Abstract
Epigenetic information refers to heritable changes in gene expression that occur without modifications at the DNA sequence level. These changes are orchestrated by different epigenetic mechanisms such as DNA methylation, posttranslational modifications of histones, and the presence of noncoding RNAs. Epigenetic information regulates chromatin structure to confer cell-specific gene expression.
The sperm epigenome is the result of three periods of global resetting during men’s life. Germ cell epigenome reprogramming is designed to allow cell totipotency and to prevent the transmission of epimutations via spermatozoa. At the end of these reprogramming events, the sperm epigenome has a very specific epigenetic pattern that is a footprint of past reprogramming events and has an influence on embryo development.
Several data demonstrate that not all regions of the epigenome are erased during the reprogramming periods, suggesting the transmission of epigenetic information from fathers to offspring via spermatozoa. Moreover, it is becoming increasingly clear that the sperm epigenome is sensitive to environmental factors during the process of gamete differentiation, suggesting the plasticity of the sperm epigenetic signature according to the circumstances of the individual’s life.
In this chapter, we provided strong evidences about the association between variations of the sperm epigenome and the exposure to environmental factors. Moreover, we will present data about how epigenetic mechanisms are candidates for transferring paternal environmental information to offspring.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Layers of Epigenetic Information in Sperm
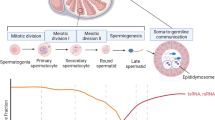
Spermatozoa are highly differentiated cells that play an essential role in reproduction by providing the haploid paternal genome to the embryo. Nevertheless, the biological relevance of sperm cells is not merely based on DNA sequence, but also on a wide range of epigenetic information such as DNA methylation, posttranslational modifications of histones, and the cargo of a specific set of RNA molecules. The orchestrated action of the different epigenetic mechanisms is essential for modulating sperm chromatin structure and gene expression, creating functional sperm able to achieve the processes of fertilization and early embryogenesis successfully.
DNA Methylation
DNA methylation mainly occurs at position 5 of cytosines (5-methylcytosine, 5mC) in 5’-CpG-3′ dinucleotides. It has been called the “fifth base” of the human genome since 4% of the cytosines are methylated. The CpG dinucleotides are present throughout the genome but concentrated in genomic regions called CpG islands (CG islands, CGI). CGI are normally found within gene promoters, being unmethylated in the case of genes that are actively transcribed and methylated in the case of inactive genes. The significance of CpG dinucleotide methylation along the transcription unit (exons, introns, and 5′ and 3′ untranslated regions) is less known.
The sperm methylome is the result of different waves of genome-wide DNA reprogramming during the differentiation of primordial germ cells (PGCs ) into spermatozoa. PGCs arise from the epiblast and migrate to colonize the genital ridge (Chuva de Sousa Lopes and Roelen 2010). They initiated their differentiation as cells with a somatic epigenetic signature exhibiting high levels of 5mC, which are passively removed during PGC migration (Guibert et al. 2012; Kagiwada et al. 2013; Seisenberger et al. 2012). PGCs enter a second stage of active DNA demethylation in the genital ridge, resulting in an almost complete loss of 5mC (Hackett et al. 2013; Tang et al. 2015). The demethylation process in PGCs also affects imprinted genes (Hackett et al. 2013; Hajkova et al. 2002; Sasaki and Matsui 2008). Although the global loss of methylation affects all methylation levels, some retrotransposon-associated and single copy regions of the genome are resistant to reprogramming (Tang et al. 2015). The establishment of new methylation marks starts in type A spermatogonia (Kota and Feil 2010) and is completed before the onset of meiosis (Davis et al. 2000; Kerjean et al. 2000).
The sperm methylome is the consequence of this process of DNA methylation erasure and reestablishment. The result is a marked hypomethylated state with a high homogeneity among sperm samples from different individuals (Camprubí et al. 2017; Krausz et al. 2012). Some authors have demonstrated that genes with hypomethylated promoter regions are functionally associated with biological processes related to embryonic development (Camprubí et al. 2017; Hammoud et al. 2009; Krausz et al. 2012; Molaro et al. 2011). In contrast, genomic regions containing repetitive DNA sequences appear to be significantly hypermethylated, probably to prevent the activation of transposable elements (Molaro et al. 2011). Authors agree that these features reflect the reprogramming phenomena occurred during spermatogenesis, a process designed to confer a pluripotent state to the sperm, which will facilitate the epigenetic reprogramming that will take place during the early stages of embryo development.
Sperm Chromatin
During the postmeiotic differentiation of round spermatids into spermatozoa, chromatin is extensively remodeled resulting in nucleoprotamine structure in 85% of the nucleus (Gatewood et al. 1987). This process allows the establishment of highly ordered and compacted toroid-chromatin structures. The remaining 15% of the sperm chromatin retain a nucleohistone structure (Gatewood et al. 1987).
In human spermatozoa, residual nucleosomes are programmatically retained in gene regulatory regions, including the promoters of developmental genes, microRNA genes, and imprinted loci (Hammoud et al. 2009). Moreover, these histones carry multiple posttranslational modifications, suggesting some degree of retained regulatory competence through histone tail modifications (Arpanahi et al. 2009; Hammoud et al. 2009). The fact that sperm histone modifications are transmitted to the embryo and are resistant to protein oocyte replacement (Van Der Heijden et al. 2008) argues in favor of an effect beyond fertilization.
Like histones, protamines also exhibited posttranslational modifications (Brunner et al. 2014; Oliva et al. 2015). Nevertheless, protamines are exchanged by the histones provided by the oocyte (Van Der Heijden et al. 2008), which argues against an effect of posttranslational protamine modifications beyond fertilization.
Noncoding RNAs
Sperm RNAs have emerged as a field of interest because of their high complexity and diversity. Beyond the relevance of coding RNAs, different populations of sperm noncoding RNAs (ncRNAs ) have been characterized in the last decade, revealing their strong contribution in processes related to cellular spermatogenesis, fertilization, and embryogenesis (Corral-Vazquez and Anton 2018). Sperm RNA transcripts mainly originated from the two transcriptional waves that take place during spermatogenesis generating specific transcripts for the correct development of spermatogenesis (de Mateo and Sassone-Corsi 2014). Moreover, some sperm ncRNAs remain intact after being released into the oocyte (Boerke et al. 2007) regulating the expression of specific oocytes transcripts (Amanai et al. 2006), which suggest their ability to introduce epigenetic modifications in the early embryo.
ncRNAs are classified, depending on their length, into long noncoding RNA (lncRNA ) and small noncoding RNA (sncRNA ). The biological functions of lncRNAs mainly comprise epigenetic regulation of single mRNA transcription or whole chromosomes (Bao et al. 2013). There are specific lncRNAs that are especially abundant in the sperm transcriptome, suggesting their role in male fertility (Jodar et al. 2013).
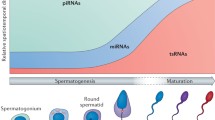
The sperm sncRNA family includes microRNA (miRNAs ), Piwi-interacting RNA (piRNAs ), and endogenous small interfering RNAs (endo-siRNAs ). MicroRNAs are a family of functional RNA molecules of 22–24 nucleotides (nt) that form complementary stem-loop structures in the 3′ untranslated region (3’ UTR) of their target messenger RNAs (mRNAs ). Usually, this association leads to mRNA degradation and/or translational repression. It is known that each miRNA has hundreds of potential mRNA targets, and it has been estimated that they can regulate up to 60% of protein-coding genes (Luo et al. 2015). Human spermatozoa show homogeneous and stable expression patterns of miRNAs, which have a significant ontological relation with processes involved in embryogenesis and spermatogenesis (Salas-Huetos et al. 2014). PiRNAs are 24–30 nt monocatenary RNA molecules. They are the most abundant sncRNA in both human and mice sperm transcriptomes (Pantano et al. 2015; Röther and Meister 2011). Their functionality is based on their attachment to PIWI proteins, which are exclusive of germ cells, to allow the posttranscriptional silencing of retrotransposons (Chuma and Nakano 2012). Accordingly, their biological function is in connection with a protective mechanism against genome modifications produced by transposable elements. Endo-siRNAs are 22 nt RNA molecules highly expressed in male germ cells (Song et al. 2011). The posttranscriptional gene regulatory function of endo-siRNAs is similar to the gene-silencing pathway of miRNAs. It is based on their attachment to 3’ UTR regions of target mRNAs (Song et al. 2011), which lead to the silencing or degradation of the mRNA sequences (Luo et al. 2015). In spermatozoa, these molecules control the expression of epigenetic regulators, such as histone methyltransferases, and promote the modification of chromatin conformation (Song et al. 2011). Additionally, some studies suggest that endo-siRNAs are necessary in postfertilization processes for the correct development of preimplantational embryos (Suh et al. 2010).
An Overview into the Concept of Transgenerational Inheritance
In human and animal models, several studies have demonstrated that the exposure to certain environmental factors in specific windows of the epigenome reprogramming affects the mechanisms that lead to the establishment of the sperm epigenome. Since the sperm epigenome is crucial for the proper fertility of the individuals, these variations have been related to male infertility (Camprubí et al. 2016). Moreover, it is becoming clear that some epimutations could be also transmitted via spermatozoa to offspring, which introduce the concept of epigenetic inheritance.
Inheritance of environmental-induced epigenetic changes is associated to the permanent transmission of epigenetic variations through the germline (Skinner 2008). In the case of exposure of a gestating F0 female, only the transmission of a phenotypic alteration until the third generation (F3) could be considered true transgenerational inheritance (Fig. 4.1). In this case, the germ cells of the F1 generation also carry the epigenetic variation induced in the gestating female (F0), which could also affect F1 gametes. Accordingly, F2 individuals could inherit the trait from an F0 gestating female. Therefore, in this model, the transmission until the F3 generation is required to assure that the results are the consequence of epigenetic transmission between cells unrelated to previous exposure effects (Fig. 4.1). When the exposure occurs in an adult male (F0), germ cells of the F1 generation could inherit the variation from the F0 spermatozoa. Thus, in this case, the first nonexposed generation involved would be F2 (Fig. 4.2). If the transmission of an epigenetic treat does not reach F3 (from a gestating female exposure) or F2 (from an adult male exposure), we talk about intergenerational or multigenerational inheritance.
Epigenetic inheritance of environmental-induced changes through the male germline. Transgenerational epigenetic inheritance from an exposed gestating female (F0) occurs when the transmission of a phenotypic alteration via spermatozoa reaches the third generation (F3); otherwise the mode of inheritance is classified as intergenerational
Epigenetic inheritance of environmental-induced changes through the male germline. Transgenerational epigenetic inheritance from the exposure in an adult individual (F0) occurs when the transmission of a phenotypic alteration via spermatozoa reaches the second generation (F2); otherwise the mode of inheritance is classified as intergenerational
In this context, it is important to remark that the only way to explain the transmission of any epigenetic variation induced by any agent between generations is the permanent reprogramming of germ cells. That is, the variation must be resistant to the resetting periods in which the epigenome is involved during the man’s life. This would guarantee stable transmission across generations.
From these premises, posttranslational modifications of histones and sncRNAs signature are epigenetic mechanisms that can hardly be associated with transgenerational epigenetic transmission. Concerning DNA methylation, it was assumed until a few years ago that the only regions that escaped from the global demethylation during epigenetic reprogramming were those regulated by genomic imprinting (Branco et al. 2008) and some repetitive noncoding DNA (Lane et al. 2003). Nevertheless, several pieces of data suggest that the number of regions is more extensive, affecting non-imprinted coding regions of the genome. For instance, it has been identified a group of CGI (Hackett et al. 2013; Seisenberger et al. 2012) and non-imprinted promoter sequences (Borgel et al. 2010) that resist the global DNA methylation reprogramming in the embryo. In human and mouse embryonic cells, it has been demonstrated the existence of single copy non-imprinted sequences resistant to reprogramming. Interestingly, these regions seem to be enriched in genes particularly active in the brain during adult life development (McGraw et al. 2015; Tang et al. 2015).
The existence of coding regions that escape DNA methylation epigenetic reprogramming points to the possibility of the existence of transgenerational epigenetic inheritance. That is, a part of the genome could be involved in the transgenerational epigenetic transmission of adult-onset disease phenotypes.
Environmental Factors Affect the Human Sperm Epigenome
There are several pieces of evidence demonstrating the influence of environmental factors over the sperm epigenome. Nevertheless, the molecular basis of this phenomenon is poorly understood and appears to be variable between inductor factors. In overall terms, the alteration of the sperm epigenetic signature has been associated to epigenetic insults in the development of PGCs that ultimately affects spermatozoa. Moreover, environmental factors could also disturb testis microenvironment that is crucial to accomplish the epigenetic mechanisms in germ cells during spermatogenesis. It is important to mention that epigenetic modifications associated to environmental factors mainly affect germ cells rather than spermatozoa since the sperm chromatin is a highly condensed structure and, therefore, highly resistant to environmental-induced perturbations.
Overall, the information provided in this section suggest that fetal, perinatal, or adult exposure of male germ cells to environmental factors has a detrimental effect on the sperm epigenome. Therefore, the fertility of the exposed individuals could be compromised. Furthermore, since some of the epimutations appear to be permanent, which is resistant to the reprogramming events, they could be transmitted to upcoming generations.
Age
Some authors have found a general increase of sperm DNA methylation with age (Camprubí et al. 2016; Jenkins et al. 2014). Since the negative influence of age on the testicular function and seminogram is well documented (Eisenberg and Meldrum 2017), it has been suggested that advanced age could alter the methylation marks of genes associated with male fertility. Actually, the influence of age over DNA methylation goes beyond male fertility. It has been described that DNA from blood of old individuals is more heterogeneous and hypomethylated in comparison with newborn DNA (Heyn et al. 2012).
It is interesting to remark that age-associated epigenome variations observed in human spermatozoa are specially associated to genes involved in neuropsychiatric disease in adult life (Jenkins et al. 2014). In a mouse model, a genome-wide DNA methylation study comparing sperm from young and old mice has revealed that the offspring of older fathers exhibited similar brain DNA methylation abnormalities than that observed in the paternal sperm (Milekic et al. 2014). Moreover, these methylation abnormalities are related to transcriptional dysregulation of developmental genes implicated in autism and schizophrenia (Milekic et al. 2014). These results suggest the possibility of transmission to the next generation of epimutations associated with brain disorders via spermatozoa.
Although the mechanisms that drive age-related methylation alterations in the sperm remain elusive, it appears that the rate of cell proliferation has a direct influence. It has been reported that highly proliferative cells exhibited a greater magnitude of age-associated DNA methylation changes (Thompson et al. 2010), while nondividing cells are less prone to these age effects (Chu et al. 2007). The high proliferation rate of spermatogonial germ cells along reproductive man lifespan made this cell type especially susceptible to age-related epigenetic alterations. It is possible that dividing cells are more prone to the accumulation of epimutations over time since they are exposed to errors during the transmission of the methylation marks in the S-phase of the cell cycle. As stated by other authors, further studies are required to determine whether the observed age-associated effects in spermatozoa are a consequence of the accumulation of epimutations in primordial germ cells or whether they are a consequence of testicular microenvironment perturbations related to advanced age (Oakes et al. 2003).
Obesity
Obesity may induce male infertility by a combination of different factors including endocrine abnormalities that ultimately affects the process of spermatogenesis and early embryogenesis (Du Plessis et al. 2010).
It is well documented that obese men had an increased incidence of sperm epimutations, which is interpreted by some authors as a contributing factor for male infertility. For instance, it has been described sperm DNA methylation differences at specific CpG of imprinted genes between overweight men and normal weight men (Soubry et al. 2016). In a sperm epigenome study from lean and obese men, a difference in small noncoding RNA expression and DNA methylation pattern was observed (Donkin et al. 2016). Moreover, morbidly obese men submitted to surgery-induced weight loss modifies the sperm epigenetic pattern (Donkin et al. 2016). In this regard, in an obesity mouse model, it has been demonstrated the differential abundance of different molecules of sperm microRNAs that have been ontologically associated with embryo development and metabolic and reproductive dysregulations in adulthood (Fullston et al. 2016).
The reason why obesity induces sperm epigenetic alterations has been related to different causes. Endocrine disruptions appear to be one of the most significant. Obesity has been associated with hypogonadism, leading to alterations of the testicular microenvironment that could interfere with the normal development of the sperm epigenome. In rat models, tamoxifen (estrogen receptor modulator) has been shown to reduce sperm DNA methylation at specific loci (Igf2/H19 differentially methylated region) through DNA methyltransferase 1 (Dnmt1 ) functional alterations in the testis. Hence, it is likely that this alteration could influence the proliferative phase of spermatogonial germ cells where Dnmt1 proteins are expressed abundantly, resulting in methylation errors in spermatozoa leading to male infertility (Pathak et al. 2009).
Other authors have related the presence of obesity-related epigenetic variations with an increased scrotal temperature, which led to testis hyperthermia and the subsequent reactive oxygen species (ROS) production. It has been described that DNA damage induced by oxidative stress could disturb the functionality of DNA methyltransferases (DNMTs ), resulting in methylome variations. DNA lesions affect the ability of DNA to function as a substrate for the DNMTs resulting in hypomethylation (Franco et al. 2008). Moreover, oxidative DNA damage leads to mutations preferably at methylated CpGs that would result in loss of epigenetic marks (Lee 2002). In this regard, ROS production has also been associated to hypermethylation of promoter regions of tumor suppression genes promoting carcinogenesis (Lim et al. 2008). Moreover, DNA damage induced by oxidative stress has also been implicated in the regulation of miRNA expression (Mateescu et al. 2011; Simone et al. 2009).
In animal models, it has been demonstrated a perturbed methylation pattern in the paternal pronuclei derived from heat-stressed spermatozoa (Rahman et al. 2014). In humans, it has been described that varicocele, which has been related to the exposure of sperm to heat, is associated with alterations of the sperm methylome (Bahreinian et al. 2015). Since an increased scrotal temperature is expected in obese men (because of sedentarism), testis heat stress and their detrimental effects on the sperm methylome are expected in obese men.
Endocrine Disruptors
Endocrine disruptors (ER) are a heterogeneous set of exogenous chemical substances capable of altering the regulation of the hormonal system. In reproduction, ER can disturb the regulation of the hypothalamic-pituitary-gonads axis and therefore alter the gonadal sex differentiation and gametogenesis, which ultimately lead to infertility.
In animal models, prenatal or perinatal exposure to relevant doses of ER leads to testis disease, ovarian disease, and pubertal abnormalities in adult individuals (Manikkam et al. 2013; Salian et al. 2011). The exposure to ER in mice causes changes in spermatogonia that result in meiotic alterations in the spermatogenesis of the adult male (Vrooman et al. 2015) that could result in a disruption in the progression of meiosis I and decreased sperm counts (Liu et al. 2013; Tiwari and Vanage 2013).
Since ER act at the time of the germ cell epigenome reprogramming, some authors have associated the exposure to ER to perturbation of the sperm epigenome, mainly by means of alterations of DNA methylation (Consales et al. 2016; Miao et al. 2014). ER would induce alterations of the testicular microenvironment and increase sperm DNA damage (Tiwari and Vanage 2013) that ultimately would perturb the epigenetic marks by affecting DNA methylation patterns.
Diet
It is well known that dietary compounds , such as phytochemicals, minerals and vitamins, can promote changes in epigenetic mechanisms of somatic as well as germ cells by influencing enzymes and other proteins responsible for epigenetic modifications (Schagdarsurengin and Steger 2016).
For instance, B vitamins must be provided by diet or supplementation and modulate the availability of methyl groups provided by the 1 Carbon Cycle, which is essential to ensure the availability of activated methyl groups for the methylation reactions of the cell. Methyl groups needed by methyltransferases are provided by S-adenosyl-L-methionine (SAM ) through the 1 Carbon Cycle. Thus, diet can influence the levels of DNA methylation and consequently affect gene expression. Other authors have reported an association between vitamin D deficiency and global dysregulation of the methylome via overexpression of DNA methyltransferase 3b (Dnmt3b ) transcripts (Xue et al. 2016).
In this regard, the influence of diet on the sperm epigenome has been demonstrated in several studies including humans (Schagdarsurengin et al. 2012), mainly through alteration of sperm DNA methylation (Aarabi et al. 2015; Lambrot et al. 2013). These variations have been associated with negative effects on the sperm quality that would affect the reproduction success of the couple.
Metabolic Disorders: Diabetes
Glucose metabolism is of great importance for sperm cell functionality. Diabetic disease has been associated with detrimental effects on male fertility, especially on sperm quality, sperm DNA integrity, and sperm epigenome dysregulations (Ding et al. 2015). In particular, alterations of the sperm methylome in paternal prediabetes individuals have been described (Wei et al. 2014).
Diabetes-induced testicular impairment due to its detrimental effect over testis microcirculation (Long et al. 2018). This detrimental effect increases the susceptibility of spermatogenic germ cells to generate ROS (Long et al. 2015). ROS generation in diabetic patients has been also associated with increased testicular temperature resulting from fat accumulation, which leads to testis hyperthermia (Wei et al. 2014). Among the collateral damage on male fertility induced by ROS, aberrant sperm DNA methylation is one of the most significant.
Chemotherapy
Those agents used to treat cancer that interfere with the process of DNA methylation or DNA replication have a severe impact over spermatogenesis (Chan et al. 2012; Doerksen et al. 2000; Doerksen and Trasler 1996; Kelly et al. 2003) and early embryo development (Doerksen et al. 2000; Kelly et al. 2003). When these treatments are of sufficient duration to affect the spermatogonia, the alterations of the sperm epigenome are permanent (Chan et al. 2012; Doerksen et al. 2000; Kelly et al. 2003). In this regard, adolescent chemotherapy exposure in patients with osteoblastoma has been related with sperm epimutations in adult life (Shnorhavorian et al. 2017). These results suggest that chemotherapy exposure causes permanent epigenetic alterations in the spermatogonial epigenome.
Alcohol
Although the association between alcohol intake and male infertility remains controversial (Martini et al. 2004), there is no doubt about the detrimental effect of alcohol consumption on DNA integrity due to the oxidative damage induced by consumption (Ellegaard and Poulsen 2016). This effect has been also found in male germ cell line (Aboulmaouahib et al. 2018). Since ROS is connected to alterations of DNA methylation, some authors have found sperm methylome variations in alcohol-exposed individuals (Liang et al. 2014; Ouko et al. 2009).
The alteration of the sperm methylome induced by alcohol intake has been also associated with decreases in the activity of DNA methyltransferase 1 (Dnmt1 ) (Bielawski et al. 2002; Garro et al. 1991) or reduced production of the methyl donor SAM (Sultana et al. 2015).
Smoking
Like alcohol intake, seminal quality is not clearly altered by cigarette consumption, although subtle modifications have been described suggesting an effect on male reproductive function (Martini et al. 2004).
There is a clear connection between tobacco and DNA oxidative damage as a consequence of the production of ROS (Ellegaard and Poulsen 2016). Moreover, smoking is known to cause ROS throughout spermatogenesis, which would affect the sperm DNA integrity (Aboulmaouahib et al. 2018), including some marginal effects on sperm DNA methylation (Al Khaled et al. 2018; Hamad et al. 2018; Laqqan et al. 2017).
Although the reason why smoking causes sperm DNA methylation variations deserves further investigation, some authors have identified a detrimental effect of nicotine on DNA methyltransferase expression (Satta et al. 2008). Moreover, cigarette smoke may alter DNA methylation via the interference of hypoxia (which is usual in smoker individuals) with the availability of SAM (Liu et al. 2011).
Epigenetic Mechanisms Are Strong Candidates for Transferring Paternal Environmental Information
In the last decade, several studies have addressed the analysis of the sperm epigenome as a vehicle for the transmission to offspring of epimutations induced by environmental factors (Table 4.1). Among the different epigenetic mechanisms, DNA methylation has been the most studied, probably because it has been proved that some sperm DNA methylation signatures escape the reprogramming events in the early embryo. Accordingly, at least a portion of the sperm DNA methylation variations induced by environmental factor has the potential to be retained in germ cells and be transmitted to the next generation.
Sperm Epimutations Affect Embryo Development
Several pieces of data suggest that sperm epigenome variations have a detrimental effect on embryo development, suggesting their fundamental role in postfertilization events. In humans, some authors have associated the presence of sperm DNA methylation variations and low pregnancy rate (Benchaib et al. 2005). Recently, Denommme et al. have described sperm DNA methylation differences at CGI contained in retained histone regions between good and poor blastocyst development groups (Denomme et al. 2017). In the case of histones, the fact that histones are retained in the promoters of developmental genes (Hammoud et al. 2009), and the fact that sperm histone modifications are transmitted to the embryo and are resistant to protein oocyte replacement (Van Der Heijden et al. 2008), argues in favor of an effect beyond fertilization. Finally, some data from sncRNAs demonstrated the importance of some sperm-borne miRNAs for early embryo development, suggesting that alteration of the sperm RNA cargo could be critical for the first cleavage events (Liu et al. 2012).
Sperm Epimutations Affect the Health of the Exposed Men and Their Offspring
Environmentally induced epigenetic inheritance refers to the transmission of epigenetic information through sperm cells in the absence of continuous exposure to the inductor agent. A great number of studies have addressed the issue of the transmission of epigenetic changes via spermatozoa through epigenetic perturbations of the germ line (Table 4.1). Most of the studies have analyzed this phenomenon using animal models, whereas in humans this phenomenon has been poorly studied. Several factors may induce epigenetic variations among which are endocrine disruptors, diet, exercise training, diabetes, alcohol, obesity, stress, smoking, dioxin, pesticide, hydrocarbon, and age.
As we stated before, the only way to explain the transmission of an induced epigenetic variation across generations is the permanent reprogramming of germ cells. That is, the variation must be resistant to the different reprogramming periods. This situation hardly will occur in the case of posttranslational modifications of histones and sncRNA signature, but it is possible for DNA methylation. The discovery of coding regions that escape DNA methylation epigenetic reprogramming points out the possibility of the participation of this mechanism in transgenerational epigenetic inheritance events. Therefore, DNA methylation is, by far, the most studied epigenetic mechanisms in transgenerational studies (Table 4.1).
It is important to mention again that only the transmission of a phenotypic alteration via spermatozoa until the third generation (in the case of exposure of a gestating F0 female; Fig. 4.1), or the second generation (when the exposure occurs in an adult individual; Fig. 4.2), could be considered true transgenerational inheritance (Skinner 2008).
Intergenerational Inheritance
Intergenerational analysis has been performed in 10 different studies, 5 from a gestating female (Ding et al. 2012; Lambrot et al. 2013; Martínez et al. 2014; Radford et al. 2014; Xue et al. 2016) and 5 from the exposure of an adult male (Carone et al. 2010; Finegersh and Homanics 2014; Fullston et al. 2016; Liang et al. 2014; Milekic et al. 2014).
In the studies from a gestating female, authors demonstrated the transmission of phenotypic alterations to the F2 generation, including metabolic and body weight alterations which in all cases were related with the inducing factor (diabetes and diet) (Ding et al. 2012; Lambrot et al. 2013; Martínez et al. 2014; Radford et al. 2014; Xue et al. 2016). In three studies, authors found that the same epimutations observed in spermatozoa were maintained, at least in part, in somatic tissues of the following generation, reinforcing the interpretation of epigenetic inheritance (Ding et al. 2012; Lambrot et al. 2013; Martínez et al. 2014).
The remaining five studies were designed from exposures of adult males (Carone et al. 2010; Finegersh and Homanics 2014; Fullston et al. 2016; Liang et al. 2014; Milekic et al. 2014). Again, the authors observed phenotypic effects in offspring related to the inducing agent. Concerning the postulation of the sperm cell as a vehicle for transmission, all studies except one (Carone et al. 2010) demonstrated sperm epigenome variations. Two works demonstrated the presence of the same sperm methylome variation in sperm and somatic tissues from next generation (Liang et al. 2014; Milekic et al. 2014).
Transgenerational Inheritance
Eight transgenerational studies have been published so far, five from the exposure to a gestating female (Anway et al. 2005; Manikkam et al. 2012a, b, Manikkam et al. 2013; Tracey et al. 2013) and three from adult male exposure (de Castro Barbosa et al. 2016; Gapp et al. 2014; Wei et al. 2014).
In the cases of gestational female exposure (Anway et al. 2005; Manikkam et al. 2012a, b, Manikkam et al. 2013; Tracey et al. 2013), authors demonstrated the transmission of phenotypic alterations (including alterations of the reproductive system, kidney disease, and obesity) until the F3 generation. Moreover, all these studies found sperm DNA methylation variations in F3 spermatozoa, suggesting that the transmission of the epigenetic phenotypic alteration is associated to sperm methylome variations that are not reprogrammed across generations. It is important to mention that, in none of them, the authors analyzed if the epimutations observed in spermatozoa were also present in somatic tissues of the subsequent generation.
Three more studies demonstrated transgenerational inheritance in adult male exposures (F2 phenotypic alterations related to the inducing agent) (de Castro Barbosa et al. 2016; Gapp et al. 2014; Wei et al. 2014). In all the cases, authors identify sperm and somatic epigenome variations associated with the inducing factor that would explain the phenotypic alteration observed in F2 individuals. These results are highly indicative of true transgenerational inheritance.
Overall, the revision of the literature performed in the present manuscript (Table 4.1) demonstrated the existence of strong evidences about the presence of epigenetic inheritance via spermatozoa. Nevertheless, we must be cautious in the interpretation of the results. It is important to mention that most of the studies only provide partial evidences about this phenomenon. In this sense, a study demonstrating unequivocally the presence of transgenerational inheritance via spermatozoa is currently lacking. This study must accomplish the following requirements: (i) to identify sperm epigenome variations induced by environmental factors, (ii) to demonstrate the transmission of epigenome variations from sperm to somatic tissues, (iii) to identify phenotypic effects associated to the presence of epimutations, and iv) to demonstrate the presence of the same epimutations in sperm and somatic tissues at least until the first nonexposed generation.
Is Multigenerational Disease Prevention a New Paradigm?
From the information provided in the preceding paragraphs, it becomes clear that the exposure to certain environmental factors in specific windows of sperm development influences the risk of developing chronic diseases and behavior disorders in adulthood. These studies support the intriguing idea that human beings could adapt the expression of genes to environmental signals. That is, epigenetic plasticity would provide the ability for adaptation to the current environment in individuals of equal genotype. Accordingly, an area of research that could be crucial in the near future regards the possibility to prevent the onset of epigenetic-based diseases through the modulation of the sperm epigenome in the previous generation. That is, the modification of lifestyle factors driving to sperm epimutations could be a powerful tool to normalize the sperm epigenome and avoid their negative consequences.
Some evidence suggests the veracity of this possibility. For instance, in a mouse model, it has been demonstrated that diet or exercise training in obese males restores insulin sensitivity and normalized adiposity in female offspring. These modifications are associated with the normalization of sperm microRNA pattern, suggesting that diet and/or exercise normalize aberrant epigenetic signals in sperm and improve the metabolic health of offspring (McPherson et al. 2015). In humans, it has been demonstrated that exercise training modified the sperm DNA methylation mark of genes related to schizophrenia and Parkinson’s disease (Denham et al. 2015). Also, surgery-induced weight loss has been associated with a remodeling of sperm DNA methylation, especially at genetic locations implicated in the central control of appetite (Donkin et al. 2016).
Concluding Remarks
The sperm epigenome is the result of the different periods of epigenome reprogramming in germ cells. These reprogramming events have the main function to develop totipotent cells and to prevent the transmission of epimutations via spermatozoa. At the end of these reprogramming events, spermatozoa carry a distinctive epigenome, which is a footprint of spermatogenesis events and is programmed to allow embryogenesis and to influence in adult life.
Since the sperm epigenome is sensitive to numerous environmental factors, it is clearly susceptible to variations. The discovery of coding regions that escape DNA methylation epigenetic reprogramming points to the possibility of the transmission of epigenetic variation between generations (induced by environmental factors) and hence, to the existence of transgenerational epigenetic inheritance. In animal models, there are strong evidences about the presence of transgenerational epigenetic inheritance via spermatozoa. Nevertheless, a complete study unequivocally demonstrating this kind of transmission is currently lacking.
The high plasticity of the sperm epigenome opens the possibility of its modulation through the modification of lifestyle factors. This is a very promising area in the field of reproductive epigenetics, that is, the analysis of the normalizer effect of changes in lifestyle factors on the sperm epigenome as a tool to overcome some types of male infertility.
References
Aarabi M, San Gabriel MC, Chan D, Behan NA, Caron M, Pastinen T, Bourque G, MacFarlane AJ, Zini A, Trasler J (2015) High-dose folic acid supplementation alters the human sperm methylome and is influenced by the MTHFR C677T polymorphism. Hum Mol Genet 24:6301–6313
Aboulmaouahib S, Madkour A, Kaarouch I, Sefrioui O, Saadani B, Copin H, Benkhalifa M, Louanjli N, Cadi R (2018) Impact of alcohol and cigarette smoking consumption in male fertility potential: looks at lipid peroxidation, enzymatic antioxidant activities and sperm DNA damage. Andrologia 50:e12926
Al Khaled Y, Tierling S, Laqqan M, Lo Porto C, Hammadeh ME (2018) Cigarette smoking induces only marginal changes in sperm DNA methylation levels of patients undergoing intracytoplasmic sperm injection treatment. Andrologia 50:e12818
Amanai M, Brahmajosyula M, Perry ACF (2006) A restricted role for sperm-borne MicroRNAs in mammalian fertilization. Biol Reprod 75:877–884
Anway MD, Cupp AS, Uzumcu M, Skinner MK (2005) Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–1469
Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D (2009) Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 19:1338–1349
Bahreinian M, Tavalaee M, Abbasi H, Kiani-Esfahani A, Shiravi AH, Nasr-Esfahani MH (2015) DNA hypomethylation predisposes sperm to DNA damage in individuals with varicocele. Syst Biol Reprod Med 61:179–186
Bao J, Wu J, Schuster AS, Hennig GW, Yan W (2013) Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol Reprod 89:107
Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, Guérin JF (2005) Influence of global sperm DNA methylation on IVF results. Hum Reprod 20:768–773
Bielawski DM, Zaher FM, Svinarich DM, Abel EL (2002) Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res 26:347–351
Boerke A, Dieleman SJ, Gadella BM (2007) A possible role for sperm RNA in early embryo development. Theriogenology 68(Suppl 1):S147–S155
Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M (2010) Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42:1093–1100
Branco MR, Oda M, Reik W (2008) Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev 22:1567–1571
Brunner AM, Nanni P, Mansuy IM (2014) Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin 7(2)
Camprubí C, Salas-Huetos A, Aiese-Cigliano R, Godo A, Pons MC, Castellano G, Grossmann M, Sanseverino W, Martin-Subero JI, Garrido N, Blanco J (2016) Spermatozoa from infertile patients exhibit differences of DNA methylation associated with spermatogenesis-related processes: an array-based analysis. Reprod Biomed Online 33:709–719
Camprubí C, Aiese-Cigliano R, Salas-Huetos A, Garrido N, Blanco J (2017) What the human sperm methylome tells us. Epigenomics 9:1299–1315
Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ (2010) Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143:1084–1096
Chan D, Delbe’s G, Landry M, Robaire B, Trasler JM (2012) Epigenetic alterations in sperm DNA associated with testicular cancer treatment. Toxicol Sci 125:532–543
Chu MW, Siegmund KD, Eckstam CL, Kim JY, Yang AS, Kanel GC, Tavaré S, Shibata D (2007) Lack of increases in methylation at three CpG-rich genomic loci in non-mitotic adult tissues during aging. BMC Med Genet 8:50
Chuma S, Nakano T (2012) piRNA and spermatogenesis in mice. Philos Trans R Soc London B Biol Sci 368:20110338
Chuva de Sousa Lopes SM, Roelen BAJ (2010) On the formation of germ cells: the good, the bad and the ugly. Differentiation 79:131–140
Consales C, Toft G, Leter G, Bonde JPE, Uccelli R, Pacchierotti F, Eleuteri P, Jönsson BAG, Giwercman A, Pedersen HS, Struciński P, Góralczyk K, Zviezdai V, Spanò M (2016) Exposure to persistent organic pollutants and sperm DNA methylation changes in Arctic and European populations. Environ Mol Mutagen 57:200–209
Corral-Vazquez C, Anton E (2018) Epigenetic and assisted reproduction experimental studies: sperm ncRNAs. In: Blanco J, Camprubí C (eds) Epigenetics and assisted reproduction an introductory guide. Taylor & Francis, Boca Raton
Davis TL, Yang GJ, McCarrey JR, Bartolomei MS (2000) The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9:2885–2894
de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, Donkin I, Sjögren R, Mudry JM, Vetterli L, Gupta S, Krook A, Zierath JR, Barrès R (2016) High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab 5:184–197
de Mateo S, Sassone-Corsi P (2014) Regulation of spermatogenesis by small non-coding RNAs: role of the germ granule. Semin Cell Dev Biol 29:84–92
Denham J, O’Brien BJ, Harvey JT, Charchar FJ (2015) Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics 7:1–15
Denomme MM, McCallie BR, Parks JC, Schoolcraft WB, Katz-Jaffe MG (2017) Alterations in the sperm histone-retained epigenome are associated with unexplained male factor infertility and poor blastocyst development in donor oocyte IVF cycles. Hum Reprod 32:1–13
Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, Leung PCK, Sheng JZ, Huang HF (2012) Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes 61:1133–1142
Ding G-L, Liu Y, Liu M-E, Pan J-X, Guo M-X, Sheng J-Z, Huang H-F (2015) The effects of diabetes on male fertility and epigenetic regulation during spermatogenesis. Asian J Androl 17:948
Doerksen T, Trasler JM (1996) Developmental exposure of male germ cells to 5-Azacytidine results in abnormal preimplantation development in rats. Biol Reprod 55:1155–1162
Doerksen T, Benoit G, Trasler JM (2000) Deoxyribonucleic acid hypomethylation of male germ cells by mitotic and meiotic exposure to 5-azacytidine is associated with altered testicular histology. Endocrinology 141:3235–3244
Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, Mortensen B, Appel EVR, Jørgensen N, Kristiansen VB, Hansen T, Workman CT, Zierath JR, Barrès R (2016) Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab 23:369–378
Du Plessis SS, Cabler S, McAlister DA, Sabanegh E, Agarwal A (2010) The effect of obesity on sperm disorders and male infertility. Nat Rev Urol 7:153–161
Eisenberg ML, Meldrum D (2017) Effects of age on fertility and sexual function. Fertil Steril 107:301–304
Ellegaard PK, Poulsen HE (2016) Tobacco smoking and oxidative stress to DNA: a meta-analysis of studies using chromatographic and immunological methods. Scand J Clin Lab Invest 76:151–158
Finegersh A, Homanics GE (2014) Paternal alcohol exposure reduces alcohol drinking and increases behavioral sensitivity to alcohol selectively in male offspring. PLoS One 9:e99078
Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI (2008) Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 266:6–11
Fullston T, Ohlsson Teague EM, Palmer NO, DeBlasio MJ, Mitchell M, Corbett M, Print CG, Owens JA, Lane M (2013) Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J 27:4226–4243
Fullston T, Ohlsson-Teague EMC, Print CG, Sandeman LY, Lane M (2016) Sperm microRNA content is altered in a mouse model of male obesity, but the same suite of microRNAs are not altered in offspring’s sperm. PLoS One 11:e0166076
Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM (2014) Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17:667–669
Garro AJ, McBeth DL, Lima V, Lieber CS (1991) Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res 15:395–398
Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW (1987) Sequence-specific packaging of DNA in human sperm chromatin. Science 236:962–964
Ge ZJ, Liang QX, Hou Y, Han ZM, Schatten H, Sun QY, Zhang CL (2014) Maternal obesity and diabetes may cause DNA methylation alteration in the spermatozoa of offspring in mice. Reprod Biol Endocrinol 12:29
Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK (2010) Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 5:e13100
Guibert S, Forne T, Weber M (2012) Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22:633–641
Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339:448–452
Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA (2002) Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117:15–23
Hamad MF, Dayyih WAA, Laqqan M, AlKhaled Y, Montenarh M, Hammadeh ME (2018) The status of global DNA methylation in the spermatozoa of smokers and non-smokers. Reprod Biomed Online 37:581–589
Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478
Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, Puca AA, Sayols S, Pujana MA, Serra-Musach J, Iglesias-Platas I, Formiga F, Fernandez AF, Fraga MF, Heath SC, Valencia A, Gut IG, Wang J, Esteller M (2012) Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci 109:10522–10527
Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Wu X, Pfeifer GP, Szabó PE (2015) Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genom Biol 16:59
Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT (2014) Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet 10:e1004458
Jenkins TG, James ER, Alonso DF, Hoidal JR, Murphy PJ, Hotaling JM, Cairns BR, Carrell DT, Aston KI (2017) Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 5:1089–1099
Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA (2013) The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update 19:604–624
Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M (2013) Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32:340–353
Kelly TLJ, Li E, Trasler JM (2003) 5-Aza-2′-Deoxycytidine induces alterations in murine spermatogenesis and pregnancy outcome. J Androl 24:822–830
Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Pàldi A, Jouannet P, Jeanpierre M (2000) Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet 9:2183–2187
Kota SK, Feil R (2010) Epigenetic transitions in germ cell development and meiosis. Dev Cell 19:675–686
Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, Esteller M (2012) Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS One 7:e44479
Lambrot R, Xu C, Saint-Phar S, Chountalos G, Cohen T, Paquet M, Suderman M, Hallett M, Kimmins S (2013) Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat Commun 4:2889
Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W (2003) Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35:88–93
Laqqan M, Tierling S, Alkhaled Y, Porto CL, Solomayer EF, Hammadeh ME (2017) Aberrant DNA methylation patterns of human spermatozoa in current smoker males. Reprod Toxicol 71:126–133
Lee DH (2002) Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG -->TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. Nucleic Acids Res 30:3566–3573
Liang F, Diao L, Liu J, Jiang N, Zhang J, Wang H, Zhou W, Huang G, Ma D (2014) Paternal ethanol exposure and behavioral abnormities in offspring: associated alterations in imprinted gene methylation. Neuropharmacology 81:126–133
Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM, Jung G (2008) Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology 135:2128–2140
Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z (2011) Hypoxia induces genomic DNA demethylation through the activation of HIF-1 and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther 10:1113–1123
Liu WM, Pang RTK, Chiu PCN, Wong BPC, Lao K, Lee KF, Yeung WSB (2012) Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci 109:490–494
Liu C, Duan W, Li R, Xu S, Zhang L, Chen C, He M, Lu Y, Wu H, Pi H, Luo X, Zhang Y, Zhong M, Yu Z, Zhou Z (2013) Exposure to bisphenol a disrupts meiotic progression during spermatogenesis in adult rats through estrogen-like activity. Cell Death Dis 4:e676
Long L, Wang J, Lu X, Xu Y, Zheng S, Luo C, Li Y (2015) Protective effects of scutellarin on type II diabetes mellitus-induced testicular damages related to reactive oxygen species/Bcl-2/Bax and reactive oxygen species/microcirculation/staving pathway in diabetic rat. J Diabetes Res 2015:252530
Long L, Qiu H, Cai B, Chen N, Lu X, Zheng S, Ye X, Li Y (2018) Hyperglycemia induced testicular damage in type 2 diabetes mellitus rats exhibiting microcirculation impairments associated with vascular endothelial growth factor decreased via PI3K/Akt pathway. Oncotarget 9:5321–5336
Luo LF, Hou CC, Yang WX (2015) Small non-coding RNAs and their associated proteins in spermatogenesis. Gene 578:141–157
Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK (2012a) Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One 7:e46249
Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK (2012b) Pesticide and insect repellent mixture (permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod Toxicol 34:708–719
Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK, Gounon P (2013) Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 8:e55387
Martínez D, Pentinat T, Ribó S, Daviaud C, Bloks VW, Cebrià J, Villalmanzo N, Kalko SG, Ramón-Krauel M, Díaz R, Plösch T, Tost J, Jiménez-Chillarón JC (2014) In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metab 19:941–951
Martini AC, Molina RI, Estofán D, Senestrari D, Fiol De Cuneo M, Ruiz RD (2004) Effects of alcohol and cigarette consumption on human seminal quality. Fertil Steril 82:374–377
Mateescu B, Batista L, Cardon M, Gruosso T, De Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F (2011) miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med 17:1627–1635
McGraw S, Zhang JX, Farag M, Chan D, Caron M, Konermann C, Oakes CC, Mohan KN, Plass C, Pastinen T, Bourque G, Chaillet JR, Trasler JM (2015) Transient DNMT1 suppression reveals hidden heritable marks in the genome. Nucleic Acids Res 43:1485–1497
McPherson NO, Owens JA, Fullston T, Lane M (2015) Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring. Am J Physiol Endocrinol Metab 308:E805–E821
Miao M, Zhou X, Li Y, Zhang O, Zhou Z, Li T, Yuan W, Li R, Li DKK (2014) LINE-1 hypomethylation in spermatozoa is associated with Bisphenol A exposure. Andrology 2:138–144
Milekic MH, Xin Y, O’Donnell A, Kumar KK, Bradley-Moore M, Malaspina D, Moore H, Brunner D, Ge Y, Edwards J, Paul S, Haghighi FG, Gingrich J a (2014) Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry 20:1–7
Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD (2011) Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146:1029–1041
Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B (2003) Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci U S A 100:1775–1780
Oliva R, Castillo J, Estanyol J, Ballescà J (2015) Human sperm chromatin epigenetic potential: genomics, proteomics, and male infertility. Asian J Androl 17:601
Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M (2009) Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes – implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res 33:1615–1627
Pantano L, Jodar M, Bak M, Ballescà JL, Tommerup N, Oliva R, Vavouri T (2015) The small RNA content of human sperm reveals pseudogene-derived piRNAs complementary to protein-coding genes. RNA 21:1085–1095
Pathak S, Kedia-Mokashi N, Saxena M, D’Souza R, Maitra A, Parte P, Gill-Sharma M, Balasinor N (2009) Effect of tamoxifen treatment on global and insulin-like growth factor 2-H19 locus-specific DNA methylation in rat spermatozoa and its association with embryo loss. Fertil Steril 91:2253–2263
Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, Peters AH, Patti ME, Ferguson-Smith AC (2014) In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 345:1255903
Rahman MB, Kamal MM, Rijsselaere T, Vandaele L, Shamsuddin M, Van Soom A (2014) Altered chromatin condensation of heat-stressed spermatozoa perturbs the dynamics of DNA methylation reprogramming in the paternal genome after in vitro fertilisation in cattle. Reprod Fertil Dev 26:1107–1116
Röther S, Meister G (2011) Small RNAs derived from longer non-coding RNAs. Biochimie 93:1905–1915
Salas-Huetos A, Blanco J, Vidal F, Mercader JM, Garrido N, Anton E (2014) New insights into the expression profile and function of micro-ribonucleic acid in human spermatozoa. Fertil Steril 102:213–222
Salian S, Doshi T, Vanage G (2011) Perinatal exposure of rats to Bisphenol A affects fertility of male offspring-an overview. Reprod Toxicol 31:359–362
Sasaki H, Matsui Y (2008) Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 2008:129–140
Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, Guidotti A (2008) Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci 105:16356–16361
Schagdarsurengin U, Steger K (2016) Epigenetics in male reproduction: effect of paternal diet on sperm quality and offspring health. Nat Rev Urol 13:584–595
Schagdarsurengin U, Paradowska A, Steger K (2012) Analysing the sperm epigenome: roles in early embryogenesis and assisted reproduction. Nat Rev Urol:1–11
Seisenberger S, Andrews S, Krueger F, Arand J, Walter JJJ, Santos FF, Popp C, Thienpont B, Dean W, Reik W (2012) The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48:849–862
Shnorhavorian M, Schwartz SM, Stansfeld B, Sadler-Riggleman I, Beck D, Skinner MK (2017) Differential DNA methylation regions in adult human sperm following adolescent chemotherapy: potential for epigenetic inheritance. PLoS One 12:e0170085
Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, DeGraff W, Cook J, Harris CC, Gius D, Mitchell JB (2009) Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One 4:e6377
Skinner MK (2008) What is an epigenetic transgenerational phenotype?. F3 or F2. Reprod Toxicol 25:2–6
Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W (2011) Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci U S A 108:13159–13164
Soubry A, Guo L, Huang Z, Hoyo C, Romanus S, Price T, Murphy SK (2016) Obesity-related DNA methylation at imprinted genes in human sperm: results from the TIEGER study. Clin Epigenetics 8:51
Stouder C, Paoloni-Giacobino A (2010) Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction 139:373–379
Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R (2010) MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20:271–277
Sultana S, Zulkifle M, Ansari A, Shahnawaz (2015) Efficacy of local application of an Unani formulation in acne vulgaris. Anc Sci Life 35:124
Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA (2015) A unique gene regulatory network resets the human germline epigenome for development. Cell 161:1453–1467
Thompson RF, Atzmon G, Gheorghe C, Liang HQ, Lowes C, Greally JM, Barzilai N (2010) Tissue-specific dysregulation of DNA methylation in aging. Aging Cell 9:506–518
Tiwari D, Vanage G (2013) Mutagenic effect of Bisphenol A on adult rat male germ cells and their fertility. Reprod Toxicol 40:60–68
Tracey R, Manikkam M, Guerrero-Bosagna C, Skinner MK (2013) Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod Toxicol 36:104–116
Van Der Heijden GW, Ramos L, Baart EB, Van Den Berg IM, Derijck AA, Van Der Vlag J, Martini E, De Boer P (2008) Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol 8:34
Vrooman LA, Oatley JM, Griswold JE, Hassold TJ, Hunt PA (2015) Estrogenic exposure alters the spermatogonial stem cells in the developing testis, permanently reducing crossover levels in the adult. PLoS Genet 11:e1004949
Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY (2014) Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A 111:1873–1878
Xue J, Schoenrock SA, Valdar W, Tarantino LM, Ideraabdullah FY (2016) Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clin Epigenetics 8:107
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Blanco Rodríguez, J., Camprubí Sánchez, C. (2019). Epigenetic Transgenerational Inheritance. In: Baldi, E., Muratori, M. (eds) Genetic Damage in Human Spermatozoa. Advances in Experimental Medicine and Biology, vol 1166. Springer, Cham. https://doi.org/10.1007/978-3-030-21664-1_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-21664-1_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-21663-4
Online ISBN: 978-3-030-21664-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)