xyloglucans, xylans, mannans, and glucomannans are the main examples of hemicelluloses (Scheller and Ulvskov 2010). These are known to strengthen the plant cell wall by filling the voids around cellulose fibrils and interacting with lignin.
Hemicelluloses are considered as the second most abundant polysaccharide in the nature (Saha 2003). Lignin is complex, aromatic, heteropolymeric, and most indigestible parts of the plant cell wall exhibiting almost complete resistance to hydrolytic degradation (Lewis and Yamamoto 1990; Janusz et al. 2017; Brink et al. 2019; Ralph et al. 2019). It contributes 15–30% of dry weight of vascular plant cell walls (Lewis and Yamamoto 1990; Gall et al. 2017). It is a phenolic polymer composed mainly of p-coumaryl, coniferyl, and sinapyl alcohols (Janusz et al. 2017; dos Santos et al. 2018; Brink et al. 2019; Ralph et al. 2019). Lignin confers rigidity to the plant cell walls and inhibits hydrolytic attacks on adjacent cellulose and hemicellulose (Lewis and Yamamoto 1990; Gall et al. 2017; Ralph et al. 2019). Lignocellulose-rich forest waste, dead woods, agro-food industry wastes, and leftover crop residues offer a sustainable, eco-friendly, and abundant resource for industrial-scale production of green energy and biofuels.
1.2 Degradation and Depolymerization of Plant Biomass
The aromatic nature of lignin is a major obstacle for biodegradation and mineralization of lignocellulose (Lewis and Yamamoto 1990; Janusz et al. 2017; Bissaro et al. 2018; Brink et al. 2019). Lignin and its phenolic derivatives are known to inhibit lignocellulolytic enzymes by adsorption or deactivation. In the biological world, fungi are considered to be the most prolific producers of lignocellulolytic enzymes (Blanchette 1991; Conesa et al. 2001; Bouws et al. 2008; Eastwood et al. 2011; Girard et al. 2013; Edwards et al. 2017; Kameshwar et al. 2019; Yadav et al. 2019a, b). These eukaryotic organisms play indispensable role in plant matter decomposition, carbon cycle, and overall nutrients recycling. Most fungi are aerobic and facultative anaerobic with the exception of strict anaerobic species present in the rumen of herbivorous animals (Sirohi et al. 2012; Edwards et al. 2017; Hooker et al. 2019). Filamentous fungi, in particular, secrete abundant enzymes to break down plant matter and complex materials in the environment which is in turn absorbed through hypha walls and utilized further for their growth and maintenance (Bouws et al. 2008; Eastwood et al. 2011; Girard et al. 2013; Edwards et al. 2017). Their involvement in plant matter decomposition, association with plant roots as mycorrhiza, association with photobionts in lichens, and presence in rumen and intestine of wood-decaying insects are well established, widely reported, and extensively reviewed earlier by several investigators.
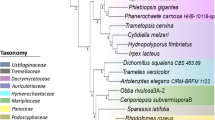
Fungi and bacteria are indispensably involved in degradation of recalcitrant plant biomass, thus contributing massively to carbon cycle in various ecosystems (Cragg et al. 2015; Janusz et al. 2017). Surprisingly, animals such as ruminants, termites, millipedes, and terrestrial isopods are dependent on their respective microbiomes for lignocellulose degradation as none of them encode the entire enzymatic repertoire required for lignocellulose degradation (Sirohi et al. 2012; Edwards et al. 2017; Kameshwar et al. 2019). In this chapter, we will emphasize the roles and secretomes of wood-degrading fungi and anaerobic rumen fungi associated with plant biomass degradation. Fungi are among the most diversified and widely studied Natural Biomass Utilization Systems (NBUS; Eastwood et al. 2011; Girard et al. 2013). Several fungi are equipped with enzymes which empower them to harvest energy and nutrition from plant biomass, which otherwise indigestible for other living organisms (Girard et al. 2013; Edwards et al. 2017; Janusz et al. 2017). Several fungal species from class Agaricomycetes (Basidiomycota) are closely associated with wood decay in different ecological niches (Krah et al. 2018). The saprotrophic members of this class cause decay of dead woods as either white rot or brown rot. The white rot fungi (WRF) and brown rot fungi (BRF) are the prominent colonizers which degrade lignocellulose cell wall components of compact wood logs, branches, and stumps (Martinez et al. 2004, 2009; Girard et al. 2013; Edwards et al. 2017; Janusz et al. 2017; SistaKameshwar and Qin 2018; Reina et al. 2019) (Fig. 1.1). In addition to white and brown rot, other wood-decaying manifestations, viz., soft rot and gray rot, are also exhibited by some members of Basidiomycota and Ascomycota (Riley et al. 2014).
Anaerobic rumen fungi (ARF) account for 8–20% of total microbial biomass of rumen and alimentary tract of herbivorous mammals (Sirohi et al. 2012; Edwards et al. 2017; Kameshwar et al. 2019; Hooker et al. 2019). First described in 1975 by Colin Orpin (Youssef et al. 2013), these obligate anaerobic, filamentous, and motile zoospore-forming fungi are assigned to phylum Neocallimastigomycota. Despite their low numbers (106 per ml of rumen fluid), ARF are key players in lignocellulose degradation in the rumen as these physically penetrate and disrupt the plant cell walls and thus facilitate rapid growth of fibrolytic bacteria leading to optimal degradation and utilization of lignocellulosic biomass (Sirohi et al. 2012; Youssef et al. 2013; Solomon et al. 2016; Haitjema et al. 2017; Edwards et al. 2017; Hooker et al. 2019). At present, only 11 genera have been cultured from rumen ecosystem of various herbivorous animals. These genera are Neocallimastix, Anaeromyces, Caecomyces, Cyllamyces, Orpinomyces, Piromyces, Buwchfawromyces, Feramyces, Oontomyces, Pecoramyces, and Liebetanzomyces (Sirohi et al. 2012; Edwards et al. 2017; Hooker et al. 2019; Li et al. 2019). In the next section, we have discussed the metabolic capabilities of wood-degrading fungi and ARF involved in plant biomass degradation and mineralization.
1.3 Secretomics and Mechanism of Lignocellulose Biodegradation
Secretome, a term coined by Tjalsma et al. (2000), represents all the proteins and cellular machineries which are secreted outside the plasma membrane into the environment or extracellular matrix by a cell (McCotter et al. 2016). Plants, bacteria, and fungi exhibit their unique and substrate-specific secretome under different environmental conditions. Fungal secretome, therefore, essentially comprises extracellular enzymes which are released exterior to the cell wall, usually in the presence of lignocellulosic plant matter (Bouws et al. 2008; Eastwood et al. 2011; Girard et al. 2013; Kameshwar et al. 2019). In the past decade, advances in protein identification techniques and genome sequencing have enabled detailed investigation of the secretomes of many saprophytic, pathogenic, and symbiotic fungal species revealing rich, diverse, and highly specific enzymatic profiles. The deconstruction and mineralization of recalcitrant and indigestible plant biomass require the synergistic and cooperative action of several hydrolytic, oxidative, and non-hydrolytic enzymes (Blanchette 1991; Girard et al. 2013; Cragg et al. 2015; Edwards et al. 2017; Janusz et al. 2017; Bissaro et al. 2018). An extensive knowledge of fungal secretomes involved in recalcitrant plant biomass degradation is of immense significance in the present scenario where increasing emphasis is devoted toward sustainable bioeconomy. In the coming sections, we describe different carbohydrate-active enzymes (CAZymes) involved in lignocellulose degradation (Table 1.1 and Fig. 1.2). According to CAZy database, there are six types of CAZymes, i.e., glycoside hydrolase (GH), carbohydrate esterase (CE), glycosyltransferase (GT), polysaccharide lyases (PL ), auxiliary activity (AA), and carbohydrate-binding domains (Lombard et al. 2014). It is estimated that the proportion of secreted proteins in fungal species ranges from 4 to 14% (Lowe and Howlett 2012). Degradation of recalcitrant plant biomass, viz., dead wood, wheat straw, fodder, etc., is accomplished by highly coordinated and synergistic actions of multiple CAZymes exhibiting a combination of oxidative, hydrolytic, and non-hydrolytic activities (Bugg et al. 2011; Lombard et al. 2014; Janusz et al. 2017; SistaKameshwar and Qin 2018; Bissaro et al. 2018). The most difficult part of plant cell wall is lignin and it needs to be degraded before enzyme can access cellulose and hemicellulose components. This activity is achieved in white rot fungi by extracellular enzymes, viz., oxidoreductases (AA2), lignin peroxidases, laccases, manganese peroxidases, versatile peroxidases, copper radical oxidases (AA5), dye-decolorizing peroxidases, and phenol-oxidizing multicopper oxidases (AA1), and a number of mediators, e.g., reactive oxygen species, free radicals, and aromatic intermediates (Martinez et al. 2004; Kuuskeri et al. 2016; Janusz et al. 2017; Bissaro et al. 2018; dos Santos et al. 2018). A detailed list of these enzymes is provided in Table 1.1. Both lignin-modifying enzymes and lignin-degrading auxiliary enzymes are involved in lignin degradation process (Bugg et al. 2011; Janusz et al. 2017; Bissaro et al. 2018). On the other hand, cellulose and hemicellulose degradation into disaccharides and monosaccharides is accomplished with the help of cellobiohydrolases (GH families GH6 and GH7), β-glucosidases (GH1 and GH3), endoglucanases (GH5, GH9, GH12, GH44, and GH45), lytic polysaccharide monooxygenases (AA9), polysaccharide lyases, and carbohydrate esterases (Kuuskeri et al. 2016; Janusz et al. 2017; Bissaro et al. 2018). Exoglucanases from GH family, GH6, GH7, and GH48, attack cellulose fibrils at the ends of the chain. In case of hemicelluloses, it is endo-hemicellulases, exo-hemicellulases, and accessory enzymes which cleave chains at various positions and locations (Table 1.1). Compositional details of lignocellulosic plant biomass, microorganisms involved in its biodegradation, and associated enzymatic repertoire have also been extensively reviewed earlier by Blanchette (1991), Eastwood et al. (2011), Girard et al. (2013), Lombard et al. (2014), Guerriero et al. (2015), Cragg et al. (2015), Janusz et al. (2017), Gall et al. (2017), Edwards et al. (2017), Bissaro et al. (2018), dos Santos et al. (2018), and Hooker et al. (2019). Fungal secretome studies are increasingly facilitated by accelerated genome sequencing availability of advanced software, databases, algorithms, analytical tools, prediction models, and improved proteomic approaches (Table 1.2).
The carbohydrate-active enzymes involved in degradation and depolymerization of recalcitrant plant biomass consisting primarily of lignin, cellulose, and hemicellulose. White rot fungi, brown rot fungi, and anaerobic rumen fungi secrete an array either all or some of essentially involved lignocellulolytic enzymes on variable substrates under in situ and in vitro conditions. Lignocellulose degradation is achieved by concerted and simultaneous action of several hydrolytic enzymes, oxidoreductases, peroxidases, free radicals, and other reaction mediators
As depicted in Fig. 1.2, lignin is acted upon by oxidative enzymes (laccase, lignin peroxidase, versatile peroxidase, manganese peroxidase) and auxillary activity redox enzymes (glyoxal oxidase, pyranose oxidase, aryl alcohol oxidase, methanol oxidase; Hori et al. 2014; Adesogan et al. 2019; Brink et al. 2019; Ralph et al. 2019). During lignin depolymerization by fungi, laccases and manganese peroxidases mainly act on phenols, and the nonphenolic lignin components are attacked by lignin peroxidase, whereas versatile peroxidases act on both (Bugg et al. 2011; Janusz et al. 2017; Brink et al. 2019; Ralph et al. 2019). In addition to numerous well-characterized hydrolytic enzymes, lignocellulolytic fungi exhibit simultaneous secretion of an array of lytic polysaccharide monooxygenases (LPMOs ) and several other oxidoreductases (Bissaro et al. 2018). LPMOs are a class of copper-dependent enzymes classified as auxiliary activities (AA) and belong to families AA9, AA10, AA11, AA13, AA14, and AA15 of CAZy (Bissaro et al. 2018). LPMOs are known to hydroxylate carbons at scissile glycosidic bonds. Interestingly, H2O2 plays a crucial role in catalytic activity of LPMOs (Bugg et al. 2011; Janusz et al. 2017; Bissaro et al. 2018).
1.4 Wood-Degrading Fungi
The number of species of potent wood-degrading fungi is more than 1000; however, the actual numbers are expected to be much higher. These fungi mainly exhibit saprotrophic mode of nutrition but sometimes may be showing parasitic attributes in forest ecosystems (Janusz et al. 2017). Ongoing research based on transcriptome and secretome has offered considerable insights on enzymatic machinery, and lignocellulose-degrading capabilities of several fungal species adapted to saprophytic, plant pathogenic, symbiotic, anaerobic, and endosymbiotic life styles (SistaKameshwar and Qin 2018). Secretomic studies offer deeper insights into mechanistic details of lignocellulose degradation by various species of bacteria and fungi in their respective natural habitats or under symbiotic associations with higher organisms. These studies have the potential to elucidate the life style adaptation and survival mechanisms of wood-degrading fungi on highly recalcitrant plant biomass as sole carbon source. In addition, secretomic analyses can facilitate our search for novel enzymes and exploitable secretary pathways essentially needed to establish industrial-scale and sustainable biofuels production. Here, we specifically focus on wood-decaying fungi and anaerobic rumen fungi well recognized for their exceptional lignocellulolytic potential. The challenging task of plant biomass depolymerization in natural environments is primarily accomplished by filamentous fungi.
Wood-degrading fungi mostly belong to the class Agaricomycetes of phylum Basidiomycota, although some members of Ascomycota are also of considerable significance. Lignocellulolytic activity of wood-degrading fungi is manifested through concerted action of oxidoreductases, peroxidases, glycoside hydrolases, exoglucanases, endoglucanases, xylanases, and a number of other CAZymes (Gaskell et al. 2016; Bissaro et al. 2018). In addition, the role of H2O2 and reactive oxygen species is also crucial. The most extensively studied (in terms of transcriptome and secretome) wood-degrading fungi are Phanerochaete chrysosporium , Phanerochaete carnosa, Phlebia tremellosa, Trametes versicolor, Phlebia radiata, Bjerkandera adusta, Irpex lacteus, Gloeophyllum trabeum, Agaricus bisporus, Stropharia coronilla, Agrocybe praecox, Chondrostereum purpureum, Heterobasidium annosum, Ceriporiopsis subvermispora, Phellinus pini, Lentinula edodes, Hericium clathroides, Pleurotus ostreatus, Obba rivulosa, Postia placenta, Piptoporus betulinus, Serpula lacrimans, Fomitopsis lilacinogilva, Ganoderma lucidum, Laetiporus portentosus, Fomitiporia mediterranea, Pycnoporus cinnabarinus, Dichomitus squalens, Punctularia strigosozonata, Botrytis cinerea, Stereum hirsutum, Pleurotus eryngii, Fibroporia radiculosa, Wolfiporia cocos, Dacryopinax primogenitus, Daedalea quercina, Laetiporus sulphurous, Neolentinus lepideus, Calocera cornea, Fistulina hepatica, Hydnomerulius pinastri, and Coniophora puteana (Riley et al. 2014; Presley and Schilling 2017; SistaKameshwar and Qin 2018).
1.4.1 Secretomes of White Rot Fungi (WRF)
WRF have the unique distinction of being the only microorganism in the biological world to completely degrade lignin, cellulose, and hemicellulose simultaneously (Manavalan et al. 2015; Xie et al. 2016). WRF are members of Basidiomycota, the largest phylum of kingdom Fungi (Blanchette 1991; Martinez et al. 2004; Krah et al. 2018). During lignin degradation by WRF, the crystalline cellulose with a bleached appearance is left behind which appears as white rot. This is why these are called white rot fungi. WRF initiate lignin depolymerization by producing large amount of free radicals mediated by oxidases and peroxidases (Martinez et al. 2004). With the help of a consortium of enzymes, WRF carry out mineralization of lignocellulose plant matter to carbon dioxide and thereby ensure continuity of carbon cycle in the habitats characterized by forest litter, fallen trees, wooden stumps, wood logs, etc. In return, WRF fulfil their energy and nutrition requirements from the same substrate (Blanchette 1991; Martinez et al. 2004; Baldrian and Valaskova 2008; Eastwood et al. 2011; Krah et al. 2018; Bissaro et al. 2018).
WRF from phylum Basidiomycota are reported to efficiently degrade lignin, cellulose, and hemicellulose present in the plant cell wall manifested through co-secretory and synergistic action of hydrolytic, oxidative, and non-hydrolytic enzymes, thus leaving a bleached fibrous residue (Krah et al. (2018). WRF can uniquely attack lignin barrier first before gaining access to cellulose and hemicelluloses of plant cell wall. WRF also produce extracellular reactive oxygen species mediated by peroxidases which facilitate physical disruption of crystalline lignocellulose biomass leading to greater access for degradative enzymes (Martinez et al. 2004; Bissaro et al. 2018). P. chrysosporium is a model organism for studying lignin degradation. Its genome sequence is published and offers greater insights on lignin-degradative enzymatic machinery and corresponding genomic organization (Martinez et al. 2004; Ohm et al. 2014). It is considered as the most prolific producer of CAZy, especially laccases, lignin peroxidases, manganese peroxidases, and LPMOs among various WRF species (Singh and Chen 2008). P. chrysosporium RP78 genome encodes >240 putative CAZymes belonging to 69 distinct families. Among these, 166 were glycoside hydrolases, 57 glycosyltransferases, 40 putative endoglucanases (GH5, GH9, GH12, GH61, and GH74), 14 carbohydrate esterases, at least 9 β-glucosidases, and 7 exocellobiohydrolases (Martinez et al. 2004). Further, ten lignin peroxidases, five manganese peroxidases, and several other lignocellulolytic enzymes were encoded by its genome (Martinez et al. 2004). P. chrysosporium is an excellent decomposer of soft- and hardwood, branches, logs, leaves, etc. in forests.
The secretomes of a number of other WRF growing in situ or on various substrates under in vitro conditions are now available in the literature. As compared to P. chrysosporium , which carry out simultaneous degradation of cellulose, hemicellulose, and lignin, Ceriporiopsis subvermispora exhibit unique and selective characteristic of lignin removal before initiating the cellulose degradation (Hori et al. 2014). Chondrostereum purpureum, a basidiomycetous fungus, produces an extensive repertoire of lignocellulolytic enzymes (Reina et al. 2019). Almost 50% of CAZy encoded by its genome belongs to GHs (GH5, GH6, GH7, GH10, GH11, GH12, GH16, GH30), CEs, and cellulose-binding domains (CBMs) which specifically target cellulose and hemicelluloses. In addition, 153 oxidoreductases, lignin-modifying enzymes, and auxillary activity (AA1, AA3) enzymes are encoded by its genome under variable culture conditions (Reina et al. 2019). Similarly, several families of CBMs and CEs were found to be encoded by C. purpureum genome. Further, expression of 81 oxidoreductases was recorded under substrate-specific culture conditions (Reina et al. 2019). Secretome analysis of Pleurotus eryngii, an edible mushroom and white rot fungi, revealed the production of seven glucanases, cellobiohydrolase, cellulose 1,4-beta-cellobiosidase, glucosidases, 22 glycoside hydrolase (GH families GH1, GH6, GH12, GH16, GH17, GH24, GH31, GH32, GH35, GH43, GH44, GH51, GH61, GH74, GH76, GH78, GH79, GH88, GH92, GH95), and CBM of family 21. These findings conclusively established the rich cellulolytic enzymes repertoire in P. eryngii under different substrate conditions (Xie et al. 2016). In another study, Kuuskeri et al. (2016) studied the secretory enzyme profile of Phlebia radiata cultured in solid state on spruce wood. The transcriptomic and secretomic analyses indicated expression and secretion of oxidoreductase, glyoxal oxidases, alcohol oxidases, cellobiohydrolases (GH6 and GH7), LPMO (AA9), lignin peroxidases, and acetyl xylan esterase. Prominent upregulation of genes whose products are involved in wood decay was observed at different growth periods.
1.4.2 Secretome of Brown Rot Fungi (BRF)
BRF are basidiomycetous fungi which occur as common pests of plants in conifer-dominated woodlands (Presley and Schilling 2017). BRF decompose wood via glycoside hydrolase-mediated saccharification and free radical oxidation (Baldrian and Valaskova 2008). However, compared to WRF, BRF preferentially degrade cellulose and hemicellulose whereas lignin is not depolymerized significantly thus leaving behind a brownish residue with fragmented appearance (Krah et al. 2018). Genomic analysis revealed evolution of BRF from WRF by gradual loss of genes which encode for ligninolytic peroxidases (Januszet al. 2017). Absence of class II peroxidases and cellobiohydrolases was reported in Wolfiporia cocos (Gaskell et al. (2016). The important species of BRF are Gloeophyllum trabeum, Postia placenta, Piptoporus betulinus, Serpula lacrimans, Fomitopsis lilacinogilva, Laetiporus portentosus, Wolfiporia cocos, Fibroporia radiculosa, Dacryopinax primogenitus, Daedalea quercina, Laetiporus sulphurous, Neolentinus lepideus, Calocera cornea, Fistulina hepatica, Hydnomerulius pinastri, and Coniophora puteana (Krah et al. 2018; SistaKameshwar and Qin 2018).
Most of BRF are aerobic in nature and contribute a little inside digestive tracts of herbivorous mammals. WRF vary significantly in terms of substrate specificity and mechanisms of action. The genomic, phenotypic, and phylogenetic basis of these variations is yet to be fully understood. Presley and Schilling (2017) studied in vitro degradation of spruce wafers by two BRF, namely, Serpula lacrymans and Gloeophyllum trabeum, using a proteomic approach. Upon initial colonization, oxidoreductase diversity was observed first followed by higher glycoside hydrolase activity at later stages. Their findings suggest significant variations in their oxidoreductase profiles as indicated by presence of putative copper radical oxidase in S. lacrymans but absence in G. trabeum. On the other hand, GMC oxidoreductase and a xyloglucan-specific AA9 family protein were produced by G. trabeum but not by S. lacrymans. S. lacrymans exhibited higher mannanase activity compared to G. trabeum which showed elevated xylanase production. Interestingly, GH6 and cellobiohydrolases (CBHs) were not detected in case of S. lacrymans. As compared to 93 proteins identified in S. lacrymans, the protein counts were 209 in G. trabeum. Overall analysis of their secretomes indicates a two-step brown rot decay mechanism manifested through entirely different biochemical routes. In another BRF species Postia placenta from phylum Basidiomycota, 242 putative CAZY-encoding genes were reported (Martinez et al. 2009). Among these, the number of GHs, CEs, glycosyltransferases, and polysaccharide lyases were 144, 10, 75, and 6, respectively. In addition, expansin-like proteins, laminarinases, chitinases, endoxylanases, β-xylosidases, and L-α-arabinofuranosidases were identified. However, the enzymes/proteins involved in lignin degradation, viz., lignin peroxidase, manganese peroxidase, exocellobiohydrolases, versatile peroxidase, cellulose-binding domains, and cellulose-binding endoglucanases, were entirely absent. This unique secretome profile substantiated the non-action of P. placenta against lignin components of plant biomass. Gaskell et al. (2016) determined 64 glycoside hydrolases from Wolfiporia cocos growing on different media containing glucose, purified crystalline cellulose, lodgepole pine, and aspen. More than fourfold upregulation of hemicellulase-, endoxylanase-, and chitinase-encoding genes was observed. Additionally, there was upregulation of genes involved in oxidative depolymerization of cellulose.
1.5 Secretome of Anaerobic Rumen Fungi (ARF)
ARF have indispensable role in digestion of recalcitrant lignocellulosic feed materials in digestive system of herbivorous animals. CAZymes in ARF exist either as free enzymes or as cellulosome, a multiprotein complex (Haitjema et al. 2017). The genome sequencing of four species of Neocallimastigomycota suggests that many of these CAZymes have been acquired by horizontal gene transfer from rumen bacteria (Youssef et al. 2013; Haitjema et al. 2017). The extensive CAZyme repertoire, cellulosome, and extracellular proteases produced by Neocallimastigomycetes may help these microbes compete with other rumen inhabitants for limited nutrients (Youssef et al. 2013; Haitjema et al. 2017). As described earlier in Sect. 1.3, only 11 genera have been cultured and exhaustively investigated for secretome analyses. Due to their strict anaerobic lifestyles, in vitro studies are limited on these fungi. Still, a number of studies have offered insights in secretome profiles of ARF. Wang et al. (2011) identified 25 families of glycosyl hydrolases (GHs) from anaerobic rumen fungus Neocallimastix patriciarum W5 culture anaerobically on substrate mixture comprising rice straw, napier grass, and sugarcane bagasse. Transcriptome and secretome analysis revealed 25 putative GH families dominated by GH6 (15%), GH10 (9.5%), GH5 (9.1%), and GH43 (9.1%). The main CAZymes were cellobiohydrolase (EC 3.2.1.91), endoglucanase (EC 3.2.1.4), and xylanases. The number of cellulases and hemicellulases was found to be higher in N. patriciarum W5 as compared to other plant matter-degrading fungi. The genome sequencing of Orpinomyces sp. strain C1A by Youssef et al. (2013) revealed an efficient lignocellulolytic enzymes repertoire comprising 357 glycoside hydrolase genes, 92 carbohydrate esterases genes, and 24 polysaccharide lyases genes. Interestingly, 220 genes with fungal dockerin domain and 103 genes harboring carbohydrate-binding module domains were also identified. Further, expansion of cellulolytic and hemicellulolytic families, viz., GH6, GH9, Gh10, GH11, Gh43, GH45, and GH48, and reduction or complete loss of families GH7, GH16, GH18, GH28, and GH61 were also observed (Youssef et al. 2013). The genes attributing for an efficient and extensive glycoside hydrolase machinery of this rumen fungus are believed to be acquired through horizontal gene transfer from multiple ruminal bacteria present in rumen. A previous study also reported presence of cellulase (GH family 48) containing two C-terminal fungal dockerin domains from Piromyces equi (Steenbakkers et al. 2002).
Kameshwar and Qin (2018) compared the genome-wide annotations of five ARF, namely, Neocallismatix californiae, Anaeromyces robustus, Orpinomyces sp., Piromyces sp. E2, and Piromyces finnis. Findings of this comprehensive analysis revealed that ARF have the highest number of CAZyme-encoding genes compared to other fungi. Moreover, the presence of genes for cellulosomes and carbohydrate transport and metabolism strongly supported their remarkable polysaccharide-degrading abilities. Surprisingly, the genes encoding for lignin-degrading auxiliary activity enzymes, such as lignin peroxidase, laccase, manganese peroxidase, versatile peroxidase, aryl alcohol oxidase, glyoxal oxidases, and glucose oxidases, were completely lacking in their genomes. Among these five ARF species, Neocallimastix californiae was found to possess the highest number of genes coding for CAZymes involved in cellulolytic, hemicellulolytic, and pectinolytic activities (Kameshwar and Qin 2018). A comparative analysis of transcriptome of four ARF, i.e., Anaeromyces mucronatus YE505, Neocallimastix frontalis 27, Orpinomyces joyonii SG4, and Piromyces rhizinflata YM600 revealed that 8.1–11.2% of the entire transcriptome were predicted CAZymes with highest in O. joyonii. About 40–44% of the CAZymes-encoding contigs had one or more carbohydrate-binding modules (Gruninger et al. 2018).
1.6 Conclusion
In spite of the availability of enormous amount of plant matter, woody substances, crop residues, and agro-food industry by-products as an attractive renewable resource, its industrial-scale utilization remains limited due to inherent structural complexity and recalcitrance. The controlled decomposition of biomass in general and of lignocellulose in particular involves a wide diversity of enzymatic activities and chemical reactions, which are far from fully elucidated. Moreover, our knowledge of the fungal secretion pathway is still at an early stage. Wood-degrading fungi and anaerobic rumen fungi can accomplish this daunting task with remarkable efficiency in natural environments using their specialized and sophisticated biomolecular machinery. The fungal secretomes have been explored to find enzymes and enzyme combinations for paper, textile, and food manufacturing industries. Similarly, low cost and sustainable processes for plant biomass conversion to biofuels can revolutionize industrial and environmental microbiology. The study of secretomes of novel fungal genera/species is interestingly poised to elucidate novel enzymes suitable for efficient plant cell wall degradation which can be exploited for commercial biotechnological applications. White rot fungi in particular have tremendous potential for biotechnological applications, bioremediation, pulp and paper industries, and effluents treatment in different industrial settings. These fungi possess remarkable potential for implementing eco-friendly, sustainable, and consolidated biological processing of lignocellulosic biomass for biofuel and biorefining industries and bioremediation processes.
References
Adesogan AT, Arriola KG, Jiang Y, Oyebade A, Paula EM, Pech-Cervantes AA, Romero JJ, Ferraretto LF, Vyas D (2019) Symposium review: technologies for improving fiber utilization. J Dairy Sci S0022-0302:30295–30294. https://doi.org/10.3168/jds.2018-15334
Baldrian P, Valaskova V (2008) Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32:501–521. https://doi.org/10.1111/j.1574-6976.2008.00106.x
Bissaro B, Várnai A, Røhr ÅK, Eijsink VGH (2018) Oxidoreductases and reactive oxygen species in conversion of lignocellulosic biomass. Microbiol Mol Biol Rev 82:e00029–18. https://doi.org/10.1128/MMBR.00029-18
Blanchette R (1991) Delignification by wood-decay fungi. Annu Rev Phytopathol 29:381–398. https://doi.org/10.1146/annurev.py.29.090191.002121
Bouws H, Wattenberg A, Zorn H (2008) Fungalsecretomes-nature’s toolbox for white biotechnology. Appl Microbiol Biotechnol 80:381–388. https://doi.org/10.1007/s00253-008-1572-5
Brink DP, Ravi K, Lidén G, Gorwa-Grauslund MF (2019) Mapping the diversity of microbial lignin catabolism: experiences from the eLignin database. Appl Microbiol Biotechnol 103:3979–4002. https://doi.org/10.1007/s00253-019-09692-4
Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R (2011) Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896. https://doi.org/10.1186/s12862-018-1229-7
Conesa A, Punt PJ, van Luijk N, van den Hondel CA (2001) The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet Biol 33:155–171. https://doi.org/10.1006/fgbi.2001.1276
Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, Etxabe AG, Goodell BS, Jellison J, McGeehan JE, McQueen-Mason SJ (2015) Lignocellulose degradation mechanisms across the Tree of Life. Curr Opin Chem Biol 29:108–119. https://doi.org/10.1016/j.cbpa.2015.10.018
dos Santos AC, Ximenes E, Kim Y, Ladisch MR (2018) Lignin-enzyme interactions in the hydrolysis of lignocellulosic biomass. Trends Biotechnol S0167-7799:30306–30308. https://doi.org/10.1016/j.tibtech.2018.10.010
Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A et al (2011) The plant cell wall decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. https://doi.org/10.1126/science.1205411
Edwards JE, Forster RJ, Callaghan TM, Dollhofer V, Dagar SS, Cheng Y et al (2017) PCR and omics based techniques to study the diversity, ecology and biology of anaerobic fungi: insights, challenges and opportunities. Front Microbiol 8:1657. https://doi.org/10.3389/fmicb.2017.01657
Gall DL, Ralph J, Donohue TJ, Noguera DR (2017) Biochemical transformation of lignin for deriving valued commodities from lignocellulose. Curr Opin Biotechnol 45:120–126. https://doi.org/10.1016/j.copbio.2017.02.015
Gaskell J, Blanchette RA, Stewart PE, BonDurant SS, Adams M, Sabat G, Kersten P, Cullen D (2016) Transcriptome and secretome analyses of the wood decay fungus Wolfiporia cocos support alternative mechanisms of lignocellulose conversion. Appl Environ Microbiol 82:3979–3987. https://doi.org/10.1128/AEM.00639-16
Girard V, Dieryckx C, Job C, Job D (2013) Secretomes: the fungal strike force. Proteomics 13:597–608. https://doi.org/10.1002/pmic.201200282
Gruninger RJ, Nguyen TTM, Reid ID, Yanke JL, Wang P, Abbott DW, Tsang A, McAllister T (2018) Application of transcriptomics to compare the carbohydrate active enzymes that are expressed by diverse genera of anaerobic fungi to degrade plant cell wall carbohydrates. Front Microbiol 9:1581. https://doi.org/10.3389/fmicb.2018.01581
Guerriero G, Hausman J, Strauss J, Ertan H, Siddiqui KS (2015) Destructuring plant biomass: Focus on fungal and extremophilic cell wall hydrolases. Plant Sci 234:180–193. https://doi.org/10.1016/j.plantsci.2015.02.010
Haitjema CH, Gilmore SP, Henske JK, Solomon KV, Groot R, Kuo A et al (2017) A parts list for fungal cellulosomes revealed by comparative genomics. Nat Microbiol 2:17087. https://doi.org/10.1038/nmicrobiol.2017.87
Hooker CA, Lee KZ, Solomon KV (2019) Leveraging anaerobic fungi for biotechnology. Curr Opin Biotechnol 59:103–110. https://doi.org/10.1016/j.copbio.2019.03.013
Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, Cullen D (2014) Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism’s strategy for degrading lignocellulose. Appl Environ Microbiol 80:2062–2070. https://doi.org/10.1128/AEM.03652-13
Janusz G, Pawlik A, Sulej J, Swiderska-Burek U, Jarosz-Wilkolazka A, Paszczynski A (2017) Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev 41:941–962. https://doi.org/10.1093/femsre/fux049
Kameshwar AKS, Qin W (2018) Genome wide analysis reveals the extrinsic cellulolytic and biohydrogen generating abilities of Neocallimastigomycota fungi. J Genomics 6:74–87. https://doi.org/10.7150/jgen.25648
Kameshwar AKS, Ramos LP, Qin W (2019) Metadata analysis approaches for understanding and improving the functional involvement of rumen microbial consortium in digestion and metabolism of plant biomass. J Genomics 7:31–45. https://doi.org/10.7150/jgen.32164
Krah F, Bässler C, Heibl C, Soghigian J, Schaefer H, Hibbett DS (2018) Evolutionary dynamics of host specialization in wood-decay fungi. BMC Evol Biol 18:119. https://doi.org/10.1186/s12862-018-1229-7
Kuuskeri J, Häkkinen M, Laine P, Smolander OP, Tamene F, Miettinen S, Nousiainen P, Kemell M, Auvinen P, Lundell T (2016) Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. Biotechnol Biofuels 9:192. https://doi.org/10.1186/s13068-016-0608-9
Lewis NG, Yamamoto E (1990) Lignin-occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41:455–496. https://doi.org/10.1146/annurev.pp.41.060190.002323
Li Y, Li Y, Jin W, Sharpton TJ, Mackie RI, Cann I, Cheng Y, Zhu W (2019) Combined genomic, transcriptomic, proteomic, and physiological characterization of the growth of Pecoramyces sp. F1 in monoculture and co-culture with a syntrophic methanogen. Front Microbiol 10:435. https://doi.org/10.3389/fmicb.2019.00435
Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
Lowe RGT, Howlett BJ (2012) Indifferent, affectionate, or deceitful: lifestyles and secretomes of fungi. PLoS Pathog 8(3):e1002515. https://doi.org/10.1371/journal.ppat.1002515
Manavalan T, Manavalan A, Heese K (2015) Characterization of lignocellulolytic enzymes from white-rot fungi. Curr Microbiol 70:485–498. https://doi.org/10.1007/s00284-014-0743-0
Martinez D, Larrondo LF, Putnam N, SollewijnGelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700. https://doi.org/10.1038/nbt967
Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP et al (2009) Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A 106:1954–1959. https://doi.org/10.1073/pnas.0809575106
McCotter SW, Horianopoulos LC, Kronstad JW (2016) Regulation of the fungal secretome. Curr Genet 62:533–545. https://doi.org/10.1007/s00294-016-0578-2
McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65:69–94. https://doi.org/10.1146/annurev-arplant-050213-040240
Ohm RA, Riley R, Salamov A, Min B, Choi I, Grigoriev IV (2014) Genomics of wood-degrading fungi. Fungal Genet Biol 72:82–90. https://doi.org/10.1016/j.fgb.2014.05.001
Presley GN, Schilling JS (2017) Distinct growth and secretome strategies for two taxonomically divergent brown rot fungi. Appl Environ Microbiol 83:e02987–e02916. https://doi.org/10.1128/AEM.02987-16
Ralph J, Lapierre C, Boerjan W (2019) Lignin structure and its engineering. Curr Opin Biotechnol 56:240–249. https://doi.org/10.1016/j.copbio.2019.02.019
Reina R, Kellner H, Hess J, Jehmlich N, García-Romera I, Aranda E, Hofrichter M, Liers C (2019) Genome and secretome of Chondrostereum purpureum correspond to saprotrophic and phytopathogenic life styles. PLoS One 14:e0212769. https://doi.org/10.1371/journal.pone.0212769
Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW et al (2014) Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. https://doi.org/10.1073/pnas.1400592111
Saha BC (2003) Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30:279–291. https://doi.org/10.1007/s10295-003-0049-x
Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61:263–289. https://doi.org/10.1146/annurev-arplant-042809-112315
Siegenthaler U, Sarmiento JL (1993) Atmospheric carbon dioxide and the ocean. Nature 365:119–125. https://doi.org/10.1038/365119a0
Singh D, Chen S (2008) The white-rot fungus Phanerochaete chrysosporium: conditions for the production of lignin-degrading enzymes. Appl Microbiol Biotechnol 81:399–417. https://doi.org/10.1007/s00253-008-1706-9
Sirohi SK, Singh N, Dagar SS, Puniya AK (2012) Molecular tools for deciphering the microbial community structure and diversity in rumen ecosystem. Appl Microbiol Biotechnol 95:1135–1154. https://doi.org/10.1007/s00253-012-4262-2
SistaKameshwar AK, Qin W (2018) Comparative study of genome-wide plant biomass-degrading CAZymes in white rot, brown rot and soft rot fungi. Mycology 9:93–105. https://doi.org/10.1080/21501203.2017.1419296
Steenbakkers P, Freelove A, Van Cranenbroek B, Sweegers B, Harhangi H, Vogels G, Hazlewood G, Gilbert H, Op den Camp H (2002) The major component of the cellulosomes of anaerobic fungi from the genus Piromyces is a family 48 glycoside hydrolase. DNA Seq 13:313–320. https://doi.org/10.1080/1042517021000024191
Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A et al (2016) Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351:1192–1195. https://doi.org/10.1126/science.aad1431
Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM (2000) Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev 64:515–547. https://doi.org/10.1128/MMBR.64.3.515-547.2000
Wang TY, Chen HL, Lu MJ, Chen YC, Sung HM, Mao CT, Cho HY, Ke HM, Hwa TY et al (2011) Functional characterization of cellulases identified from the cow rumen fungus Neocallimastix patriciarum W5 by transcriptomic and secretomic analyses. Biotechnol Biofuels 4:24. https://doi.org/10.1186/1754-6834-4-24
Xie C, Yan L, Gong W, Zhu Z, Tan S, Chen D, Hu Z, Peng Y (2016) Effects of different substrates on lignocellulosic enzyme expression, enzyme activity, substrate utilization and biological efficiency of Pleurotus eryngii. Cell Physiol Biochem 39:1479–1494. https://doi.org/10.1159/000447851
Yadav AN, Mishra S, Singh S, Gupta A (2019a) Recent advancement in white biotechnology through fungi Volume 1: diversity and enzymes perspectives. Springer International Publishing, Cham
Yadav AN, Mishra S, Singh S, Gupta A (2019b) Recent advancement in white biotechnology through fungi. Volume 2: perspective for value-added products and environments. Springer International Publishing, Cham
Youssef NH, Couger MB, Struchtemeyer CG, Liggenstoffer AS, Prade RA, Najar FZ, Atiyeh HK, Wilkins MR, Elshahed MS (2013) The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl Environ Microbiol 79:4620–4634. https://doi.org/10.1128/AEM.00821-13
Acknowledgments
NS is grateful to The Chancellor, Eternal University, for their financial support and infrastructural facilities. The authors are thankful to Dr. Sumit Singh Dagar for his valuable and expert suggestions.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Singh, N., Singh, J. (2019). Secretomics of Wood-Degrading Fungi and Anaerobic Rumen Fungi Associated with Biodegradation of Recalcitrant Plant Biomass. In: Yadav, A., Singh, S., Mishra, S., Gupta, A. (eds) Recent Advancement in White Biotechnology Through Fungi. Fungal Biology. Springer, Cham. https://doi.org/10.1007/978-3-030-25506-0_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-25506-0_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-25505-3
Online ISBN: 978-3-030-25506-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)