Abstract
Alzheimer Disease (AD) is progressive mental deterioration disease results in memory loss issues. Alzheimer is not treatable at the severe stage, but an accurate and earlier identification can aid signs and have great clinical significance. In this chapter, Machine Learning techniques are utilized for the identification purpose for Alzheimer patients. The proposed method extract features from Magnetic Resonance Images (MRI) without segmentation. An accuracy of 94.2% is attained for multiclass classification using random forest approach. The results prove that there is no need for segmentation and hence the process gets robust while keeping the accuracy high. The proposed method is evaluated on OASIS dataset. The computational time for Alzheimer detection is 205 ms and 56 ms for segmentation-based detection and the proposed method respectively.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Alzheimer’s disease (AD) is the type of dementia, which damages the brain’s cells and causes memory loss. This ultimately results that a patient is not able to do simple daily errands [1]. There is no appropriate medical remedy has been found yet and the reason of AD is insufficiently comprehended [2, 3]. As per an approximation, in 2015 about 5.3 million American is going through the AD. In 2050, the digit is probably going high up to 16 million [4]. The life quality of AD patients can be improved by early identification of this disease. At present time for medical diagnosis, MRI is extensively utilized in hospitals for the AD because of its remarkable resolution, good contrast and high availability [5, 6]. The region of interest (ROI), Volume of interest (VOI) [7,8,9], the medial temporal lobe, Gray matter (GM) voxels in the image segmentation [10], and hippocampus measurement and morphometric methods, structural MRI has been utilized for the extraction and classification of AD features [11,12,13]. Regardless of the enhancement in Initial Identification of AD, structural MRI remains a challenging task for the prediction of progression of ailment and needs further exploration.
A cautious medical assessment is needed for the finding of the AD. This includes physical, neuro-biological exams, and clinical dementia rating (CDR) which is a 5-point scale used to characterize six domains of cognitive and functional performance and mini-mental state examination (MMSE) questionnaire. Moreover, examining structural and functional variation in the cerebrum, non-invasive approaches like resting-state functional MRI (rs-fMRI) and structural MRI have been utilized [14, 15]. The tasks including reminiscence, preparation, thinking, and ruling gets effect because of the changes in the brain and hippocampus. The stage of diseases progression changes the number of different cerebrum areas. Essentially expanded ventricles and an extreme volume decrease of the hippocampus and cerebral cortex can be easily detected using MRI images.
Developing an algorithm, which can distinguish between an AD patient and normal person is of extraordinary significance to the clinicians [16]. In this work, we present a statistical-based analysis, in which MRI images are divided into three different sections Axial, Coronal, and Sagittal. Histogram of oriented gradient (HOG) descriptor, local binary pattern, and Gray-level co-occurrence matrix method (GLCM) techniques are utilized without segmentation MRI images to extract features. For AD diagnosis, multiclass classification is performed by providing these enhanced features as an input to classifiers. Our contribution to this chapter has described in the following points:
-
Features based classification methods are proposed using statistical features
-
Using the whole image without segmentation and produces enhanced results in multiclass classification.
The remaining chapter is structured as Sect. 20.2 presents related work, Sect. 20.3 shows methodology and Sect. 20.4 shows results section followed by a conclusion.
2 Related Work
Many contributions have been done by the researcher in the detection of Alzheimer’s disease. Ateeq et al. [17] proposed an efficient approach for the detection of cerebral microbleeds (CMB) in brain MRI, as this CMB is the essential indicator for the detection of dementia. This approach is comprised of three phases including brain extraction, extraction of the initial candidate based on the threshold and the last one is feature extraction and classification using Support Vector Machine, Quadric Discriminant Analysis (QDA) and ensemble methods. They attained the best sensitivity of 93.7% by utilizing QDA classifier.
In [1], five phases were used, wherein the first stage images were pre-processed, and segmentation was used to divide the image into a gray and cerebrospinal fluid and white matter. The second phase is to build similarity, the GM segmented ROIs were utilized through features and separated in the next phase. To categorize ADs patients and healthy subjects, 3-Dimensional displacement field was utilized in [2]. Features were extracted using three approaches i.e. Bhattacharya distance, Welch’s t-test. Voxel-based morphometry (VBM) is used to find Global and Local atrophy in AD patients vs normal subjects [18].
In [19], Sections having remarkable GM atrophy variations were chosen as the volume of interest (VOIs) and GM volume was perceived by using VBM for the AD and normal subjects. The voxel values separated from the sections that were perceived as raw features are next assessed using seven distinct features grading approaches i.e. Fisher’s criterion, T-test score, statistical dependency, mutual information, information gain, the Gini-index, and Pearson correlation coefficient. SVM was used for classification and achieved 92.4% accuracy. To classify the AD a Laplace Beltrami eigenvalue form descriptor was utilized in [20]. Segmenting T1 weighted MRI scans, the shape change of corpus callosum were examined through reaction diffusion level set technique. Information gain ranking was used to select the significant features and then subjected to classification using SVM and K-nearest neighbor (KNN). KNN classifier gives 93.37% accuracy. On the other hand, the difficulty of quantifying differences in the micro arrangements of corpus callosum makes this the technique less beneficial.
In [21], an outline for the extraction of features from low aspect sub-spaces was proposed. To build the manifold sub-space, Data-driven ROI were used. To learn these regions, MMSE score with sparse regression was used. For performing the variable selection, the sampling bias was abridged accompanied by resampling arrangement using sparse regression. The accuracy of classification achieved was 71%. A classification technique for the AD and cognitively normal (CN) classification, named sulcal medial surface was also proposed [22]. To take out 24 different sulci from every individual subject, Brain-VISA sulcal identification pipeline was utilized and SVM was used to classify AD and CN and attained 87.9% accuracy. To make a bio-marker, different dimensions like hippocampus texture, cortical thickness, and shape were combined, that utilized information from MRI data [23]. The accuracy of multiclass classification using LDA was 62.7% for ADNI [24].
In [25], a deep learning-based method was proposed to classify AD subjects. To foresee the output classes as AD patients and healthy, auto-encoders were used together with a convolutional neural network (CNN) and reported an accuracy of 98.7%. CNN has been used to excerpt discriminative features for classification of the AD and healthy persons [26, 27]. In big data analysis, deep learning methods have attained remarkable outcomes. However, a lot of training and a large number of computational power is required for gathering useful information from a large collection of unstructured data [28]. This is also a difficult task to select optimal-hyper parameters and best structural design.
Most strategies reported in the literature highlights that the limitation in classification is due to features taken out directly from the brain images. Both binary and multiclass classification is still an open research problem. In this study, the binary and multiclass classification performance is improved by using clinical information along with features.
3 Proposed Methodology
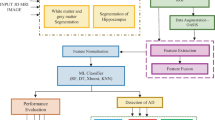
Three main segments are proposed in this method, specifically the preprocessing phase, feature, and results. The complete process is shown in Fig. 20.1.
3.1 Preprocessing
In this phase, MR images are firstly sliced to get three different angles of that image—Axial, Coronal, and Sagittal. Features of all these three angles are extracted separately to make a feature vector.
3.2 Feature Extraction
3.2.1 Gray-Level Co-occurrence Matrix
The Gray-level co-occurrence matrix (GLCM) was firstly analyzed by Haralick in 1973 [1]. The algorithm then analyzed and fourteen texture based features were proposed known as Gray-level co-occurrence features [2]. Normally in images, pixels are highly correlated because of the same levels of gray in a specific region. In a specified area, the frequency of pixels that occur with value ‘j’ having spatial relation with pixels value ‘p’ was computed by GLCM [3, 4].
3.2.1.1 Contrast
The contrast measures the local difference in GLCM and calculated as:
Contrast is the ‘sum of square variance’. As (i − j) values increases, the contrast also continues to increase but there is no increase in contrast when the values go equivalent.
3.2.1.2 Correlation
Correlation measures the joint probability and is calculated as:
Images that are correlated have μ 1, μ 2 . as the means and σ 1, σ 2 as the variance value.
3.2.1.3 Homogeneity
It measures the closeness of elements i.e. how close the GLCM elements are to its diagonal. The weight of homogeneity value is opposite to the weight of contrast and can be measured as:
3.2.1.4 Entropy
It measures image disorder, when the image has non-uniform texture, entropy value is high. Entropy is calculated as:
3.3 Classification
A multiclass classification of ADs without segmentation has been proposed. Different classifiers including the SVM, Ensembles method, and Random Forest were trained to classify the data and to validate the efficiency of the extracted features. All these classifiers are separately trained on normal, very mild, mild and severe AD according to their CDR values.
4 Experimental Results and Discussion
In this section, the details for dataset, experimental setup and results are reported in detail.
4.1 Dataset
The dataset has been taken from the OASIS database which is openly accessible to be utilized for research objective for AD classifications.
This set contains a total of 416 subject’s stage adult (18) to old age (96) years old. For every subject, three to four persons contained MRI (T1-weighted) scans captured in single sessions. We have not used the images with missing information. Around 100 of subjects are found from with very mild to the mild AD. We limited our research to right-handed participants, both men, and women. These participants were categorized based on CDR values i.e. clinical dementia rating, as having normal cognition (CDR = 0) i.e. healthy subject or early-stage AD (CDR = 0.5). The set also has age, sex (male/female), socioeconomic status (SES), mini-mental state examination (MMSE) and estimated total interaction volume (eTIV). Demographic physiognomies of the individuals are presented in Table 20.1.
4.2 Result and Discussion
This proposed approach is evaluated using accuracy and precision [5,6,7, 29,30,31,32]. Models were trained and then validated by using fivefold cross-validation. Different classifiers were utilized to evaluate the performance of the obtained features. The main classifiers utilized were supported vector machine (SVM), ensemble methods, naïve Bayes, and random forest.
Random Forest was trained with 100 numbers of iterations and gave the best accuracy of 94.2%. Three different SVM kernels—Linear, Poly-linear, and RBF were used to evaluate the results. The output accuracy for polylinear kernel function, linear kernel and RBF is 92.9%, 92.7%, and 90.5% respectively. Ensemble methods were trained by using Adaboost (boosting) technique and attained 83% accuracy.
A good accuracy has been achieved for classification of binary class but multiclass classification is still tough errand as multiclass classification has been a major issue in Alzheimer’s subjects. The results for multiclass classification are presented in the table and obtained by using features extracted from MR images. It is evident that from all the classifiers used, Random Forest generated best results.
Table 20.2 shows that the highest classification accuracy was achieved using Random Forest classifier. We compared the computational time for a clustering-based method and our proposed algorithm. The results in terms of computational time are 205 ms and 56 ms respectively.
4.2.1 Importance in Multimedia Applications
Multimedia applications are based on both image-based and audio-based applications. Image processing has seen an enormous growth in the last few decades. Medical imaging is one of the important research areas because of its critical nature. The innovations in medical imaging can help to improve computer diagnostic based applications. There is an enormous increase in the volume of medical imaging and diversity in datasets, automated tools are required to analyze the collection of images and do predictions based on information extracted. This study further helps to find the efficacy of multimedia tools. Furthermore, it also helps to build mobile technology to aid patients with Alzheimer decease like memory test applications.
5 Conclusion and Future Directions
In this research, statistical based features extraction is proposed to classify Alzheimer’s using MRI images automatically. The output has shown that these statistical features produce good results in comparison with other features including texture based. It is evident from the results that the multiclass classification utilizing Random Forest gives best results on publicly available OASIS dataset. By correcting diagnose the AD in patients will enhance the efficiency of treatments. The accuracy of multiclass classification was significantly increased. In the future, we are exploring the effect of class imbalance issue in the available datasets. There is also a need to explore the effect of more sophisticated machine learning approaches like deep learning for Alzheimer detection.
References
Beheshti, I., et al., Histogram-Based Feature Extraction from Individual Gray Matter Similarity-Matrix for Alzheimer’s Disease Classification. Journal of Alzheimer’s Disease, 2017. 55(4): p. 1571-1582.
Wang, S., et al., Detection of Alzheimer’s disease by three-dimensional displacement field estimation in structural magnetic resonance imaging. Journal of Alzheimer’s Disease, 2016. 50(1): p. 233-248.
Belleville, S., et al., Detecting early preclinical Alzheimer’s disease via cognition, neuropsychiatry, and neuroimaging: qualitative review and recommendations for testing. Journal of Alzheimer’s disease, 2014. 42(s4): p. S375-S382.
Beheshti, I., H. Demirel, and A.s.D.N. Initiative, Feature-ranking-based Alzheimer’s disease classification from structural MRI. Magnetic resonance imaging, 2016. 34(3): p. 252-263.
Zhang, Y., et al., Detection of Alzheimer’s disease and mild cognitive impairment based on structural volumetric MR images using 3D-DWT and WTA-KSVM trained by PSOTVAC. Biomedical Signal Processing and Control, 2015. 21: p. 58-73.
Altaf, T., et al. Multi-class Alzheimer disease classification using hybrid features. in IEEE Future Technologies Conference. 2017.
Liu, Y., et al. Discriminative MR image feature analysis for automatic schizophrenia and Alzheimer’s disease classification. in International conference on medical image computing and computer-assisted intervention. 2004. Springer.
Lao, Z., et al., Morphological classification of brains via high-dimensional shape transformations and machine learning methods. Neuroimage, 2004. 21(1): p. 46-57.
Fung, G. and J. Stoeckel, SVM feature selection for classification of SPECT images of Alzheimer’s disease using spatial information. Knowledge and Information Systems, 2007. 11(2): p. 243-258.
Klöppel, S., et al., Automatic classification of MR scans in Alzheimer’s disease. Brain, 2008. 131(3): p. 681-689.
Chincarini, A., et al., Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage, 2011. 58(2): p. 469-480.
Westman, E., et al., Sensitivity and specificity of medial temporal lobe visual ratings and multivariate regional MRI classification in Alzheimer’s disease. PloS one, 2011. 6(7): p. e22506.
Ahmed, O.B., et al., Classification of Alzheimer’s disease subjects from MRI using hippocampal visual features. Multimedia Tools and Applications, 2015. 74(4): p. 1249-1266.
Vemuri, P., D.T. Jones, and C.R. Jack, Resting state functional MRI in Alzheimer’s Disease. Alzheimer’s research & therapy, 2012. 4(1): p. 2.
He, Y., et al., Regional coherence changes in the early stages of Alzheimer’s disease: a combined structural and resting-state functional MRI study. Neuroimage, 2007. 35(2): p. 488-500.
Tripoliti, E.E., D.I. Fotiadis, and M. Argyropoulou. A supervised method to assist the diagnosis and classification of the status of Alzheimer’s disease using data from an fMRI experiment. in Engineering in Medicine and Biology Society, 2008. EMBS 2008. 30th Annual International Conference of the IEEE. 2008. IEEE.
Ateeq, T., et al., Ensemble-classifiers-assisted detection of cerebral microbleeds in brain MRI. Computers & Electrical Engineering, 2018.
Beheshti, I., et al., Classification of Alzheimer’s disease and prediction of mild cognitive impairment-to-Alzheimer’s conversion from structural magnetic resource imaging using feature ranking and a genetic algorithm. Computers in biology and medicine, 2017. 83: p. 109-119.
Beheshti, I., et al., Structural MRI-based detection of Alzheimer’s disease using feature ranking and classification error. Computer methods and programs in biomedicine, 2016. 137: p. 177-193.
Ramaniharan, A.K., S.C. Manoharan, and R. Swaminathan, Laplace Beltrami eigen value based classification of normal and Alzheimer MR images using parametric and non-parametric classifiers. Expert Systems with Applications, 2016. 59: p. 208-216.
Guerrero, R., et al., Manifold population modeling as a neuro-imaging biomarker: application to ADNI and ADNI-GO. NeuroImage, 2014. 94: p. 275-286.
Plocharski, M., L.R. Østergaard, and A.s.D.N. Initiative, Extraction of sulcal medial surface and classification of Alzheimer’s disease using sulcal features. Computer methods and programs in biomedicine, 2016. 133: p. 35-44.
Sørensen, L., et al., Differential diagnosis of mild cognitive impairment and Alzheimer’s disease using structural MRI cortical thickness, hippocampal shape, hippocampal texture, and volumetry. NeuroImage: Clinical, 2017. 13: p. 470-482.
Ahmed, O.B., et al., Alzheimer’s disease diagnosis on structural MR images using circular harmonic functions descriptors on hippocampus and posterior cingulate cortex. Computerized Medical Imaging and Graphics, 2015. 44: p. 13-25.
Sarraf, S. and G. Tofighi, DeepAD: Alzheimer’s Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI. bioRxiv, 2016: p. 070441.
Payan, A. and G. Montana, Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks. arXiv preprint arXiv:1502.02506, 2015.
Farooq, A., et al. Artificial intelligence based smart diagnosis of Alzheimer’s disease and mild cognitive impairment. in Smart Cities Conference (ISC2), 2017 International. 2017. IEEE.
Chen, X.-W. and X. Lin, Big data deep learning: challenges and perspectives. IEEE access, 2014. 2: p. 514-525.
Mishra, S., et al., Gray level co-occurrence matrix and random forest based acute lymphoblastic leukemia detection. Biomedical Signal Processing and Control, 2017. 33: p. 272-280.
Kalsoom, A., et al., A dimensionality reduction-based efficient software fault prediction using Fisher linear discriminant analysis (FLDA). The Journal of Supercomputing, 2018: p. 1-35.
Khan, S., et al., Optimized Gabor feature extraction for mass classification using cuckoo search for big data e-healthcare. Journal of Grid Computing, 2018: p. 1-16.
Nazir, F., et al., Social media signal detection using tweets volume, hashtag, and sentiment analysis. Multimedia Tools and Applications, 2018: p. 1-34.
Acknowledgement
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1D1A1A09919551).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Afzal, S., Javed, M., Maqsood, M., Aadil, F., Rho, S., Mehmood, I. (2019). A Segmentation-Less Efficient Alzheimer Detection Approach Using Hybrid Image Features. In: Singh, A., Mohan, A. (eds) Handbook of Multimedia Information Security: Techniques and Applications. Springer, Cham. https://doi.org/10.1007/978-3-030-15887-3_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-15887-3_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-15886-6
Online ISBN: 978-3-030-15887-3
eBook Packages: Computer ScienceComputer Science (R0)